Abstract
Background
Although, there is a variable burden of human papillomavirus (HPV) in women infected with HIV in developing countries, there are few studies that attempted to surmise such variable evidences. This review aimed to estimate the pooled prevalence of HPV genotype distribution and risk factors contributing to HPV infection among women infected with HIV in low- and middle-income countries.
Methods
We conducted a systematic review and meta-analysis of studies conducted in developing countries and reported HPV prevalence. We searched electronic databases: PubMed/Medline, SCOPUS, ScienceDirect, Excerpta Medical Database from Elsevier, Web of science, Cumulative Index of Nursing and allied Health Sciences and Google scholar databases to retrieve primary studies published in English language till 11th August 2019. We used random-effects model to estimate the pooled prevalence of HPV genotypes, and funnel plot to assess publication bias. The registration number of this review study protocol is CRD42019123549.
Results
We included nineteen studies with a total of 8,175 participants in this review. The prevalence of HPV was extremely heterogeneous across the studies (χ2= 3782.80, p value < 0.001, I2 = 99.6%). The estimated pooled prevalence of all HPV genotypes was 63.0% (95% CI: 48.0–78.0) while the pooled prevalence of high risk and low risk HPV genotypes were 51.0% (95% CI: 38.0–63.0) and 28.0% (95% CI: 12.0–43.0), respectively. The pooled prevalence of HPV genotype 16 was 20%, while genotype 18 and 52 were 15% and 13%, respectively. Different risk factors reported for HPV infection and the frequently reported were low CD4 count below 200 cells/mm3 and high HIV viral load.
Conclusion
The pooled prevalence of HPV among HIV infected women in low- and middle-income countries was considerable and the proportion of high risk HPV genotypes were high when compared with low risk genotypes. Therefore, it is essential for the HPV prevention program to prevent the double burden of HPV and HIV in women.
Similar content being viewed by others
Background
The papillomavirus is a heterogeneous group of DNA virus with circular, non-enveloped, double-stranded DNA genomes [1, 2]. This virus infects humans and different species of animals [2]. The virus is discovered from the horn of Cottontail rabbit at the beginning of 1930s [3] and also revealed as a main cause of human cervical cancer in 1970s [4]. More than 300 papillomaviruses have been identified and completely sequenced, including over 200 human papillomaviruses [5]. The high-risk carcinogenic types of HPV currently designated by the International Agency for Research on Cancer (IARC) are HPV16, HPV18, HPV31, HPV33, HPV35, HPV39, HPV45, HPV51, HPV52, HPV56, HPV58, and HPV59. The HPV68 is classified as probably carcinogenic, and HPV26, HPV30, HPV34, HPV53, HPV66, HPV67, HPV 69, HPV70, HPV73, HPV82, HPV85, and HPV97 have been associated with rare cases of cervical cancer and are considered probable carcinogens [6, 7]. Genotype 6 and 11 are low-risk types that cause genital and skin warts [8]. Genital HPV infections are very common and prevalent in the age range of 18 to 30 years [9, 10]. Infection of the cervix with HPV is necessary to cause cervical neoplasia and cervical cancer [11, 12], and integration of viral DNA into the host genome is necessary for persistent infection which could lead to the development of cervical dysplasia [11].
The prevalence of HPV is variable across the world. The study reported from developed countries indicate that the prevalence of HPV was 11 to 12% [13]. The recent global estimate indicates 11.7% of the HPV infection burden in the world [14]. The occurrence of about 85% of infected cases and 88% of the deaths due to cervical cancer is in developing countries [11].
The highest prevalence was reported in sub-Saharan Africa (24%), Eastern Europe (21%) and Latin America (16%) [15].
The burden of HPV infection is higher in HIV infected women (50.8%) than un-infected (22.6%) [16] and 78.8% among HIV infected than 34.4% of un-infected women [17]. Similarly, high-risk oncogenic HPV types is higher among HIV infected than un-infected women (48.4% vs. 17.3%) [16]. Other studies reported a prevalence of 68.0% [18] and 33.2% [19]. Moreover, the study reported from developing countries indicated extremely variable prevalence of HPV that ranges from 20 to 70% [20]. The prevalence of low-risk HPV types were 3.6 to 5.6 times higher in HIV-sero-positive women when compare to HIV sero-negative's [8].
Several risk factors are reported to be associated with HPV infection and these include HIV infection, other STIs (e.g., chlamydia, herpes simplex virus), and multiple sexual partners [11, 21]. There are also other factors that mediate HPV infection such as cigarette smoking, oral contraceptive or hormonal contraceptive use, chronic inflammation and immunosuppressive conditions [10, 11, 21, 22]. Dietary factors, socioeconomic status, race/ethnicity, geographic disparity and polymorphisms in the human leukocyte antigen system are additional factors that could mediate HPV infection [10, 11, 21, 22]. Being young age and having active sexual behavior are key risk factors for HPV acquisition and persistence of the infection [22].
