Abstract
Background
Plant viruses in agricultural crops are of great concern worldwide, and over 75% of them are transmitted from infected to healthy plants by insect vectors. Tomato yellow leaf curl virus (TYLCV) is a begomovirus, which is the largest and most economically important group of plant viruses, transmitted by the whitefly Bemisia tabaci. The circulation of TYLCV in the insect involves complex insect-virus interactions, whereas the molecular mechanisms of these interactions remain ambiguous. The insect gut as a barrier for viral entry and dissemination is thought to regulate the vector specificity. However, due to its tiny size, information for the responses of whitefly gut to virus infection is limited.
Methods
We investigated the transcriptional response of the gut of B. tabaci Middle East-Asia Minor 1 species to TYLCV infection using Illumina sequencing.
Results
A total of 5207 differentially expressed genes (DEGs) between viruliferous and non-viruliferous whitefly guts were identified. Enrichment analyses showed that cargo receptor and ATP-binding cassette (ABC) transporters were enriched in DEGs, and might help the virus to cross gut barrier. TYLCV could perturb cell cycle and DNA repair as a possible result of its replication in the whitefly. Our data also demonstrated that TYLCV can activate whitefly defense responses, such as antimicrobial peptides. Meanwhile, a number of genes involved in intracellular signaling were activated by TYLCV infection.
Conclusions
Our results reveal the complex insect-virus relationship in whitefly gut and provide substantial molecular information for the role of insect midguts in virus transmission.
Similar content being viewed by others
Background
Plant viral diseases have received great attention worldwide because of their tremendous economic impact [1]. The majority of plant viruses are transmitted by insects of hemipteran families, such as aphids, whiteflies, leafhoppers, planthoppers, and thrips [2]. As a consequence, vector control is currently the only practical and effective strategy for disease prevention. Over the past few decades, a number of research have investigated the interactions between plant viruses and insect vectors because of its importance in viral epidemiology and disease management [2,3,4]. A detailed understanding of the genetic and molecular basis of insect-virus interaction will lead the discovery of novel and specific molecular targets for whitefly and whitefly-transmitted virus control.
Tomato yellow leaf curl virus (TYLCV) (Geminiviridae; Begomovirus) causes one of the most devastating emerging diseases of tomato worldwide [5]. TYLCV is transmitted by the whitefly Bemisia tabaci in a circulative persistent manner [2, 6]. B. tabaci is a cryptic species complex composed of at least 36 species [7]. In this species complex, the Middle East-Asia Minor 1 (Herein called MEAM1) species is highly invasive and a superior, co-adapted vector for begomoviruses. The epidemics of begomoviruses are usually associated with outbreaks of MEAM1 and the relationships between begomoviruses and whiteflies are complex [8]. When the MEAM1 viruliferous whiteflies were transferred onto non-host plants of the virus, the longevity and fecundity of the viruliferous whiteflies decreased [9]. This indicates that begomoviruses, in some cases, are insect pathogens. The transcriptional response of MEAM1 whiteflies to Tomato yellow leaf curl China virus (TYLCCNV) demonstrated that TYLCCNV can activate whiteflies’ immune response [10]. Further results showed that whiteflies use a variety of defense mechanisms to combat virus infection, such as autophagy and antimicrobial peptides (AMPs) [11, 12].
Circulative plant viruses move through the insect vector, from the gut lumen into the haemolymph or other tissues and finally into the salivary glands from where viruses are disseminated to new host plants during insect feeding [13]. In this process, midgut and salivary glands are the two major barriers that viruses have to overcome before successfully transmitted [2]. In fact, the gut barrier is the principal determinant for the ability of an insect species to transmit a virus. For instance, the greenhouse whitefly Trialeurodes vaporariorum is a non-vector of TYLCV, because the viruses can’t cross midgut into haemolymph [14]. Persistent viruses, whether propagative or nonpropagative, can be transmitted to plants after injection into the insect hemolymph [15]. In many cases, injected viruses are transmitted at higher rates than orally acquired viruses [16, 17]. Microscopic studies have shown that TYLCV virions is extensively localized in the filter chamber and cross the epithelial cells in the midgut [6, 18]. Compared to the whole body of whiteflies, TYLCV has a longer retention and higher quantity in the midgut [19]. Nevertheless, TYLCV infection can activate the autophagy pathway in whitefly midguts, which inhibits the efficiency of virus transmission [11]. These studies show that midguts are major reservoir where virions accumulate during acquisition and are critical in insect-virus interaction. However, due to the small size of whitefly midgut, the transcriptional responses of whitefly gut to virus infection remains unknown.
