Abstract
Background
Since H7N9 influenza A virus (H7N9) was first reported in 2013, five waves of outbreaks have occurred, posing a huge threat to human health. In preparation for a potential H7N9 epidemic, it is essential to evaluate the efficacy of anti-H7N9 drugs with an appropriate model.
Methods
Well-differentiated pseudostratified human airway epithelium (HAE) cells were grown at the air–liquid interface, and the H7N9 cell tropism and cytopathic effect were detected by immunostaining and hematoxylin-eosin (HE) staining. The H7N9 replication kinetics and anti-H7N9 effect of recombinant human α2b (rhIFN-α2b) and rhIFN-λ1 were compared with different cell lines. The H7N9 viral load and interferon-stimulated gene (ISG) expression were quantified by real-time PCR assays.
Results
H7N9 could infect both ciliated and non-ciliated cells within the three-dimensional (3D) HAE cell culture, which reduced the number of cilia and damaged the airways. The H7N9 replication kinetics differed between traditional cells and 3D HAE cells. Interferon had antiviral activity against H7N9 and alleviated epithelial cell lesions; the antiviral activity of rhIFN-α2b was slightly better than that of rhIFN-λ1. In normal cells, rhIFN-α2b induced a greater amount of ISG expression (MX1, OAS1, IFITM3, and ISG15) compared with rhIFN-λ1, but in 3D HAE cells, this trend was reversed.
Conclusions
Both rhIFN-α2b and rhIFN-λ1 had antiviral activity against H7N9, and this protection was related to the induction of ISGs. The 3D cell culture model is suitable for evaluating interferon antiviral activity because it can demonstrate realistic in vivo-like effects.
Similar content being viewed by others
Background
In addition to the seasonal influenza virus, some avian influenza viruses, such as H7N9 and H5N1 avian influenza A, can also infect humans. Since the first human infection with a novel H7N9 influenza virus (H7N9) was confirmed in China in the spring of 2013 [1], there have been five epidemic waves of human H7N9 infections through 2017 [2]. Vaccination is the most effective way to prevent influenza infection. However, there is currently no commercial human vaccine available that is specific for H7N9. Antiviral treatment is another critical strategy for controlling infection with H7N9, and neuraminidase (NA) inhibitors are the most widely used drugs against influenza infection [3]. However, with the increase in drug-resistance-conferring mutations, other measures are also needed to treat infection with H7N9. Previous studies have shown that type I interferon (IFN) was active against the influenza 2009 pandemic H1N1 and highly pathogenic H5N1 strains [4, 5]. Additionally, the natural IFN Alferon N, was shown to inhibit the replication of oseltamivir-sensitive and -resistant H7N9 isolates [6].
Primary human airway epithelium (HAE) cells can be further differentiated into polarized HAE cells when they are subjected to air–liquid interface (ALI) culture for 4 to 6 weeks. The morphology and functionality of these cells resembles the in vivo human pseudostratified mucociliary epithelium, and this system is a promising tool for the study of respiratory viruses. Many common and emerging respiratory viruses, such as influenza A [7, 8], respiratory syncytial virus (RSV) [9], adenovirus (ADV) [10], parainfluenza virus (PIV) [11], and human coronavirus (HCoV) [8, 12], can replicate in these three-dimensional (3D) HAE cells. Moreover, some newly described viruses, including HBoV [13, 14] and HCoV HKU1 [15], that could not be cultured on traditional cell lines can be cultured on these cells. Of the currently available cell culture models, 3D HAE cells reconstruct the morphological and physiological characteristics of the respiratory tract to the greatest extent, therefore, it is a robust cellular model for respiratory virus research and can also be used to evaluate the therapeutic effect of drugs and transgenic strategies [16].
Despite type I and type III IFN binding to different receptors, they both use similar JAK–STAT signal pathways and induce the expression of an overlapping set of IFN-stimulated genes (ISGs) [17]. Thus, the type III IFNs shares some properties with the type I IFNs, such as a role in antiviral defense as well as antiproliferative and immunoregulative activities [18].
In this study, we used 3D HAE cell cultures to study the target cell tropism and the infection and proliferation features of H7N9. The antiviral activities of type III and type I recombinant human IFNs (rhIFNs) were compared on A549 cells, 2D HAE cells, and 3D HAE cells, and the expression of antiviral genes in different cell models was also analyzed.