HIV infection increases the risk of cervical infection due to high-risk HPV genotypes that induces high-grade cervical squamous intraepithelial lesions (HSILs), which in turn leads to the development of pre-invasive cervical lesions and invasive cervical cancer (ICC) [23,24,25]. HIV infection could alter the natural history of HPV infection through decreasing the self-clearance rate of infection and increasing progression to high grade and invasive lesions [24]. Furthermore, the incidence of HPV infection is three times higher in HIV-positive women [25], and can cause cervical cancer than their counterparts [26]. Nonetheless, with the exception of the systematic review and meta-analysis done in Kenya [27], evidences in this regard showing the burden and molecular distribution of HPV in low and middle income countries (LMICs) is limited [28]. Therefore, this review aims to fill the identified gaps by estimating the pooled prevalence of HPV, and investigating the factors associated with HPV infection among HIV infected women in LMICs.
Methods
Search strategy and screening of papers
We conducted a systematic review and meta-analysis of published articles to estimate the pooled prevalence of HPV in LMICs. We systematically searched the papers published in the following electronic databases; PubMed/MEDLINE, SCOPUS, Science Direct, Excerpta Medical Database from Elsevier (EMBASE), Web of science, Cumulative Index of Nursing and Allied Health Sciences (CINAHL) and Google scholar. The review was conducted in accordance with Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA) standard [29]. We used a search strategy by combining the following key terms: molecular, molecular epidemiology, human papillomavirus, or HPV, papillomavaridae, Human immunodeficiency virus (HIV), AIDS (acquired human immunodeficiency syndrome), HIV infected, HIV positive, HIV sero-reactive, women, female and girl. We used Truncation(*) to manage spelling variation during search: infect* or positive, wom*n or female* or girl*. We used both free text and Medical subject heading [MeSH] terms during electronic database search.
PubMed database search strategy was:((((molecular[tiab] OR "Molecular Epidemiology"[Mesh]) AND ((Human papillomavirus[tiab] OR HPV[tiab]) OR "papillomaviridae"[MeSH Terms])) AND (((Human immunodeficiency virus[tiab] OR HIV[tiab]) OR "hiv"[MeSH Terms]) OR "hiv"[MeSH Terms])) AND (infected[tiab] OR positive[tiab])) AND (((women[tiab] OR females[tiab]) OR "women"[MeSH Terms]) OR "female"[MeSH Terms]) AND (("1966/01/01"[PDAT]: "2019/08/11"[PDAT]) AND "humans"[MeSH Terms] AND English[lang] AND "female"[MeSH Terms]).
The search was repeated to identify the consistency of search terms and results. Two authors independently reviewed the titles, abstracts and full articles of the retrieved studies.
Study inclusion and exclusion criteria
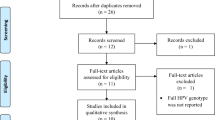
We included a cross sectional and cohort studies conducted in LMICs based on World Bank Country Classifications, 2018 [30] and that reported prevalence of HPV genotypes. The inclusion was restricted to the papers published in English language without limiting publication year till 11th August 2019. We excluded studies that did not clearly state the study design, outcome measured, the study conducted on HIV negative women alone, conducted in high-income countries, and the study reported HPV genotype from anal and oral organ types (Fig. 1).
Study quality assessment
We assessed the quality of included studies by using the 14 items Quality Assessment Tool for Observational Cohort and Cross-Sectional studies NHLBI, NIH [31]. This assessment tool mainly focused on research question, study population, eligibility criteria (inclusion and exclusion criteria of study participants), sample size justification, exposure measures and assessment, sufficient time frame to see an effect, outcome measures and blinding of outcome assessors, follow up rate, and statistical analysis. The quality assessment was rated as good, fair and poor based on the quality assessment tool criteria. The maximum score indicating high quality was 14 and the lowest possible score was zero. The rating values of the included studies in terms of their quality were based on their design. Cross-sectional type do not consider the items which fit for cohort and taken as not-applicable (NA) and thus, the rating values were not taken from the possible maximum score (i.e. 14). In this review, all scores are written in percentage.
Data extraction
Data from eligible abstract and/or full text of the articles were extracted by considering the outcome variables (i.e. prevalence or proportion of HPV genotypes, magnitude of cancer causing HPVs or high risk (HR) HPV genotypes and low-risk HPV types), and factors that could potentially be associated with these outcomes. The characteristics of study participants of an eligible paper such as age range, mean or median age, sex, HIV sero-status, the prevalence of HPV genotype were also extracted. Study characteristics such as first author, year of publication, study duration, study setting, study location or country, study design, sample size were also extracted (Table 1). Other extracted data include the prevalence of different HPV genotypes (Table 2), factors which could potentially be associated with HPV infection and diagnostic methods applied to detect HPV infection (Table 3).