With the development of sequencing technique, next generation sequencing have provided us a valuable tool for exploring transcriptional changes using less than 1 μg RNA samples. In this study, we extracted 700 ng RNA from approximately 1000 whitefly guts for RNA-Seq to examine changes in gene transcription between viruliferous and nonviruliferous whiteflies. Transcriptome analysis revealed that a number of genes involved in the material transport, cell cycle, DNA repair, defense responses, signaling molecules and pathways were regulated in the viruliferous whitefly guts. These data provide a valuable source of molecular information for the research of guts in TYLCV-whitefly interactions. To our knowledge, this is the first report to study the direct effect of a plant virus on global gene expression profile of vector’s gut using a high-throughput sequencing method.
Results
Sequence assembly and functional annotation
Two cDNA libraries of viruliferous and nonviruliferous guts were sequenced and generated 51,471,400 and 51,554,406 raw reads, respectively. After filtering for high quality sequences, 44,362,142 and 45,610,802 clean reads were obtained for each sample (Table 1). Subsequently, the clean reads from the two samples were combined to do de novo assembly, resulting in 69,836 contigs with a mean length of 1121 bp and an N50 of 2407 bp. The lengths of contigs ranged from 200 bp to over 25,000 bp. After clustering, these contigs generated 55,124 unigenes (Fig. 1a). From these unigenes, a total of 18,574 predicted ORFs was obtained and the lengths ranged from 297 bp to 15,858 bp, with a mean length of 1049 and an N50 of 1491 (Fig. 1b). To annotate the unigenes, we searched reference sequences against NT, NR, PFAM and Uniprot databases, using Trinotate and Blast with a cut-off E-value of 10− 5. 21,259 (38.57%) unigenes were annotated at least in one database, including 6623 in NT, 18,556 in NR, 12,457 in BLASTP and 17,772 in BLASTX against Uniprot (Fig. 2a). On the basis of NR annotation, 15.53% (n = 2882) of unigenes matched to Zootermopsis nevadensis, followed by Acyrthosiphon pisum: 8.40% (n = 1560); Ceratitis capitata: 5.53% (n = 1026); Diaphorina citri: 5.15% (n = 956); Tribolium castaneum: 4.07% (n = 755) (Fig. 2b).
Global patterns of gene expression in response to TYLCV infection
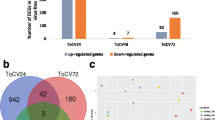
Subsequently, the differentially expressed genes (DEGs) between viruliferous and nonviruliferous guts were identified using DEGseq [20]. A total of 5207 differentially expressed genes were identified, with 4014 genes upregulated and 1193 genes downregulated in viruliferous whitefly guts (Fig. 3a). The detected fold changes (log2 ratio) of gene expression ranged from − 13.31 to 6.45, and more than 90% of the genes (4696) were up- or downregulated between 1.0- and 4.0-fold (Fig. 3b).
Analysis of differentially expressed genes between two libraries. a Summary of the numbers of differentially expressed genes in the TYLCV viruliferous whitefly guts. Genes with q ≤ 0.05 (adjusted p-value) and log2 ratio ≥ 1 were considered differentially expressed. b Correlation analysis of fold change and p-value
Assignment of DEGs to Gene Ontology (GO) terms and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways
To further reveal their functions, GO assignments were used to classify the DEGs. A total of 1216 DEGs (840 upregulated and 376 downregulated) were categorized into 54 secondary GO categories under the ‘biological process’, ‘cellular component’ and ‘molecular functions’ divisions. While ‘binding’ and ‘catalytic’ were among the most represented ‘molecular functions’ categories, the ‘cellular component’ most represented were ‘cell part’ and ‘membrane part’. Interestingly, in ‘biological process’ category, viral infections dramatically changed the expression of genes in ‘response to stimulus’, ‘reproductive process’ and ‘immune system process’ (Fig. 4). The GO enrichment analysis showed that ‘Cargo receptor activity’ was significantly enriched with DEGs in ‘molecular functions’ (11 out of 30 genes). Exactly 6 genes were upregulated while a gene was downregulated in ‘scavenger receptor activity’ under the ‘cargo receptor activity’ (Table 2).
