Abstract
Background
Human papillomavirus (HPV) 33 belongs to the Alphapapillomavirus 9 (α-9 HPV) species group, which also contains types 16, 31, 35, 52, 58 and 67. The purpose of this study was to investigate the genetic variations of HPV33 and to explore its carcinogenicity among women in Taizhou, Southeast China.
Methods
Exfoliated cervical cells were collected for HPV genotyping. Only single HPV33 infection cases were selected, and their E6 and E7 genes were sequenced using the ABI 3730xl sequencer and then analysed using MEGA X.
Results
From 2014 to 2020, a total of 185 single HPV33-positive specimens were successfully amplified. We obtained 15 distinct HPV33 E6/E7 variants, which were published in GenBank under accession numbers OQ672665-OQ672679. Phylogenetic analysis revealed that all HPV33 E6/E7 variants belonged to lineage A, of which 75.7% belonged to lineage A1. Compared with CIN1, the proportion of sublineage A1 in CIN2/3 was higher, but there was no significant difference (76.5% vs. 80.6%, P > 0.05). Altogether, 20 single nucleotide substitutions were identified, of which 6 were novel substitutions, including T196G (C30G), A447T, G458T (R117L), G531A, A704A, and C740T. In addition, no significant trends were observed between the nucleotide substitutions of HPV33 E6/E7 variants and the risk of cervical lesions.
Conclusion
This study provides the most comprehensive data on genetic variations, phylogenetics and carcinogenicity of HPV33 E6/E7 variants in Southeast China to date. The data confirmed that cervical lesions among women in Taizhou are attributable to HPV33, which may be due to the high infection rate of sublineage A1 in the population.
Similar content being viewed by others
Background
Human papillomavirus (HPV) is the most common pathogen of cervical cancer, especially the high-risk HPV types [1]. HPV16 and 18 are the two most carcinogenic types, accounting for approximately 70% of cervical cancers worldwide [2]. Types 31, 33, 35, 39, 45, 51, 52, 56, 58, 59, 66, and 68 are related to the remaining cases of cervical cancers. Notably, HPV type distribution varies with geographical location and ethnic group. Moreover, these types differ in biological functions due to differences in genetic variations, which may become a key risk factor in cervical cancer.
Based on the American Society for Colposcopy and Cervical Pathology (ASCCP) guidelines, colposcopy is recommended immediately in women with HPV16 or 18 infections [3]. However, our previous study showed that women infected with HPV16, 18, 31, 33, 52, or 58 should be recommended for colposcopy immediately, which can improve the detection rate of CIN2 + lesions in the Taizhou area of southeast China [4]. HPV types 33, 52 and 58 were detected less frequently than HPV16 in terms of association with severe cervical intraepithelial neoplasia (CIN) lesions but more frequent than HPV18 in China [4, 5]. Notably, HPV33 has a higher absolute risk for CIN2 + than other non-HPV16 high-risk types [6,7,8]. Therefore, our research team should devote more attention to these common carcinogenic types, such as HPV33.
HPV33 was originally cloned from cervical cancer in 1986 [9]. HPV33 variants are clustered into three main phylogenetic lineages (A, B, and C) and five sublineages (A1, A2, A3, B1, and C1) [10, 11]. These lineages exhibit a distribution pattern that varies across the world. Unfortunately, the data available on HPV33 genetic variations and their carcinogenicity are still limited in China. Therefore, the purpose of this study was to investigate the genetic variations in the E6 and E7 oncogenes of HPV33 and to explore their potential role in cervical cancer risk among Chinese women in the Taizhou area.
Materials and methods
Study population and specimen collection
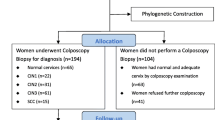
From February 2014 to December 2020, exfoliated cervical cells were collected from women who underwent cervical screenings at Taizhou Hospital, Zhejiang Province. Subsequently, HPV genotyping was performed, which has been described in detail in our previous study [4]. Only single HPV33 infection cases were selected for this study. The specimens were stored in cell preservation buffer at -20 °C.