The majority of the studies included in our review had more than eighty percent and the lowest score observed was 62.5% in terms of quality. There was however one abstract included in the review, which was difficult to assess the quality of the article (Table 3).
Statistical analysis
We estimated the pooled prevalence of HPV with its 95% Confidence Interval (CI) using random effects meta-analysis model assuming the true effect size varies between studies [32]. The proportion of HPV reported in each study is multiplied by its sample size to express patients with HPV infection in number, and data presented in forest plot. Heterogeneity in the prevalence of different studies was assessed using Chi-square (χ2) based Q test with significant level of p value < 0.1 and I2. The I2 value of25% indicates low heterogeneity while 50% moderate and 75% high [33]. The potential publication bias was assessed using a funnel plot. If the 95% of the point estimate of studies lie within the funnel plot defined by straight lines, then that indicates the absence of heterogeneity [34]. The potential sources of heterogeneity were assessed by doing subgroup analysis and moment based meta regression. Meta-regression extends subgroup analyses and allows to estimate effect size. Data analysis was conducted using STATA version 14.
Results
Study characteristics
We included 19 studies in our review (Fig. 1). These studies were reported from Rwanda [36, 37], Brazil [38,39,40], Nigeria [41, 42] Thailand [43], South Africa [44,45,46,47], Zambia [48], Burkinafaso [35, 49], Senegal [23] and Colombia [50, 51]. There was one study conducted in two countries Burkinafaso and South Africa [52]. Five studies were from South America (three from Brazil and two from Colombia), one study from Asia (Thailand) and the rest were from African countries. All of the studies were from health facilities (Hospital and clinic) and the majority were cross sectional studies. The publication year varied from 2003 to 2017 while the majorities (13 articles) were published after 2009. Eight studies were published in 2013 and 2014. The maximum sample size was 1371 [44] and the minimum was 98 [41]. The age of participants ranged from 14 to 73 years [39, 50]. Three studies didn't mention the upper age range of the participants [23, 36, 42] (Table 1).
Pooled prevalence of HPV
We pooled data from 8175 HIV infected women to estimate the pooled prevalence of HPV infection using meta-analysis. The overall pooled prevalence of all types of HPV infection was 63.0% (Fig. 2) with high heterogeneity across the studies (χ2 = 3,782.80 (d.f. = 15); p value < 0.001 and I2 = 99.6%). The pooled prevalence of high risk HPV was 51.0% (Fig. 3) with heterogeneity of χ2 = 1069.84 (d.f. = 12); p value < 0.001 and I2 = 98.88%. Similarly, the estimated pooled prevalence of low risk HPV was 28.0% (Fig. 4) with heterogeneity of χ2 = 296.83; (d.f. = 5); p value < 0.001 and I2 = 98.32%.
Forest plot to estimate the pooled prevalence of human papillomavirus infection among HIV infected women with 95% CI (the estimate weighted based on random effects model): ES-Effect Size equivalent to prevalence, CI—confidence interval. In the plot, the diamond shows the pooled result and the boxes show the effect estimates from the single studies. The purple dotted vertical line indicates pooled estimate. The purple solid vertical line indicates the reference line at zero indicating no effect. The horizontal line through the boxes illustrate the length of the confidence interval and the boxes show the effect estimates from the single studies
Forest plot to estimate the pooled prevalence of high risk human papillomavirus infection among HIV infected women (the estimate weighted based on random effects model): ES—effect Size equivalent to prevalence, CI—confidence interval. In the plot, the diamond shows the pooled result and the boxes show the effect estimates from the single studies. The purple dotted vertical line indicates pooled estimate. The purple solid vertical line indicates the reference line at zero indicating no effect. The horizontal line through the boxes illustrate the length of the confidence interval and the boxes show the effect estimates from the single studies
Forest plot to estimate the pooled prevalence of low risk human papillomavirus infection among HIV infected women with 95% CI (the estimate weighted based on random effects model): ES—effect size equivalent to prevalence, CI—confidence interval. In the plot, the diamond shows the pooled result and the boxes show the effect estimates from the single studies. The purple dotted vertical line indicates pooled estimate. The purple solid vertical line indicates the reference line at zero indicating no effect. The horizontal line through the boxes illustrate the length of the confidence interval and the boxes show the effect estimates from the single studies
The pooled prevalence of high risk HPV genotypes was also estimated in the studies (i.e. HPV genotypes 16, 18, 26, 31, 33, 35, 39, 45, 51, 52, 53, 56, 58, 59, 66, 67, 68, 69, 70, 73, and 82). The highest prevalence was observed for genotype 16 (20%) followed by 18 (15%) and 52 (13%). Almost all genotypes indicated heterogeneity and the highest heterogeneity was observed in genotype 16 (I2 = 98.53%) followed by 18 (I2 = 97.18%) and 31 (I2 = 96.17%) (Table 4). The HPV genotypes 26,67,69, 70, 73 and 82 reported less frequently in the included studies which is difficult to interpret.