Materials and methods
Virus and cells
H7N9 A/Anhui/1/2013 was obtained from the Chinese National Influenza Center, and all experiments with this virus were performed in approved enhanced biosafety level 3 (BSL-3) laboratories. A549 cells were cultured in Dulbecco’s modified Eagle medium (DMEM; Gibco, NY, USA) with 10% fetal bovine serum (Gibco).
Human airway epithelial cell culture
Primary HAE cells were isolated from patients who underwent surgical lung resection for pulmonary diseases in Nanjing Children’s Hospital, as described previously [19]. HAE cells were plated onto type I and III collagen-coated six-well tissue culture plates and cultured in BEGM media (Lonza, Germany) supplemented with the required additives (Lonza). When the cells reached 80–90% confluence, traditional monolayer two-dimensional (2D) HAE cells were dissociated with trypsin, and 3 × 105 cells were seeded on type IV collagen-coated 12-well transwell inserts (Costar, ME, USA). Medium was renewed for both the apical and basolateral surfaces every other day. The medium was then changed to air–liquid interface (ALI) medium (BEGM+DMEM+additives) until the HAE cells reached full confluence. After 5 days, the HAE cells were exposed to air, and only the basolateral compartment was cultured with ALI medium. The ALI culture was continued for 4–6 weeks, during which time the cells differentiated into 3D pseudostratified HAE cells. Prior to the experiments, all cultures were maintained at 37 °C in a 5% CO2 incubator.
Immunostaining for H7N9 virus on 3D HAE cells
3D HAE cells that had been infected with 100 tissue culture infective doses (TCID50) of H7N9 for 24 h were fixed with 4% paraformaldehyde (PFA) for 30 min at room temperature, followed by washing of both the apical and basolateral sides three times each with PBS. Subsequently, the fixed cells were permeabilized with 0.2% Triton X for 2 h and blocked with 5% bovine serum albumin for 1 h. For the simultaneous detection of H7N9 protein M2 and ciliated cells or tight junctions, anti-influenza A M2 rabbit polyclonal antibody (PA5–32233, 1:500, Thermo Fisher), and mouse monoclonal anti-β tubulin antibody (T4026, 1:100, Sigma) or mouse monoclonal anti-ZO-1 antibody (33–9100, 1:500, Invitrogen, CA, USA), respectively, were applied as primary antibodies. Goat-derived Dylight 594-labeled anti-mouse IgG (H + L) (35,511, 1:500, Thermo Fisher) and Dylight 488-labeled anti-rabbit IgG (H + L) (35,553, 1:500, Thermo Fisher) were applied as secondary antibodies. Nuclei were counterstained with DAPI. Finally, the filters with cells were excised from the insert and mounted under coverslips on glass slides with mounting medium. Confocal images were taken with an UltraView VoX confocal microscope (PerkinElmer, Boston, MA, USA).
Inoculation of H7N9 virus on different cell lines
The apical surface of 3D HAE cells was rinsed three times with PBS and then incubated with 100 μl of 0.25 μg/ml rhIFN-λ1 (R&D, USA) or 60 IU/ml rhIFN-α2b (Yuance, China) for 20 h at 37 °C in a 5% CO2 incubator. After discarding the cell culture supernatant, the 3D HAE cells were inoculated on the apical surface with 100 μl of 1 TCID50 of H7N9 per well and incubated for 1 h. Virus and rhIFNs diluents were prepared with ALI medium. At the end of the incubation, the unbound H7N9 viral particles were removed by rising with Hank’s buffer, and the basolateral compartment was supplemented with 1.5 ml of ALI medium for 24 h at 37 °C in a 5% CO2 incubator. Subsequently, 200 μl of ALI medium was added to the apical surface, and the supernatants were collected after 30 min. Virus RNA was extracted using the QIAamp MinElute Virus Spin Kit (Qiagen, Hilden, Germany), and quantification of H7N9 viral yields was performed using AgPath-ID™ One-Step RT-PCR Reagents (ABI, USA) to analyze the anti-H7N9 effect of different rhIFNs.
For A549 cells, the experimental method was similar to that used for 3D HAE cells, except that the A549 cells were cultured on 24-well plates and the applied virus medium was DMEM with added TPCK-trypsin (0.5 μg/ml). Nucleic acids were extracted and quantified once per day for 10 days. Real-time PCR assays were used for the detection of the HA gene from H7N9 A/Anhui/1/2013. The primer sequences were H7N9-F 5′-AGAAATGAAATGGCTCCTGTCAA-3′ and H7N9-R 5′-GGTTTTTTCTTGTATTTTTATATGACTTAG-3′, and the probe sequence was H7N9-P FAM-AGATAATGCATTCCCGCAGAT-TAMRA.