PCR and sequencing
DNA was extracted from stored specimens using a DNA Extraction Kit (#GK0122, GENEray, China) according to the manufacturers’ guidelines. Specific primer pairs were designed for the entirety of the E6 and E7 regions of HPV33 using the NCBI Primer-BLAST tool. The primers were as follows: 33E6E7F 5’-AGGGTGTAACCGAAAGCGG-3’ and 33E6E7R 5’-TTGCAGCACGATCAACAACG-3’. The PCR system and conditions were described in our previous HPV study [12]. The length of the PCR product was 1164 bp (nucleotide sites [nt] 31-1175, including the E6 gene nt 109–558 and E7 gene nt 573–866). Subsequently, PCR products were purified and sequenced at BGI company (Hangzhou, China), and all data were confirmed by repeating PCR and sequencing reactions at least twice.
Phylogenetic analysis
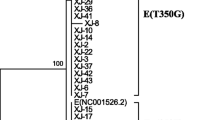
All acquired nucleotide sequences were aligned by BioEdit software using the reference sequence (GenBank accession no. M12732.1) as a standard for HPV33 nucleotide position numbering. The phylogenetic tree was generated using 15 complete HPV33 E6/E7 genes obtained in this study and 6 complete HPV33 E6/E7 genes available in NCBI (GenBank accession no. M12732.1 (A1), HQ537697 (A1), HQ537698 (A2), EU918766 (A3), HQ537705 (B1), and KF436865 (C1)). A maximum-likelihood tree was constructed by MEGA X software, with one thousand bootstrap replicates.
Statistical analysis
All analyses were performed using SPSS 16.0 software. The association of cervical lesion risk with HPV33 E6/E7 variants was analysed using the chi-square test or Fisher’s exact test. A two-sided P value < 0.05 was regarded as statistically significant.
Results
Characteristics of the study population
From February 2014 to December 2020, a total of 189 women with a single HPV33 infection were selected. The average age was 42.3 years (range 19–73 years). A total of 185 (97.9%) sequences of the entire E6 and E7 genes from HPV33 were successfully obtained. Due to the small number of HPV copies, the remaining 4 (2.1%) sequences were excluded. Out of 185 cases, 110 (59.5%) underwent colposcopy biopsy for histological diagnosis, including 57 with normal cervices after biopsy, 17 with CIN1, 16 with CIN2, 20 with CIN3, and no cases of cervical cancer. The characteristics of the study population are shown in Table 1.
Variations in the E6 and E7 genes
Compared with M12732.1, 59 of the 185 HPV33 samples (31.9%) showed nucleotide variations. Figure 1 visually distinguishes all changes in nucleotide and amino acid sequences in the entire E6 and E7 fragments of the HPV33 lineage/sublineages. In this study, 15 distinct variation patterns were detected in the HPV33 E6/E7 variants, denoted 33CNTZ01-33CNTZ15, which were published with GenBank accession nos. OQ672665 to OQ672679. 33CNTZ01 (68.1%, 126/185) was the most common variant in the Taizhou-based population, with complete E6 and E7 sequence homology with M12732.1. Ten (66.7%, 10/15) belonged to the novel HPV33 E6/E7 variants, which are highlighted in bold in Fig. 1.
Genetic variability of HPV33 E6 and E7 nucleotide sequences in Taizhou area, Southeast China. Numbering refers to the first nucleotide of the HPV33 prototype reference sequence (GenBank: M12732.1). Each row indicates the variant identification and the nucleotide sequence alignment compared to the reference. Novel HPV33 variants are highlighted in bold and novel nucleotide substitutions are highlights in gray
Altogether, 20 single nucleotide substitutions were identified, with 6 (30.0%) novel substitutions and 10 (50.0%) nonsynonymous substitutions. Nonsynonymous substitutions included T196G (C30G), A213C (K35N), C245T (A46V), A364C (N86H), A387C (K93N), A446G (Q113R), and G458T (R117L) in the E6 sequence and C706T (A45V), C706A (A45E), and A862T (Q97L) in the E7 sequence. To our knowledge, the nucleotide substitutions of T196G (C30G), A447T, G458T (R117L), G531A, A704A, and C740T have not been reported in previous studies.
Phylogenetic construction
The phylogenetic tree was constructed from 21 complete HPV33 E6 and E7 sequences (15 obtained from our study and 6 from GenBank) (Fig. 2). Based on the phylogenetic tree, sublineages A1, A2, and A3 were detected in 75.7% (140/185), 1.1% (2/185), and 23.2% (43/185) of samples, respectively. All HPV33 E6/E7 variants belonged to lineage A, whereas no variants belonged to lineage B or lineage C in the present study.