Subgroup analysis
The result of subgroup analysis based on the continent from where the studies were include shows significant heterogeneity between and within the group. The pooled prevalence of HPV in African was 69.0% (95% CI: 49.0–89.0) with heterogeneity of I2 = 99.74% and p value < 0.001. The pooled prevalence of HPV in South America was 57.0% (95% CI: 44.0–71.0) with heterogeneity of I2 = 95.93% and p value < 0.001) (Fig. 5).
Meta-regression analysis
We assessed the effects of sample size and year of the study on heterogeneity between the studies using meta-regression model. Both sample size and publication years significantly predicted the heterogeneity of the effect sizes (Table 5). In the adjusted model, both the sample size and publication year indicated heterogeneity in the effect size which is equivalent to the prevalence (p < 0.001). When we interpret the finding using β-coefficient, one unit increase in the sample size increases the effect size or the outcome of 1.04 points and the outcome decreases by 11.8 points for every one unit increase in the publication year (Table 5).
Publication bias
The funnel plot (widely used to examine bias in the results of meta-analysis) for the pooled prevalence of all genotypes HPV, high risk HPV and low risk HPV indicated that there is a publication bias (Fig. 6a–c). Figure 6a, indicates the funnel plot of the 16 estimates of the HPV types available for meta-analysis (SE-Standard error, ES-Effect size: prevalence), (b) Funnel plot of the 13 estimates of high risk HPV types available for meta-analysis (SE-Standard error, ES-Effect size: prevalence), (c) Funnel plot of the 6 estimates of low risk HPV types available for meta-analysis (SE-Standard error, ES-Effect size: prevalence). The majority of included studies were out of 95% confidence limit. The outer dashed lines indicate the triangular region within which 95% of studies are expected to lie in the absence of both biases and heterogeneity. In this funnel plot, scatter properties of the included studies made by medium small size with white background color and the scale of 1.
Publication bias assessment: a funnel plot of the 16 estimates of HPV types available for meta-analysis (SE—standard error, ES—effect size: prevalence), b funnel plot of the 13 estimates of high risk HPV types available for meta-analysis, c funnel plot of the 6 estimates of low risk HPV types available for meta-analysis. In this plot, the blue broken line indicates Pseudo 95% CI, the solid red line indicates pooled estimate of the prevalence of HPV, and the scattered circle dots indicates included studies in the meta-analysis. The scale on the X-axis indicates Effect size estimate or proportion and the Y-axis indicates the precision estimate using standard Error
Laboratory techniques applied to detect HPV infection in the included studies
Molecular genotyping and HPV detection techniques applied for selected studies were Linear Array HPV Genotyping Test (LA), careHPV, genotyping using PCR-restriction fragment length polymorphism analysis, Reverse line-blot hybridization, INNO-LiPA HPV genotyping Extra® assay (Table 3).
Factors associated with HPV infection
High HIV viral load and low CD4 count were the most frequently reported factors that associated with high-risk HPV infection [23, 47, 48, 50]. Hormonal contraceptive use, CD4count < 200 cells/mm3, history of three or more sexual partners were reported as the factors associated with HPV infection [37, 38] (Table 3).
Discussion
In the current review, the pooled estimate of HPV infection prevalence was 63.0%. The estimates of high risk and low risk genotypes were 51.0% and 28.0%, respectively. Of high risk genotypes, HPV genotype 16 was high (20%) followed by 18 (15%) and 52 (13%), respectively. Low CD4count and high HIV viral load were the risk factors that most frequently reported in this review.
This finding was lower than the findings in Kenyan which reported 68.0% overall pooled prevalence of high risk HPV among HIV positive women [27]. Genotype 16 was the most prevalent HPV genotype (20.0%) in our review. This finding, however, was different from previous review which reported HPV 52 with pooled estimate prevalence of 26% among HIV infected women with normal cytology and HPV 16 which was 26% among women with abnormal cytology [27]. This difference is likely to be due to the number of included studies and the difference in the data included in the analysis, study setting and participants exposure to risk factors including HIV.