Anti-H7N9 activity of different types of rhIFNs
2D HAE cells were rinsed three times with PBS and then incubated with 100 μl of successive 40- to 49-fold dilutions of 4 μg/ml rhIFN-λ1 (R&D) or 1000 U/ml rhIFN-α2b (Yuance) for 20 h at 37 °C in a 5% CO2 incubator. Next, the cells were challenged with 100 μl of 100 TCID50 of H7N9 per well. After incubation for 1 h, the unbound H7N9 viral particles were removed by rising with Hank’s buffer, and the cells were supplemented with 100 μl of BEGM medium. Harvests were collected at 72 h post-inoculation, and H7N9 RNA extraction and nucleic acid quantification were performed as described above to analyze the anti-H7N9 effect of different rhIFNs. For 3D HAE cells and A549 cells, the experimental protocols were the same as that described for 2D HAE cells, but the medium used for cell culture was ALI medium and DMEM, respectively.
HE staining
The apical surface and the basolateral chamber of 3D HAE cells were cultured with 4 μg/ml rhIFN-λ1 (R&D, USA) or 1000 IU/ml rhIFN-α2b (Yuance, China) for 20 h at 37 °C in a 5% CO2 incubator (500 μl and 1.5 ml for the apical surface and basolateral chamber, respectively). The apical surface was washed three times with PBS, followed by inoculation with 100 μl of 100 TCID50 of H7N9, and the basolateral chamber was maintained with 1.5 ml of ALI medium. After incubation for 1 h, the apical chamber was washed twice with PBS and maintained continuously with an ALI for 24 h. The cells were fixed in 4% PFA along with the entire transwell insert for hematoxylin-eosin (HE) staining.
Detection of ISGs
A549 cells, 2D HAE cells, and well-differentiated pseudostratified 3D HAE cells were each incubated with 60 IU/ml rhIFN-α2b (Yuance) or 0.25 μg/ml rhIFN-λ1 (R&D) for 18 h at 37 °C in a 5% CO2 incubator. After rinsing with PBS, the cells were lysed with Trizol, and the total RNA was extracted. Reverse transcription was performed using the Superscript® III First-Strand Synthesis System (Invitrogen). The relative expression levels of ISGs, including ISG15, MX1, and OAS1, were analyzed by a Taqman real-time PCR assay using the 2-ΔΔCT method, and GADPH was used as the internal reference [20]. The sequences of primers and probes are shown in Table 1.
Results
Immunofluorescence analysis
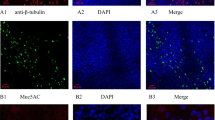
β-tubulin is an important component of the cytoskeleton and is a marker protein of cilia, so it can be used to distinguish between ciliated cells and non-ciliated cells. The tight junction protein zonula occludens-1 (ZO-1) is located at the apex of epithelial cells; it is a major component of tight junctions and plays an important role in maintaining pseudostratified cell layer integrity and barrier function. As shown by immunofluorescence staining with anti-β-tubulin IV (Fig. 1a) and anti-ZO1 (Fig. 1b) antibodies, H7N9 could infect both non-ciliated and ciliated cells. In contrast with the state of the cilia in the mock control, 3D HAE cells that were infected with H7N9 showed an obvious reduction of cilia. Additionally, the H7N9-infected 3D HAE cells showed a disassociation of the ZO-1, suggesting airway epithelial damage.
Immunofluorescence of 3D HAE cells infected with H7N9. At 24 h post-inoculation with H7N9, the 3D HAE cells were fixed, followed by double immunostaining with anti-influenza A M2 polyclonal antibody (green) and mouse monoclonal anti-β tubulin antibody (red, a) or mouse monoclonal anti-ZO-1 antibody (red, b). Confocal images were taken with a magnification of 100×. Nuclei were stained with DAPI
Replication characteristics of H7N9 influenza virus on different cells
A549 cells and 3D HAE cells were each challenged with 1 TCID50 of H7N9 after being treated with rhIFN-α2b (60 IU/ml) or rhIFN-λ1 (250 ng/ml) for 20 h. The viral load was then detected by real-time PCR for 10 consecutive days using the method described above. In the control group without rhIFN treatment, the viral load in A549 cells reached a peak of 3.9 × 109 copies/ml on the sixth day and decreased to 3.5 × 106 copies/ml on the tenth day (Fig. 2a). In control 3D HAE cells, the viral load reached a peak of 1.29 × 109 copies/ml on the third day and was reduced to 1.1 × 106 copies/ml on the tenth day (Fig. 2b). In contrast, the viral load of A549 cells treated with rhIFN peaked on the second day and then gradually decreased (Fig. 2a). For 3D HAE cells, viral replication was inhibited during the first few days after rhIFN treatment; the viral load was reduced to 1.58 × 104 copies/ml (rhIFN-α2b) or 9.55 × 103 copies/ml (rhIFN-λ1) on the third day. Later, it gradually rose to a peak of 4.26 × 108 copies/ml (rhIFN-α2b) or 6.31 × 108 copies/ml (rhIFN-λ1) on the sixth day and then gradually decreased again (Fig. 2b).