Risk association with cervical lesions
Our data suggested that the proportion of sublineage A1 in CIN2/3 was higher when compared with CIN1, but it was not significant (76.5% vs. 80.6%, P > 0.05). There was no significant difference in the risk of cervical lesions between sublineage A1 and other HPV33 E6/E7 variants. No significant difference was observed in the relative risk for cervical lesions among the nucleotide variations in the HPV33 E6 and E7 genes (Table 1).
Discussion
Cervical cancer remains a serious public health problem in developing countries, especially in China. The distribution of HPV types may differ among specific populations in different countries. Their genetic variation data can be of considerable importance to assess the impact of local HPV vaccines and strategies for cancer prevention. To our knowledge, the Taizhou Area HPV Study is a comprehensive study to assess the genetic variations of HPV types in southeast China, which is helpful for local HPV vaccine development. According to our previous findings, genetic variation in the E6 and E7 genes of α-9 HPV types was found to be highly associated with cervical carcinogenesis risk [13,14,15]. Therefore, we continue to investigate the genetic variation of HPV33 and to explore its carcinogenicity among women in Taizhou, Southeast China.
In this study, we obtained 185 complete sequences of HPV33 E6 and E7 genes among Chinese women. Our data showed that all HPV33 E6/E7 variants belonged to lineage A. Most of the HPV33 E6/E7 variants (75.7%) belonged to sublineage A1, which was similar to the results (92.1%) obtained in Southwest China [16]. Sublineage A1 was observed to be distributed throughout the world, but the relative frequency varied by region. Sublineage A2 (59.7%) was the most common lineage in the Asia and Oceania region [10]. However, only 1.1% of the HPV33 variants belonged to A2, and 23.2% of the sublineage belonged to A3 in Taizhou, Southeast China. Lineage A predominates in Asia and Europe, while lineage B or C predominates in Africa [10, 17]. Compared with other sublineage variants, the risk of CIN2/3 or cervical cancer in sublineage A1 was significantly increased [18,19,20]. Our data suggested that the proportion of sublineage A1 in CIN2/3 was higher when compared with CIN1, but it was not significant (76.5% vs. 80.6%, P > 0.05). There was no significant difference in the risk of cervical lesions between sublineage A1 and other HPV33 E6/E7 variants. Notably, 33CNTZ01 accounts for 90% (126/140) of A1 and 33CNTZ11 accounts for 86% (37/43) of A3. Other (sub)lineages were not included in these analyses due to having only one or two samples.
Compared with the HPV33 reference sequence (GenBank accession no. M12732.1), the six most prevalent nucleotide substitutions were A213C (K35N) (25.9%), A364C (N86H) (24.3%), A273G (23.2%), A387C (K93N) (24.3%), and A446G (Q113R) (23.2%) in the E6 gene and A862T (Q97L) (24.3%) in the E7 gene, which were specific to HPV33 lineage A3. He et al. [21, 22] found 15.3% G542T (R145I) of the E6 gene in Southwest China; however, this nucleotide substitution has not been detected in Taizhou, Southeast China. It is hypothesized that the substitution of G542T(R145I) may increase the infection rate of the HPV33 variant in Southwest China by reducing its immunogenicity and enhancing its adaptability to the environment [22]. In addition, the 93rd residue is the common nonsynonymous substitution in the E6 gene of α-9 HPV types [14, 15, 22]. These findings are valuable for the development of therapeutic vaccines and cancer immunotherapies.
Conclusions
In summary, this study provides the most comprehensive data on the genetic variations, phylogenetics, and carcinogenicity of HPV33 E6/E7 variants in Southeast China to date. These findings can make an important contribution to future epidemiological, HPV vaccination, and cancer immunotherapy studies.
Data Availability
All data generated during this study are included in this published article. The supplementary materials included the nucleotide variations of the E6 and E7 genes from HPV33 and the follow-up data of patients. In addtion, these sequences have been released to GenBank database with the accession codes of OQ672665-OQ672679. The links are https://www.ncbi.nlm.nih.gov/nuccore/OQ672665 ~ https://www.ncbi.nlm.nih.gov/nuccore/OQ672679.