Lower CD4 count most frequently reported in this review is concordant with previous reviews in which low CD4 was strongly associated with HPV infection [53,54,55]. Previous study also revealed that the most frequent high risk genotype observed in HIV positive women (i.e. 46.7%) [54] which is closer to the current pooled estimate for high risk HPV (51.0%), indicating that HIV infection might increase the susceptibility to latent HPV infection [55].
The review conducted to estimate prevalence of HPV genotype among African women, including Ethiopia revealed that HPV16, 52, 35, 18, 58, 51, 45, 31, 53, and 56 were the ten most common genotypes in the normal cervical cytology while HPV 16, 18, 35, and 52 were the four common types [56]. Another review from Ethiopian women depicted that HPV 16, 52, 18, 58, 45 were top five genotypes with the proportion of 45.3%, 9.4%, 8.2%, 6.9%, 5.2%, respectively [57]. Correspondingly, the pooled estimates of about 21 high risk HPV genotypes among HIV infected women were reported in this review with the estimated prevalence of genotype 16 (20%), 18 (15%), 52 (13%), 31 and 58 (11% each), 35 and 53 (10% each), 33, 51, and 66 (8% each), 45 (7%), and the remaining genotypes had the pooled estimate of less than seven percent.
The original research article conducted in Korea reported prevalence of 16.7% with the high risk HPV type of 12.5% [58] which is too far up when compared with the pooled estimates of the current review which focused on HIV positive women. In addition, the study among Arab women indicated 6.2% among Qatari women and 5.9% non-Qatari women [59] somewhat concordant with the study conducted in Lebanon which reported HPV prevalence of 6.7% [60]. This variation is probably attributed to the differences in the study settings, sample sizes used and the studied population.
Our finding indicated heterogeneity on the outcome variable which is the effect size equivalent to the prevalence of HPV genotypes. Therefore, careful interpretation of the heterogeneity chi-square test (variation in effect estimates beyond chance) is required, since it has low power in the situation of a meta-analysis when studies have a small sample size or are few in number. It is worth noting at this junction that while a statistically significant result may indicate a problem with heterogeneity, a non-significant result must not be taken as evidence of no heterogeneity.
Strength and limitations of the study
This review is conducted by searching more than five biomedical databases and a large number of pooled participants are involved in the study. Another strength is that this review assessed HPV prevalence studies among HIV infected women in developing countries at large and reported pooled estimates of all HPV genotypes, high risk HPV genotype and low risk HPV. The limitation of this study was inclusion of studies published only in English language. This could be one of the possible causes for observed publication bias and heterogeneity of the estimated effects.
Conclusion
This review indicated that the pooled prevalence of all genotypes HPV and high risk HPV among HIV infected women in LMICs were considerable. To enhance the well-being of HPV/HIV co-infected women it is necessary to strengthen programs for diagnosis, treatment, and provide HPV vaccination based on common high-risk genotypes.
Availability of data and materials
All data generated or analyzed during this review are included in this article.
Abbreviations
- CI:
-
Confidence interval
- df:
-
Degree of freedom
- DNA:
-
Deoxyribonucleic acid
- ELISA:
-
Enzyme linked immuno-sorbant assay
- ES:
-
Effect size
- HAART:
-
Highly active anti retroviral therapy
- HIV:
-
Human Immunodeficiency virus
- HPV:
-
Human papillomavirus
- HSILs:
-
High grade squamous intraepithelial lesions
- I2 :
-
Heterogeneity
- ICC:
-
Invasive cervical cancer
- LMICs:
-
Low and middle income countries
- MeSH:
-
Medical subject heading
- PCR:
-
Polymerase chain reaction
- SILs:
-
Squamous intraepithelial lesions
- STI:
-
Sexually transmitted infection
- tiab:
-
Title and abstract
- VLP:
-
Virus like particle
References
Hoory T, Monie A, Gravitt P, Wu TC. Molecular epidemiology of human papillomavirus. J Formos Med Assoc. 2008;107(3):198–217.
Terai M, Burk RD. Complete nucleotide sequence and analysis of a novel human papillomavirus (HPV 84) genome cloned by an overlapping PCR method. Virology. 2001;279:109–15.
Shope RE, Weston Hurst E. Infectious papillomatosis of rabbits. J Exp Med. 1933;58:607–24.
Hausen HZ. Human papilloma viruses and their possible role in squamous cell carcinomas. Curr Top Microbiol Immunol. 1977;78:1–30.
Egawa N, Egawa K, Griffin H, Doorbar J. Human papillomaviruses; epithelial tropisms, and the development of neoplasia. Viruses. 2015;7(7):3863–90.
Arbyn M, Tommasino M, Depuydt C, Dillner J. Are 20 human papillomavirus types causing cervical cancer? J Pathol. 2014;234(4):431–5.