Replication characteristics of H7N9 on A549 cells and 3D HAE cells. A549 cells (a) and 3D HAE cells (b) were each incubated with 0.25 μg/ml rhIFN-λ1 or 60 IU/ml rhIFN-α2b for 20 h and then infected with 1 TCID50 of H7N9 virus. Viral RNA was extracted once per day for 10 days. The H7N9 HA gene was quantitatively detected by real-time PCR
Anti-H7N9 activity of rhIFNs on different cells
A549 cells, 2D HAE cells and 3D HAE cells were each incubated with serial dilutions of rhIFNs (initial concentration of rhIFN-λ1 and rhIFN-α2b was 4 μg/ml and 1000 IU/ml, respectively) for 20 h, after which these cells were challenged with 100 TCID50 of H7N9. Viral RNA was extracted 72 h later, and quantitative real-time PCR assay targeting the H7N9 HA gene was conducted to quantify the viral load. Among the three cell types, viral proliferation was the fastest in 3D HAE cells, reaching 1 × 1010 copies/ml, while the HA gene copy number in A549 cells and 2D HAE cells was 2.5 × 107 copies/ml and 8 × 107 copies/ml, respectively. The antiviral activity of rhIFN-α2b was superior to that of rhIFN-λ1 in all three cell types, A549 cells (Fig. 3a), 2D HAE cells (Fig. 3b), and 3D HAE cells (Fig. 3c). As the dose of rhIFN decreased, the antiviral activity also decreased, which demonstrates a dose-response relationship. Compared with the control group, the viral load was decreased to 2.8 × 106 copies/ml and 2.3 × 107 copies/ml, which means 3571-fold and 435 -fold (1 × 1010 copies/ml) reduction, respectively, when the 3D HAE cells were treated with 4 μg/ml rhIFN-λ1 or 1000 IU/ml rhIFN-α2b. Because of the biggest drop in viral load, the antiviral activity of rhIFN was the strongest when it was applied to the 3D cells (Fig. 3c).
Anti-H7N9 bioactivity of different rhIFN types. A549 cells (a), 2D HAE cells (b), and 3D HAE cells (c) were each incubated with successive 40- to 49-fold dilutions of 4 μg/ml rhIFN-λ1 or 1000 IU/ml rhIFN-α2b for 20 h and then challenged with 100 TCID50 of H7N9. At 72 h post-inoculation, anti-H7N9 bioactivity of different rhIFNs were compared as described above
Cytopathic effect of H7N9 on 3D HAE cells
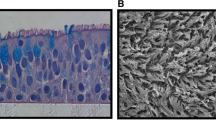
HE staining of ALI-cultured HAE cells revealed that the nuclear position was uneven, there appeared to be multi-layered cells with cilia distributed at the top, and the morphological structure was similar to the pseudostratified ciliated columnar epithelium of human tracheal tissue. In the H7N9 infection group, there were many vacuoles, the number of cilia was reduced, and the cell layer was thinner. However, when 3D HAE cells were incubated with rhIFNs and then challenged with H7N9, the cytopathic effect of H7N9 infection was alleviated (Fig. 4).
rhIFNs-induced gene expression
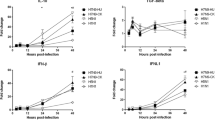
The relative expression levels of OAS1, MX1, ISG15, and IFITM3 were detected by real-time PCR using GADPH as the internal reference. Compared with the control group without rhIFNs treatment, cells treated with either rhIFN-α2b or rhIFN-λ1 had significantly upregulated expression levels of all four genes. For A549 cells and 2D HAE cells, the expression levels of these four genes were significantly higher in the rhIFN-λ1 group than in the rhIFN-α2b group. In contrast, the ISG expression levels were significantly higher in the rhIFN-α2b group than in the rhIFN-λ1 group for the 3D HAE cells (Fig. 5). These results demonstrate that the gene expression induced by rhIFNs was different between 3D HAE cells and ordinary 2D-cultured cells.