Histogram presentations of GO classificarion of differentially expressed genes.The functions of genes indentified cover three main categories: biological process, cellular component and molecular function. GO analysis showed that the distributions of gene functions for the differentially expressed genes
Among the 5207 DEGs, 414 genes were mapped to 266 pathways in KEGG database. While ‘metabolic pathways’ (169), ‘biosynthesis of secondary metabolites’ (50) and ‘microbial metabolism in diverse environments’ (37) contained most of the DEGs. Enrichment analysis demonstrated that the DEGs were only significantly enriched in ‘ATP-binding cassette (ABC) transporters’, with 16 genes upregulated and 17 genes downregulated in viruliferous whitefly guts.
Based on GO and KEGG analysis, a total of 31 genes (24 upregulated and 7 downregulated) involved in cell cycle were found in our DEGs (Table 3). For DNA repair, 2 genes were upregulated and 6 genes downregulated in viruliferous whitefly guts (Table 4). Additionally, 4 genes of cell cycle checkpoints were found in DEGs and all of them were upregulated (Table 4). We also found that 10 upregulated genes were related to ubiquitin (Table 4).
Interestingly, among the DEGs in viruliferous whitefly guts, 14 genes related to cellular and humoral immune response were upregulated (Table 5). Six genes encoding AMPs were significantly upregulated including 1 defensin and 4 knottin. For cellular responses, a total of 8 upregulated genes were found, 6 of them involved in the phagocytosis and 2 in encapsulation. In addition, 9 genes (6 upregulated and 3 downregulated) were altered in the lysosome pathway (Additional file 1: Table S1). For regulators of defense responses, 1 gene encoding secreted molecule and 9 genes encoding putative transmembrane receptors (surface receptors) were found in our DEGs (Table 5). Meanwhile, the intracellular signaling molecules or pathways, downstream of these receptors, were also activated. In viruliferous whitefly gut cells, a number of genes involved in the mitogen-activated protein kinase (MAPK), Notch, transforming growth factor beta (TGF-β), PI3K-Akt, Jak-STAT, protein kinase C (PKC), and Ras signaling molecules or pathways were upregulated (Table 6).
qRT-PCR validation and expression profiling of DEGs
To validate our data, the expression levels of 10 DEGs were verified using qRT-PCR. We analyzed the expression of 5 antimicrobial peptides (Btk-1, Btk-2, Btk-3, Btk-4, DEF), 1 scavenger receptor class B (CD36), 1 regucalcin (RGN), 1 ankyrin repeat protein (Ank), 1 E3 ubiquitin-protein ligase, and 1 unnamed protein (categorized into GO biological process, defense response to virus, DRV) in both guts and whole body. Six genes were confirmed to be significantly upregulated in guts samples (Fig. 5). Moreover, the fold changes obtained by the DEG were generally more extreme than those obtained by qRT-PCR. The inconsistency may be partially due to the lower sensitivity of qRT-PCR compared tosignificant DEG. In addition, the fold changes in guts were more significant than those in whole body. These results indicate that the transcriptome profiles of whitefly guts in response to TYLCV infection are a little different from the whitefly whole body. Nevertheless, qRT-PCR analyses confirmed the direction of change detected by DEG analysis, suggesting that DEG results are reliable.