Abbreviations
- bp:
-
Base pair
- CIN:
-
Cervical intraepithelial neoplasia
- DNA:
-
Deoxyribonucleic acid
- HPV:
-
Human papillomavirus
- LCR:
-
Long control region
- LEEP:
-
Loop electrosurgical excision procedure
- nt:
-
Nucleotide sites
- OR:
-
Odds ratios
- ORF:
-
Open reading frames
- PCR:
-
Polymerase chain reaction
- SNP:
-
Single nucleotide polymorphism
- WHO:
-
World Health Organization
References
Cohen PA, Jhingran A, Oaknin A, Denny L. Cervical cancer. Lancet. 2019;393(10167):169–82. https://doi.org/10.1016/S0140-6736(18)32470-X
Alejo M, Alemany L, Clavero O, Quiros B, Vighi S, Seoud M, et al. Contribution of human papillomavirus in neuroendocrine tumors from a series of 10,575 invasive cervical cancer cases. Papillomavirus Res. 2018;5:134–42. https://doi.org/10.1016/j.pvr.2018.03.005
Huh WK, Ault KA, Chelmow D, Davey DD, Goulart RA, Garcia FA, et al. Use of primary high-risk human papillomavirus testing for cervical cancer screening: interim clinical guidance. Gynecol Oncol. 2015;136(2):178–82. https://doi.org/10.1016/j.ygyno.2014.12.022
Xu H, Lin A, Shao X, Shi W, Zhang Y, Yan W. Diagnostic accuracy of high-risk HPV genotyping in women with high-grade cervical lesions: evidence for improving the cervical cancer screening strategy in China. Oncotarget. 2016;7(50):83775–83. https://doi.org/10.18632/oncotarget.11959
Purut YE, Uçkan K, Could HPV. Type 33 be more Risky Than we thought? Int J Surg Pathol. 2023;31(1):4–10. https://doi.org/10.1177/10668969221134692
Schiffman M, Glass AG, Wentzensen N, Rush BB, Castle PE, Scott DR, et al. A long-term prospective study of type-specific human papillomavirus infection and risk of cervical neoplasia among 20,000 women in the Portland Kaiser Cohort Study. Cancer Epidemiol Biomarkers Prev. 2011;20(7):1398–409. https://doi.org/10.1158/1055-9965.EPI-11-0206
Kjær SK, Frederiksen K, Munk C, Iftner T. Long-term absolute risk of cervical intraepithelial neoplasia grade 3 or worse following human papillomavirus infection: role of persistence. J Natl Cancer Inst. 2010;102(19):1478–88. https://doi.org/10.1093/jnci/djq356
Matsumoto K, Oki A, Furuta R, Maeda H, Yasugi T, Takatsuka N, et al. Predicting the progression of cervical precursor lesions by human papillomavirus genotyping: a prospective cohort study. Int J Cancer. 2011;128(12):2898–910. https://doi.org/10.1002/ijc.25630
Beaudenon S, Kremsdorf D, Croissant O, Jablonska S, Wain-Hobson S, Orth G. A novel type of human papillomavirus associated with genital neoplasias. Nature. 1986;321(6067):246–9. https://doi.org/10.1038/321246a0
Chen AA, Heideman DA, Boon D, Chen Z, Burk RD, De Vuyst H, et al. Human papillomavirus 33 worldwide genetic variation and associated risk of cervical cancer. Virology. 2014;448:356–62. https://doi.org/10.1016/j.virol.2013.10.033
Chen Z, Schiffman M, Herrero R, Desalle R, Anastos K, Segondy M, et al. Evolution and taxonomic classification of human papillomavirus 16 (HPV16)-related variant genomes: HPV31, HPV33, HPV35, HPV52, HPV58 and HPV67. PLoS ONE. 2011;6(5):e20183. https://doi.org/10.1371/journal.pone.0020183
Xu HH, Zheng LZ, Lin AF, Dong SS, Chai ZY, Yan WH. Human papillomavirus (HPV) 18 genetic variants and cervical cancer risk in Taizhou area, China. Gene. 2018;647:192–7. https://doi.org/10.1016/j.gene.2018.01.037
Dai MZ, Qiu Y, Di XH, Shi WW, Xu HH. Association of cervical carcinogenesis risk with HPV16 E6 and E7 variants in the Taizhou area, China. BMC Cancer. 2021;21(1):769. https://doi.org/10.1186/s12885-021-08531-y
Yang Z, He ZH, Zhang Y, Di XH, Zheng DF, Xu HH. Genetic variability in the E6 and E7 oncogenes of HPV52 and its prevalence in the Taizhou area, China. Virol J. 2022;19(1):194. https://doi.org/10.1186/s12985-022-01929-5