Effect of rhIFN treatment on ISG induction in different cells. After treatment of A549 cells, 2D HAE cells, and 3D HAE cells with rhIFN-α2b or rhIFN-λ1, the total RNA was extracted and reverse transcribed. The relative expression levels of ISG15, MX1, OAS1, and IFITM3 were measured using Taqman real-time PCR assays and quantified by the 2−ΔΔCT method. *P < 0.05; **P < 0.01
Discussion
The human population has little or no immunity to H7N9 because the H7 subtype of influenza has never previously infected humans; thus, infection with H7N9 may have a high mortality rate [21]. Since 2013, H7N9 has caused five outbreak waves, and 1561 laboratory-confirmed cases of H7N9 infection have been reported [22]. H7N9 has also been detected in animals and the environment; therefore, it poses a significant threat to public health as well as a potential pandemic risk. Currently, no universal influenza vaccine is available, and NA inhibitors are the only FDA-approved drugs for the treatment of H7N9 infection [23]. With the rapid emergence of NA inhibitor-resistant strains after treatment with oseltamivir, the utility of NA inhibitors in the clinical treatment of influenza-infected patients is limited, so other drugs will be required to control H7N9 infection.
In 1988, a tracheal epithelial cell model with a polarity of differentiation was established via ALI culture [24]. Subsequently, this model has been widely used for the amplification, isolation, and identification of respiratory viruses, as well as for the study of respiratory viral infection and therapeutic mechanisms. Most importantly, the 3D HAE system greatly facilitates the isolation and characterization of emerging viruses that are unable to proliferate in traditional 2D-cultured cells. Notably, the antiviral effect of drugs observed using 3D HAE may reflect the actual activity of those drugs in vivo. Here, we tested the anti-H7N9 activity of type I and type III rhIFNs in different culture systems using H7N9 human clinical isolate A/Anhui/1/2013.
The advantage of immunostaining for virus culture is that the cell tropism of the virus can be detected. Ciliated cells possess both α-2,6- and α-2,3-linked sialic acid receptors, whereas non-ciliated cells present mostly α-2,6-linked sialic acid receptors. We observed that H7N9 can infect both ciliated cells and non-ciliated cells because the H7N9 viral particles bound to both avian-type (α-2,3) and human-type (α-2,6) [25]. Confocal images of H7N9-infected HAE cells clearly displayed a reduction in the amount of cilia and the destruction of tight junctions compared with uninfected controls.
In 1986, rhIFN-α2b was approved by US FDA for the clinical treatment of diseases such as hair cell leukemia, AIDS-related kaposi’s sarcoma, and chronic hepatitis B and C, achieving great social and economic benefits. However, long-term use of rhIFN-α2b may cause side effects such as fatigue, fever, anorexia, and depression, which are associated with the wide distribution of rhIFN-α2b receptors. In contrast, type III IFN receptors have a limited distribution, mainly in the epithelial tissues of the lungs and intestines, so the side effects are relatively minor. Previous studies in our lab have shown that the antiviral activity of rhIFN-λ1 is higher than that of rhIFN-λ2 and rhIFN-λ3. Moreover, type I IFN is effective against the influenza 2009 pandemic H1N1 and highly pathogenic H5N1 strains. For the above reasons, rhIFN-α2b and rhIFN-λ1 were chosen to study the antiviral activity against H7N9 influenza A virus in different cell models.
The viral yield peaked on days 6 and 3 post-infection with 1 TCID50 of H7N9 virus in A549 cells and 3D HAE cells, respectively. The viral proliferation profiles in these cells were also different after rhIFNs treatment. In addition, there was a dose-effect relationship between the dose of rhIFN-α2b or rhIFN-λ1 and its activity against H7N9. In all three cell types, the antiviral activity of rhIFN-α2b was slightly better than that of rhIFN-λ1 to varying degrees. Together, these results highlight the inconsistencies between the replication kinetics of H7N9 in 2D-cultured cells and 3D HAE cells as well as the differences in the antiviral activity of rhIFNs between different cells.