Result of RT-PCR. 10 selected genes were measured the expression in both guts and whole body using comparative CT (ΔΔCT) qPCR with β-actin as the internal control gene. 10 genes were 5 antimicrobial peptides (Btk-1, Btk-2, Btk-3, Btk-4, DEF), 1 scavenger receptor class B (CD36), 1 regucalcin (RGN), 1 ankyrin repeat protein (Ank), 1 E3 ubiquitin-protein ligase, and 1 unnamed protein (categorized into go biological process, defense response to virus, DRV)
Discussion
Altered receptors and transporters in whitefly gut after TYLCV infection
The GO and KEGG enrichment analysis showed that DEGs were significantly enriched in ‘Cargo receptor activity’ and “ATP-binding cassette (ABC) transporters”. Cargo receptor can selectively bind to an extracellular substance and deliver it into cell via endocytosis [21]. For example, cargo receptor ERGRC-53 is required for cellular glycoprotein trafficking of arenavirus, hantavirus, coronavirus, orthomyxovirus, and filovirus [22]. In human, the scavenger receptor class B type I and scavenger receptor B2 have been reported to be a receptor for the hepatitis C virus and enterovirus 71 separately [23, 24]. ABC transporters are members of a transport system superfamily and involved in diverse cellular processes such as maintenance of osmotic homeostasis, nutrient uptake, resistance to xenotoxins, antigen processing, bacterial immunity and pathogenesis [25]. Interestingly, one ABC superfamily transporter, multidrug resistance protein1 (MRP1), which is well-known to confer multidrug resistance to cancer cells through enhanced drug efflux [26], was also found in our DEGs. (c18467_g1: multidrug resistance-associated protein 1-like [Diaphorina citri], log2 ratio = 1.87) In addition, the downregulation of MRP2, which shows structural similarity to MRP1, suppresses the transport of various genotoxic substances in the liver during hepatitis virus infection [27]. As we know, virus receptors play essential roles in the early steps of viral infection. These regulated receptors and transporters might help TYLCV cross the whitefly epithelial cells.
Perturbance of the cell cycle and DNA repair in response to TYLCV infection
Viruses depend on hosts machineries to replicate and express their genomes, and altering the host cell cycle is a common strategy of viruses inside the cell [28]. Perturbation of host cell cycle has been shown to be caused by geminivirus infection. For example, Cabbage leaf curl virus (CaLCuV) can activate genes expressed during S and G2 phases and inhibit genes active in G1 and M phases [29]. We noticed that one gene encoding cyclin D2 was upregulated in viruliferous guts. Cylin D2 (CCND2) is a member of the D-type cyclin family, which associates the extracellular signaling environment with cell-cycle progression [30]. Moreover, cyclin D2 was also upregulated in Hepatitis B virus-infected cells and has a positive role in HBV replication [31].
Virus infection can produce a large amount of exogenous DNA [32]. Several mammalian viruses have been shown to induce a cellular DNA damage response during replication, and in some cases, this response is required for optimal virus replication [33]. In addition, cell cycle checkpoints govern cell cycle progression in the presence of DNA damage and incomplete DNA replication [34]. The upregulated genes in cell cycle checkpoints suggest that TYLCV infection may induce DNA damage and affect cell cycle in whitefly gut cells. Interestingly, we also noted that all of the 10 upregulated genes encoding ubiquitin regulated E3 ubiquitin ligase activity. Furthermore, 5 of these genes accumulated in the G2 phase of the cell cycle. G2/M phase-specific E3 ubiquitin-protein ligase (G2E3) as a nucleo-cytoplasmic shuttling protein was implicated in the DNA damages response and cell cycle regulation [35], and then attenuated replicative stress [36]. Previous study also showed that Beet severe curly top virus (BSCYV) can induce RKP (a functional ubiquitin E3 ligase), which is able to interact with cell cycle inhibitor ICK/KRP proteins to regulate the cell cycle [37].
Defense responses in whitefly gut in response to TYLCV
In our DEGs, 6 genes encoding AMPs were significantly upregulated. AMPs are evolutionarily ancient weapons that resist bacteria, fungi, viruses and any conceivable substance [38]. Previous study has shown that knottin genes are responsive to various stresses and related to TYLCV circulative transmission in the whitefly [10, 12]. Therefore, the observed upregulation of genes encoding the knottin proteins in our study are consistent with previous reports. For cellular responses, phagocytosis is a widely conserved cellular response and refers to the recognition, engulfment and intracellular destruction of invading pathogens and apoptotic cells [39]. Previous study in our laboratory has demonstrated that lysosome participate in autophagy, which could inhibit the efficiency of TYLCV transmission by whiteflies [11]. As stated above, all genes involved in humoral and cellular responses and most genes associated with lysosome were significantly upregulated, strongly suggesting that innate immune systems were activated in viruliferous whitefly guts.