Groves IJ, Coleman N. Pathogenesis of human papillomavirus-associated mucosal disease. J Pathol. 2015;235(4):527–38.
Silverberg MJ, Ahdieh L, Munoz A, Anastos K, Klein RS, Massad S, et al. The impact of HIV infection and immunodeficiency on human papillomavirus type 6 or 11 infection and on genital warts. Sex Transm Dis. 2002;29(8):427–35.
Stanley M. Gynecologic oncology pathology and epidemiology of HPV infection in females. Gynecol Oncol. 2010;117(2):5–10.
Steben M, Duarte-Franco E. Human papillomavirus infection: epidemiology and pathophysiology. Gynecol Oncol. 2007;107:2–5.
Echelman D, Feldman S. Management of cervical pre cancers: a global perspective. Hematol Oncol Clin N Am. 2012;26(1):31–44.
Bosch FX, Lorincz A, Munoz N, Meijer CJ, Shah KV. The causal relation between human papillomavirus and cervical cancer. J Clin Pathol. 2002;55(4):244–65.
Forman D, de Martel C, Lacey CJ, Soerjomataram I, Lortet-Tieulent J, Bruni L, et al. Global burden of human papillomavirus and related diseases. Vaccine. 2012;30:12–23.
Tota JE, Chevarie-davis M, Richardson LA, de Vries M, Franco EL. Epidemiology and burden of HPV infectionand related diseases: implications for prevention strategies. Prev Med (Baltim). 2011;53:12–21.
UNAIDS. HPV, HIV and cervical cancer; 2016.
Remis RS, Liu J, Loutfy M, Tharao W, Rebbapragada A, Perusini SJ, et al. The epidemiology of sexually transmitted coinfections in HIV-positive and HIV-negative African-Caribbean women in Toronto. BMC Infect Dis. 2013;13:550.
Couture M, Page K, Stein ES, Sansothy N, Sichan K, Kaldor J, et al. Cervical human papillomavirus infection among young women engaged in sex work in Phnom Penh, Cambodia: prevalence, genotypes, risk factors and association with HIV infection. BMC Infect Dis. 2012;12:166.
Lillo FB, Ferrari D, Veglia F, Origoni M, Grasso MA, Lodini S, et al. Human papillomavirus infection and associated cervical disease in human immunodeficiency virus—infected women: effect of highly active antiretroviral therapy. J Infect Dis. 2001;184:547–51.
Stuardo V, Agustí C, Godinez JM, Montoliu A, Torné A, Tarrats A, et al. Human papillomavirus infection in HIV-1 infected women in catalonia (Spain): implications for prevention of cervical cancer. PLoS ONE. 2012;7(10):1–9.
Moodley M, Mould S. Invasive cervical cancer and human immunodeficiency virus ( HIV ) infection in KwaZulu-Natal, South Africa. J Obstet Gynaecol. 2005;25(7):706–10.
Zhu Y, Wang Y, Hirschhorn J, Zhao Z, Davis MR, Feldman S, et al. Human papillomavirus and its testing assays, cervical cancer screening, and vaccination, vol. 81. 1st ed. Amsterdam: Elsevier; 2017.
Trottier H, Franco EL. The epidemiology of genital human papillomavirus infection. Vaccine. 2006;1:4–15.
Hawes SE, Critchlow CW, Niang MA, Diouf MB, Diop A, Touré P, et al. Increased risk of high-grade cervical squamous intraepithelial lesions and invasive cervical cancer among african women with human immunodeficiency virus type 1 and 2 infections. J Infect Dis. 2003;188:555–63.
Clarke B, Chetty R. Postmodern cancer: the role of human immunodeficiency virus in uterine cervical cancer. Mol Pathol. 2002;55:19–24.
Gheit T, Simoes RT, Tommasino M, Donadi EA, Gonçalves MA. Brief report HPV16 variants in squamous intraepithelial lesions in human immunodeficiency virus-negative and -positive Brazilian Women. Viral Immunol. 2006;19(2):340–5.
Dreyer G. Best practice & research clinical obstetrics and gynaecology clinical implications of the interaction between HPV and HIV infections. Best Pract Res Clin Obstet Gynaecol. 2018;47:95–106.
Menon S, Wusiman A, Boily MC, Kariisa M, Mabeya H, Luchters S, et al. Epidemiology of HPV genotypes among HIV positive women in Kenya: a systematic review and meta-analysis. PLoS ONE. 2016;11(10):e0163965. https://doi.org/10.1371/journal.pone.0163965S.
Human papillomavirus and related diseases report, Ethiopia. June 2019.
PRISMA 2009 Checklist Section / topic PRISMA 2009 Checklist; 2009. pp. 1–2.