A549 cells are adenocarcinomic human alveolar basal epithelial cells, and the 3D HAE cells used in our study was derived from human tracheal tissue. So, the tissue sources of the two cell lines are basically the same. The two chains of type I IFNs receptors, IFNAR1 and IFNAR2, and a component of the type III IFN receptor, IL10R2, was expressed in all cells, while IFNLR1, another component of type III IFN receptors, was only expressed in certain cells. It was reported the expression of IFNLR1 in A549 cells and lung tissue were almost similar using northern blot analysis [26], so the expression levels of type I and type III IFN receptors were also consistent. When binding to receptors located on the cell surface, IFN triggers the downstream JAK–STAT signaling pathway and eventually induces the expression of a series of ISGs. These genes target multiple stages of the viral life cycle and exhibit various antiviral activities due to their diverse structures and functions. MX1 [27, 28] and IFITM3 [29, 30] mainly effect viral invasion, while OAS1 [31] and ISG15 [32, 33] act primarily on the viral replication and transcription stages.
In the present study, the expression levels of MX1, OAS1, IFITM3, and ISG15 in A549 cells, 2D HAE cells, and 3D HAE cells were all clearly increased after incubation with rhIFN-α2b or rhIFN-λ1. In the A549 and 2D HAE cells, the expression levels of these four genes were significantly higher in the rhIFN-λ1 group than in the rhIFN-α2b group. However, it is noteworthy that the corresponding result was exactly the opposite in the 3D HAE cells. This indicates that the induction of ISGs by rhIFN-α2b and rhIFN-λ1 is inconsistent between 2D-cultured cells and 3D HAE cells.
It was reported that the expression of the MX1 can be significantly upregulated in A549 cells when treated with rhIFN-λ1 at 10 ng/ml, but treatment with rhIFN-β at 0.01 pg/ml did not increase the MX1 expression level [34]. In this study, we found that MX1 expression increased markedly following treatment with rhIFN-λ1 or rhIFN-α2b in 2D HAE and A549 cells compared with the results for 3D HAE cells. In addition, knockdown of MX1 expression resulted in a remarkable decrease in the expression of ISGs in A549 cells after infected with influenza A virus, thereby increased virus replication [28]. Although the upregulation of IFN-induced IFITM3 observed in this study was relatively low, the key role of this ISG cannot be ignored. Remarkably, IFITM3 can inhibit H7N9 replication early in viral infection [35]. Additionally, it was reported that monomethylation of IFITM3 at Lys-88 is downregulated after treatment rhIFN-α, thereby enhancing IFITM3 activity and inhibiting influenza virus [36]. Furthermore, the overexpression of IFITM proteins can rigidify the cell membrane, thus markedly inhibiting viral membrane fusion and entry [37]. IFITM3 plays an important role in the pathogenesis of the influenza disease, and it may serve as a potential target for treatment and management of influenza-associated infection.
A recent study by Ilyushina and colleagues [34] showed that a combination of the NA inhibitor oseltamivir carboxylate and rhIFN-λ1 had a strong synergistic interaction. This discovery suggests that rhIFN combined with NA inhibitors might be a more effective therapeutic strategy against H7N9 infection. Our future work will investigate this combination therapy in appropriate cell lines and animal models.
Conclusions
The H7N9 cell tropism on 3D HAE cells was observed and anti-H7N9 bioactivity of rhIFN-α2b and rhIFN-λ1 on different cell culture systems was tested. Our results indicate that H7N9 can infect both ciliated cells and non-ciliated cells, causing the cilia and tight junction damage. Additionally, the replication kinetics of H7N9 in 2D-cultured cells and 3D HAE cells was different. Both rhIFN-α2b and rhIFN-λ1 have antiviral activity against H7N9 on A549 cells, 2D HAE cells, and 3D HAE cells, and the protective effects were related to the induction of ISGs.
Abbreviations
- 3D:
-
Three-dimensional
- ALI:
-
Air–liquid interface
- CPE:
-
Cytopathic effect
- HAE:
-
Human airway epithelium
- HE:
-
Hematoxylin-eosin
- IFN:
-
Interferon
- ISG:
-
Interferon-stimulated gene
- NA:
-
Neuraminidase
- ZO-1:
-
Zonula occludens-1
References
Gao R, Cao B, Hu Y, Feng Z, Wang D, Hu W, et al. Human infection with a novel avian-origin influenza A (H7N9) virus. N Engl J Med. 2013;368(20):1888–97.
Zhu W, Dong J, Zhang Y, Yang L, Li X, Chen T, et al. A gene constellation in avian influenza A (H7N9) viruses may have facilitated the fifth wave outbreak in China. Cell Rep. 2018;23(3):909–17.
Beigel J, Bray M. Current and future antiviral therapy of severe seasonal and avian influenza. Antivir Res. 2008;78(1):91–102.