For regulators of defense responses, the beta 1, 3-glucan recognition protein (βGRP), which can lead to melanotic encapsulation in insects [40], was upregulated in viruliferous gut. This result suggested that encapsulation might be activated after TYLCV infection. The altered genes encoding surface receptor were classified into three major classes; a) CD36 family (n = 4), b) epidermal growth factor (EGF)-like (n = 3), and c) Toll receptor (n = 2). CD36 family is class B scavenger receptor that participates in apoptotic cell removal in Drosophila embryos and in phagocytosis of S. aureus [41]. Interestingly, scavenger receptor has been noticed in GO enrichment analysis, but just 1 out of 4 of these CD36 genes was categorized into scavenger receptor activity. The EGF, a transmembrane protein, mediates phagocytosis, has been demonstrated with knockdown experiments, in a broad range of bacteria in Drosophila [42]. Spaetzle (markedly upregulated) is the ligand for Toll receptor, leading to Toll signally pathway controlling the potent resistances to bacterial, fungal, and viral infections [43]. Toll is one of the major signal transduction pathways (another one is Imd), used by antimicrobial responses in Drosophila [44].
The above results clearly show that following TYLCV infection, certain of immune responses of whitefly including humoral (antibacterial peptides) and cellular (phagocytosis, encapsulation) were activated. The regulators of phagocytosis (CD36, EGF) and encapsulation (βGRP, Toll receptor) were also activated. These immune responses may result in the degradation of virions, therefore, the activation of immune responses is probably the evolved strategy of the whitefly to protect itself from the deleterious effects of TYLCV infection.
Intracellular signaling molecules and pathways involved in defense response
We also observed that a number of genes involved in signal transduction of the defense response were upregulated after TYLCV invasion. Ras is a key regulator of cell growth in all eukaryotic cells and positioned centrally in signal transduction pathways that respond to diverse extracellular stimuli [45]. The medfly homologues of Ras proteins have been shown to affect phagocytosis in response to lipopolysaccharide (LPS) and E. coli [46]. PI3K-Akt pathway is involved in apoptosis and autophagy in Drosophila [47, 48], as well as in medfly apoptosis [49]. Decreases in phagocytosis produced by pathway inhibitor indicated that PI3-kinase was required for internalization of bacteria [50]. Several studies have implicated involvement of MAPKs in insect innate immune responses [51, 52], and the activation of MAPK pathway contributes to efficient infection by baculoviruses [53]. Otherwise, several studies have documented that PKC plays an important role in regulating the receptor-mediated endocytosis of virus-receptors complexes [54,55,56], and specific inhibitor for PKC is highly effective in blocking West Nile virus (WNV) entry into mosquito cell line (C6/36) [57]. Here we found that all genes involved in the MAPK pathway and PKC were upregulated in DEGs.
The intracellular signaling molecules and pathways are organized as communication networks that process, encode and integrate internal and external signals. For example, activated Ras can directly interact with MAPK kinase kinase and activates distinct MAPK cascades (ERK, JNK, p38) [58]. Upon activation, the ERKs can stimulate the activity of Elk-1 transcription factor [59]. Blockage of Elk-1 like protein phosphorylation indicated that an Elk-1 like protein was candidate for the regulation of phagocytosis of bacteria [60]. Moreover, JNK was also reported to be involved in phagocytosis in mosquito cell line [61]. In our results, Ras, MAPKK7, JNK, Elk-1, even phagocytosis were upregulated. Evidently, this signaling network in whiteflies was activated by TYLCV.
Conclusions
In summary, we present, for the first time, an analysis of the global transcription response of whitefly guts to TYLCV infection using high-throughput sequencing. Our data show that TYLCV can perturb the material transport and cell cycle of whitefly gut cells. This virus can also activate the humoral and cellular immune responses of whiteflies. Overall, our results reveal the complex interactions between begomovirus and whiteflies in the gut tissue. This study will provide a road map for future investigation of guts in plant virus-vector insect interactions and may offer hints for the discovery of novel and specific molecular targets for the control of whitefly transmitted virus.
Methods
Whitefly cultures and virus clone
The culture of B. tabaci MEAM1 (mtCO1 GenBank accession no. GQ332577) was maintained on cotton (Cossypium hirsutum cv. Zhe-Mian 1793), which is a non-host plant of TYLCV. Whiteflies were reared in a climate chamber at 26 ± 1 °C, LD 14:10 h and 60 ± 10% relative humidity. To obtain virus-infected plants, clones of TYLCV isolate SH2 (GenBank accession no. AM282874.1) were inoculated into tomato (Solanum lycopersicom L.cv. Hezuo903) at 3–4 true-leaf stage as previously described [62], TYLCV-infected and uninfected tomato plants were cultivated to 6–7 true-leaf stage when used in experiments. All plants were grown in a greenhouse at 20–30 °C, LD14:10 h and 50–70% relative humidity.