World Bank Country Classifications. 2018. https://datahelpdesk.worldbank.org/knowledgebase/articles/906519
Quality Assessment Tool for Observational Cohort and Cross-Sectional Studies; 2017. pp. 1–4
Borenstein M, Hedges LV, Higgins JPT, Rothstein HR. Introduction to meta analysis. New York: Wiley; 2009.
Higgins JPT, Thompson SG, Deeks JJ, Altman DG. Measuring inconsistency in meta-analyses. BMJ. 2003;327:557–60.
Sterne JAC, Harbord RM. Funnel plots in meta-analysis. Stata J. 2004;4(2):127–41.
Sagna T, Djigma F, Zeba M, Bisseye C, Karou SD, Ouermi D, et al. Human papillomaviruses prevalence and genital coinfectionsin HIV-seropositive women in Ouagadougou (Burkina Faso). Pak J Biol Sci. 2010;13(19):951–5.
Veldhuijzen NJ, Braunstein SL, Vyankandondera J, Ingabire C, Ntirushwa J, Kestelyn E, et al. The epidemiology of humanpapillomavirus infection in HIV-positive and HIV-negative high-risk women in Kigali, Rwanda. BMC Infect Dis. 2011;11:333.
Sinayobye JDA, et al. Prevalence and risk factors for High-Risk Human Papillomavirus (hrHPV) infection among HIV-infected and Uninfected Rwandan women: implications for hrHPV-based screening in Rwanda. Infect Agents Cancer. 2014;9:40.
Rocha-BrischiliariI SC, Gimenes F, de Abreu AL, Irie MM, Souza RP, Santana RG, et al. Risk factors for cervical HPV infection and genotypes distribution in HIV-infected South Brazilian women. Infect Agents Cancer. 2014;9:6. https://doi.org/10.1186/1750-9378-9-6.
de Mattos AT, de Freitas LB, Lima BMC, Miranda AE, Spano LC. Diversity and uncommon HPV types in HIV seropositive and seronegative women attending an sti clinic. Braz J Microbiol. 2011;42:786–93.
Nicola AF, Grinsztejnb B, Friedmanb RK, Velosob VG, Cunhab CB, Georgb I, et al. Seroprevalence of HPV vaccine types 6, 11, 16 and 18 in HIV-infected and uninfected women from Brazil. J Clin Virol. 2013;57:147–51.
Nweke IG, Banjo AAF, Abdulkareem FB, Nwadike VU. “Prevalence of human papilloma virus DNA in HIV positive women in Lagos University Teaching Hospital (LUTH) Lagos, Nigeria. Br Microbiol Res J. 2013;3(3):400–13.
Akarolo-Anthony, et al. HIV associated high-risk HPV infection among Nigerian women. BMC Infect Dis. 2013;13:521.
Bollen LJM, Chuachoowong R, Kilmarx PH, Mock PA, Culnane M, Skunodom N, et al. Immunodeficiency virus-infected pregnant thai women: implications for future HPV Immunization. Sex Transm Dis. 2006;33(4):259–64.
Mcdonald AC, Tergas AI, Kuhn L, Denny L, WrightJr TCW. Distribution of human papillomavirus genotypes among HIV-positive and HIV-negative women in Cape Town, South Africa. Front Oncol. 2014;4:1–11.
Firnhaber C, Van Le H, Pettifor A, Schulze D, Michelow P, Sanne IM, et al. Association between cervical dysplasia and human papillomavirus in HIV seropositive women from Johannesburg South Africa. Cancer Causes Control. 2010;21:433–43.
Firnhaber C, Zungu K, Levin S, Michelow P, Montaner LJ, McPhail P, et al. Diverse and high prevalence of human papillomavirus associated with a significant high rate of cervical dysplasia in human immunodeficiency virus-infected women in Johannesburg, South Africa. Acta Cytol. 2009;53:10–7.
Denny L, Boa R, Williamson A, Allan B, Hardie D, Stan R, et al. Human papillomavirus infection and cervical diseasein human immunodeficiency virus-1-infected women. Obstet Gynecol. 2008;111(6):1380–7.
Sahasrabuddhe VV, Mwanahamuntu MH, Vermund SH, Huh WK, Stringer JSA, Parham GP, et al. Prevalence and distribution of HPV genotypes among HIV-infected women in Zambia. Br J Cancer. 2007;96:1480–3.
Rousseau MND, Nagot N, Martineau VC, Ouedraogo A, Konate I, Weiss HA, et al. Human papillomavirus genotype distribution and cervical squamous intraepithelial lesions among high-risk women with and without HIV-1 infection in Burkina Faso. Br J Cancer. 2006;95(3):355–62.