Haasbach E, Droebner K, Vogel AB, Planz O. Low-dose interferon type I treatment is effective against H5N1 and swine-origin H1N1 influenza a viruses in vitro and in vivo. J Interf Cytokine Res. 2011;31(6):515–25.
Osterlund P, Pirhonen J, Ikonen N, Ronkko E, Strengell M, Makela SM, et al. Pandemic H1N1 2009 influenza A virus induces weak cytokine responses in human macrophages and dendritic cells and is highly sensitive to the antiviral actions of interferons. J Virol. 2010;84(3):1414–22.
Liu Q, Ma J, Strayer DR, Mitchell WM, Carter WA, Ma W, et al. Emergence of a novel drug resistant H7N9 influenza virus: evidence based clinical potential of a natural IFN-alpha for infection control and treatment. Expert Rev Anti-Infect Ther. 2014;12(2):165–9.
Thompson CI, Barclay WS, Zambon MC, Pickles RJ. Infection of human airway epithelium by human and avian strains of influenza a virus. J Virol. 2006;80(16):8060–8.
Farsani SM, Deijs M, Dijkman R, Molenkamp R, Jeeninga RE, Ieven M, et al. Culturing of respiratory viruses in well-differentiated pseudostratified human airway epithelium as a tool to detect unknown viruses. Influenza Other Respir Viruses. 2015;9(1):51–7.
Zhang L, Peeples ME, Boucher RC, Collins PL, Pickles RJ. Respiratory syncytial virus infection of human airway epithelial cells is polarized, specific to ciliated cells, and without obvious cytopathology. J Virol. 2002;76(11):5654–66.
Pickles RJ, McCarty D, Matsui H, Hart PJ, Randell SH, Boucher RC. Limited entry of adenovirus vectors into well-differentiated airway epithelium is responsible for inefficient gene transfer. J Virol. 1998;72(7):6014–23.
Zhang L, Bukreyev A, Thompson CI, Watson B, Peeples ME, Collins PL, et al. Infection of ciliated cells by human parainfluenza virus type 3 in an in vitro model of human airway epithelium. J Virol. 2005;79(2):1113–24.
Sims AC, Baric RS, Yount B, Burkett SE, Collins PL, Pickles RJ. Severe acute respiratory syndrome coronavirus infection of human ciliated airway epithelia: role of ciliated cells in viral spread in the conducting airways of the lungs. J Virol. 2005;79(24):15511–24.
Dijkman R, Koekkoek SM, Molenkamp R, Schildgen O, van der Hoek L. Human bocavirus can be cultured in differentiated human airway epithelial cells. J Virol. 2009;83(15):7739–48.
Deng X, Li Y, Qiu J. Human bocavirus 1 infects commercially available primary human airway epithelium cultures productively. J Virol Methods. 2014;195:112–9.
Pyrc K, Sims AC, Dijkman R, Jebbink M, Long C, Deming D, et al. Culturing the unculturable: human coronavirus HKU1 infects, replicates, and produces progeny virions in human ciliated airway epithelial cell cultures. J Virol. 2010;84(21):11255–63.
Moss RB, Milla C, Colombo J, Accurso F, Zeitlin PL, Clancy JP, et al. Repeated aerosolized AAV-CFTR for treatment of cystic fibrosis: a randomized placebo-controlled phase 2B trial. Hum Gene Ther. 2007;18(8):726–32.
Schneider WM, Chevillotte MD, Rice CM. Interferon-stimulated genes: a complex web of host defenses. Annu Rev Immunol. 2014;32:513–45.
O'Shea JJ, Schwartz DM, Villarino AV, Gadina M, McInnes IB, Laurence A. The JAK-STAT pathway: impact on human disease and therapeutic intervention. Annu Rev Med. 2015;66:311–28.
Li MZ, Pang LL, Wang H, Jin Y, Duan ZJ. Isolation of primary human bronchial epithelial cells and culture at an air-liquid interface. Chin J Biologicals. 2016;29(01):65–9. (In Chinese).
Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) method. Methods. 2001;25(4):402–8.
De Groot AS, Ardito M, Terry F, Levitz L, Ross T, Moise L, et al. Low immunogenicity predicted for emerging avian-origin H7N9: implication for influenza vaccine design. Hum Vaccin Immunother. 2013;9(5):950–6.
Wang X, Jiang H, Wu P, Uyeki TM, Feng L, Lai S, et al. Epidemiology of avian influenza A H7N9 virus in human beings across five epidemics in mainland China, 2013-17: an epidemiological study of laboratory-confirmed case series. Lancet Infect Dis. 2017;17(8):822–32.