Yu JH, Shi WW, Zhou MY, Liu JM, Han QY, Xu HH. Genetic variability and oncogenic risk association of human papillomavirus type 58 E6 and E7 genes in Taizhou area, China. Gene. 2019;686:171–6. https://doi.org/10.1016/j.gene.2018.11.066
Chen Z, Jing Y, Wen Q, Ding X, Wang T, Mu X, et al. E6 and E7 gene polymorphisms in human papillomavirus Types-58 and 33 identified in Southwest China. PLoS ONE. 2017;12(1):e0171140. https://doi.org/10.1371/journal.pone.0171140
Kovacevic G, Milosevic V, Knezevic P, Knezevic A, Knezevic I, Radovanov J, et al. Prevalence of oncogenic human papillomavirus and genetic diversity in the L1 gene of HPV16 HPV 18 HPV31 and HPV33 found in women from Vojvodina Province Serbia. Biologicals. 2019;58:57–63. https://doi.org/10.1016/j.biologicals.2019.02.001
Xi LF, Schiffman M, Koutsky LA, Hughes JP, Winer RL, Mao C, et al. Lineages of oncogenic human papillomavirus types other than type 16 and 18 and risk for cervical intraepithelial neoplasia. J Natl Cancer Inst. 2014;106(10):dju270. https://doi.org/10.1093/jnci/dju270
Xin CY, Matsumoto K, Yoshikawa H, Yasugi T, Onda T, Nakagawa S, et al. Analysis of E6 variants of human papillomavirus type 33, 52 and 58 in japanese women with cervical intraepithelial neoplasia/cervical cancer in relation to their oncogenic potential. Cancer Lett. 2001;170(1):19–24. https://doi.org/10.1016/s0304-3835(01)00570-5
Godínez JM, Heideman DA, Gheit T, Alemany L, Snijders PJ, Tommasino M, et al. Differential presence of Papillomavirus variants in cervical cancer: an analysis for HPV33, HPV45 and HPV58. Infect Genet Evol. 2013;13:96–104. https://doi.org/10.1016/j.meegid.2012.09.011
He J, Li Q, Ma S, Li T, Chen Y, Liu Y, et al. The polymorphism analysis and epitope predicted of Alphapapillomavirus 9 E6 in Sichuan, China. Virol J. 2022;19(1):14. https://doi.org/10.1186/s12985-021-01728-4
He J, Yang Y, Chen Z, Liu Y, Bao S, Zhao Y, et al. Identification of variants and therapeutic epitopes in HPV-33/HPV-58 E6 and E7 in Southwest China. Virol J. 2019;16(1):72. https://doi.org/10.1186/s12985-019-1168-y
Acknowledgements
We appreciate all the patients for their contribution to this study.
Funding
This work was supported by grants from National Natural Science Foundation of China (No.81901625). None of the funders had any influence on the study design; in the collection, analysis, and interpretation of data; in the writing of the report; and in the decision to submit the article for publication.
Author information
Authors and Affiliations
Contributions
H.H.X. designed the experiments, performed analysis and drafted the manuscript. Y.Q., J.G., and Y.Y.Y. performed HPV genotyping. X.H.D. and Z.Y.Y. carried out the sample collection and PCR amplification. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
This study was approved by the Institutional Medical Ethics Review Board of Taizhou Hospital of Zhejiang Province (approval #MERB-2017-020), and was carried out in line with the Helsinki Declaration. All participants provided written informed consent for study participation before specimen collection, and the patients’ privacy is strictly protected.
Consent for publication
Written informed consent was obtained from all patients for the publication of their medical data.
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Additional file 1.
Clinical data for HPV33 study in Taizhou area, China
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Yan, ZY., Di, XH., Qiu, Y. et al. Cervical carcinogenesis risk association of HPV33 E6 and E7 genetic variations in Taizhou, Southeast China. Virol J 20, 156 (2023). https://doi.org/10.1186/s12985-023-02125-9
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12985-023-02125-9