Camargo, et al. Human papillomavirus detection in women with and without human immunodeficiency virus infection in Colombia. BMC Cancer. 2014;14:451.
Munoz M, Camargo M, De Leon SCS, Sanchez R, Pineda-Peña AC, Perez-Prados A, et al. Classical molecular tests using urine samples as a potential screening tool for human papillomavirus detection in human immunodeficiency virusinfected women. J Clin Microbiol. 2013;51:3688–93.
Kelly HA. The epidemiology of Human Papillomavirus (HPV) infection and epigenetic factors associated with the development of cervical cancer precursor lesions in women living with HIV in Africa. 2017.
Boccalon M, Tirelli U, Sopracordevole F, Vaccher E. Intra-epithelial and invasive cervical neoplasia during HIV infection. EurJ Cancer. 1996;32A(13):2212–7.
Freitas BC, Suehiro TT, Consolaro MEL, Silva VRS. (2015) MINI-REVIEW HPV infection and cervical abnormalities in HIV positive women in different regions of Brazil, a Middle-Income Country. Asian Pac J Cancer Prev. 2015;16(18):8085–91.
Denny LA, Franceschi S, De Sanjosé S, Heard I, Moscicki AB, Palefsky J. Human papillomavirus, human immunodeficiency virus and immunosuppression. Vaccine. 2012;30:168–74.
Ogembo RK, Gona PN, Seymour AJ, Park HS-M, Bain PA, Maranda L, et al. Prevalence of human papillomavirus genotypes among African women with normal cervical cytology and neoplasia: a systematic review and meta-analysis. PLoS ONE. 2015;10(4):e0122488. https://doi.org/10.1371/journal.pone.0122488.
Derbie A, Mekonnen D, Yismaw G, Biadglegne F. Human papillomavirus in Ethiopia. Virus Disease. 2019.
Ouh YT, Min KJ, Cho HW, Ki M, Oh JK, Shin SY, et al. Prevalence of human papillomavirus genotypes and precancerous cervical lesions in a screening population in the Republic of Korea, 2014–2016. J Gynecol Oncol. 2018;29(1):1–10.
Bansal D, Elmi AA, Skariah S, Haddad P, Abu-Raddad LJ, Al Hamadi AH, et al. Molecular epidemiology and genotype distribution of Human Papillomavirus (HPV) among Arab women in the state of Qatar. J Transl Med. 2014;12:300.
Hanna J, Yassine R, El-Bikai R, Curran MD, Azar M, Yeretzian J, et al. Molecular epidemiology and socio- demographic risk factors of sexually transmitted infections among women in Lebanon. BMC Infect Dis. 2020;20:375.
Acknowledgements
The authors acknowledge the contribution of Aklilu Lemma Institute of Pathobiology, Addis Ababa University, Department of Tropical and Infectious Diseases and Ethiopian Public Health Institute for the opportunity to access an internet. In particular, authors like to thank Minilik Demesie and Habteyes Tolla, EPHI staff for their unreserved support for this review protocol.
Funding
There is no fund for this review.
Author information
Authors and Affiliations
Contributions
Conceptualization: AL. Data curation: AL, GM. Formal analysis: AL, GM. Investigation: AL and JH. Methodology: AL, GM, NB, JH. Project administration: AL. Writing—original draft: AL. Writing—review and editing: GM, NB, JH. All authors read and approved the final manuscript.
Authors’ information
The first author Agajie Likie Bogale (AL) is staff of Ethiopian Public Health Institute and currently PhD student at Aklilu Lemma Institute of Pathobiology, Addis Ababa University, Department of Tropical and Infectious Diseases; Nega Berhe Belay (NB) is senior instructor at Aklilu Lemma Institute of Pathobiology, Addis Ababa University, Department of Tropical and Infectious Diseases; Girmay Medhin (GM) is senior instructor of the course Advanced Research Methods and Biomedical Data Analysis; Professor Jemal Haidar Ali (JH) is senior consultant and instructor at school of public health, Addis Ababa University, Ethiopia.
Corresponding author
Ethics declarations
Ethical approval and consent to participate.
This review was based on previous published articles indicating that there is no need of ethical clearance. However, the protocol of this study was pre-registered on PROSPERO (International prospective register of systematic reviews) University of York, Centre for Reviews and Dissemination with registration number CRD42019123549.
Consent for publication
Not applicable.
Competing interest
There is no any competing interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Bogale, A.L., Belay, N.B., Medhin, G. et al. Molecular epidemiology of human papillomavirus among HIV infected women in developing countries: systematic review and meta-analysis. Virol J 17, 179 (2020). https://doi.org/10.1186/s12985-020-01448-1
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12985-020-01448-1