Hay AJ, Hayden FG. Oseltamivir resistance during treatment of H7N9 infection. Lancet. 2013;381(9885):2230–2.
Whitcutt MJ, Adler KB, Wu R. A biphasic chamber system for maintaining polarity of differentiation of cultured respiratory tract epithelial cells. In Vitro Cell Dev Biol. 1988;24(5):420–8.
Zhou J, Wang D, Gao R, Zhao B, Song J, Qi X, et al. Biological features of novel avian influenza a (H7N9) virus. Nature. 2013;499(7459):500–3.
Sheppard P, Kindsvogel W, Xu W, Henderson K, Schlutsmeyer S, Whitmore TE, et al. IL-28, IL-29 and their class II cytokine receptor IL-28R. Nat Immunol. 2003;4(1):63–8.
Nigg PE, Pavlovic J. Oligomerization and GTP-binding requirements of MxA for viral target recognition and antiviral activity against influenza a virus. J Biol Chem. 2015;290(50):29893–906.
Schattgen SA, Oguin TH, Thomas PG. The antiviral molecule Mx1 positively regulates the induction of type I IFN in response to influenza infection. J Immunol. 2016;196(1 Supplement):202.7.
Brass AL, Huang IC, Benita Y, John SP, Krishnan MN, Feeley EM, et al. The IFITM proteins mediate cellular resistance to influenza A H1N1 virus, West Nile virus, and dengue virus. Cell. 2009;139(7):1243–54.
Huang IC, Bailey CC, Weyer JL, Radoshitzky SR, Becker MM, Chiang JJ, et al. Distinct patterns of IFITM-mediated restriction of filoviruses, SARS coronavirus, and influenza A virus. PLoS Pathog. 2011;7(1):e1001258.
Floyd-Smith G, Slattery E, Lengyel P. Interferon action: RNA cleavage pattern of a (2′-5′)oligoadenylate--dependent endonuclease. Science. 1981;212(4498):1030–2.
Andersen JB, Hassel BA. The interferon regulated ubiquitin-like protein, ISG15, in tumorigenesis: friend or foe? Cytokine Growth Factor Rev. 2006;17(6):411–21.
Villarroya-Beltri C, Baixauli F, Mittelbrunn M, Fernandez-Delgado I, Torralba D, Moreno-Gonzalo O, et al. ISGylation controls exosome secretion by promoting lysosomal degradation of MVB proteins. Nat Commun. 2016;7:13588.
Ilyushina NA, Donnelly RP. In vitro anti-influenza A activity of interferon (IFN)-lambda1 combined with IFN-beta or oseltamivir carboxylate. Antivir Res. 2014;111:112–20.
Wang Z, Zhang A, Wan Y, Liu X, Qiu C, Xi X, et al. Early hypercytokinemia is associated with interferon-induced transmembrane protein-3 dysfunction and predictive of fatal H7N9 infection. Proc Natl Acad Sci U S A. 2014;111(2):769–74.
Shan Z, Han Q, Nie J, Cao X, Chen Z, Yin S, et al. Negative regulation of interferon-induced transmembrane protein 3 by SET7-mediated lysine monomethylation. J Biol Chem. 2013;288(49):35093–103.
Li K, Markosyan RM, Zheng YM, Golfetto O, Bungart B, Li M, et al. IFITM proteins restrict viral membrane hemifusion. PLoS Pathog. 2013;9(1):e1003124.
Acknowledgements
The authors would like to thank Katie Oakley, PhD, from Liwen Bianji, Edanz Editing China (www.liwenbianji.cn/ac), for editing the English text of a draft of this manuscript.
Funding
This work was supported by the National Key Research and Development Program of China (2016YFC1200902) from the Ministry of Science and Technology.
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Author information
Authors and Affiliations
Contributions
Conceived and designed the experiments: AJC, ZJD and LSZ. Performed the experiments: AJC, JD, XHY, HB and SZL; Analyzed the data: AJC and CW; Contributed reagents/materials/analysis tools: XHY and CW; Wrote the manuscript: AJC, ZJD and LSZ. All authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Ethics approval and consent to participate
The project was approved by the Ethical Committee of National Institute for Viral Disease Control and Prevention, China CDC.
Consent for publication
No applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Chen, Aj., Dong, J., Yuan, Xh. et al. Anti-H7N9 avian influenza A virus activity of interferon in pseudostratified human airway epithelium cell cultures. Virol J 16, 44 (2019). https://doi.org/10.1186/s12985-019-1146-4
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12985-019-1146-4