Whitefly treatments and sample collection
To prepare viruliferous and nonviruliferous whiteflies, approximately 2000 newly emerged (0–48 h) adult whiteflies were separately collected and released onto healthy tomato plants in different cages for 48 h. Then the whiteflies were transferred onto virus-infected and uninfected tomato plants for 48 h, respectively. After that, they were separately transferred onto another tomato (Solanum lycopersicom L.cv. Zheza502), which is a resistant variety to TYLCV [11], to eliminate effects of host differences on whiteflies. After 24 h, the whiteflies were used for gut dissection. For sample collection, approximately 1000 guts were dissected from viruliferous and nonviruliferous whiteflies in PBS respectively, and frozen in liquid nitrogen, stored at − 80 °C until RNA isolation. Additionally, all the guts were dissected in 36 h to minimize the impact of time on the results.
RNA isolation and cDNA library preparation
Total RNA was extracted from pools of approximately 1000 guts using the Absolutely RNA Nanoprep kit (Agilent, USA) according to the manufacturer’s manual with slight modifications [63]. RNA quantity and quality were assessed using 1% agarose gels, Qubit® 3.0 Flurometer (Life Technologies, CA, USA) and the RNA Nano 6000 Assay Kit of the Agilent Bioanalyzer 2100 system (Agilent Technologies, CA, USA). RNA (700 ng per sample) was used to generate adaptor-ligated double- stranded cDNA libraries for RNA-Seq using the NEBNext® UltraTM RNA Library Prep Kit for Illumina® (New England Biolabs, USA) following the manufacturer’s protocol. The fragment size and concentration of resultant libraries were carried out using the Agilent Bioanalyzer 2100 system (Agilent Technologies, CA, USA) and StepOnePlus™ Real-Time PCR System (Library valid concentration > 10 nM).
Transcriptome sequencing and assembly
The cDNA libraries were sequenced for 150 bp paired-end reads using Illumina Hiseq 4000 platform at Annoroad Gene Technology Company (Beijing, China). The total sequencing amount was 8 G for each library. To ensure the quality of data used in further-analysis, raw data were transformed into clean data through removing adaptor-polluted reads (more than 5 adaptor-polluted bases), low-quality reads (more than 15% bases with Phred threshold score ≤ 19), and reads with unknown sequences ‘N’ accounting for more than 5% using Perl scripts. As for paired-end sequencing data, both reads were filtered out if any read of the paired-end reads are adaptor-polluted. The assembler Trinity was used for de novo assembly [64]. We quantified the proportion of the clean reads that mapped to each assembly using Bowtie (v2.2.3). To test the completeness of the assemblies, we mapped the presence of highly conserved genes with 2748 core proteins from conserved regions of eukaryotes. TransDecoder was used to identify the candidate coding regions within transcript sequences, and Trinotate was used for performing the functional annotation of unigenes and ORFs.
Analysis of differential gene expression
The clean reads from viruliferous and nonviruliferous guts were separately mapped back to the assembled unigenes. For gene expression analysis, reads per kilobase million Mapped Reads (RPKM) was calculated to estimate the expression level of genes in each sample. RPKM can eliminate the effect of sequencing depth and gene length on gene expression levels, which facilitates the comparison of the number of transcript levels generated between samples. DEGseq (v1.18.0) was used to identify differentially expressed genes (DEGs) between the viruliferous and nonviruliferous guts. Genes with q ≤ 0.05 (adjusted p-value) and log2 ratio ≥ 1 were considered differentially expressed.
GO and KEGG pathway analysis
To obtain an overview and further understand the biological functions of genes, all DEGs were subjected to GO functional annotation using Blast2GO and mapped to terms in KEGG pathway database using KOBAS. Enrichment analysis was used to identify the GO terms and significantly regulated KEGG pathways. We selected a corrected q ≤ 0.05 as the threshold to determine significant enrichment of the gene sets.
qRT-PCR analysis
To confirm the result of the DEG analyses, we measured the expression of 10 selected genes using comparative CT (ΔΔCT) Real-time Quantitative PCR with β-actin as the internal control gene. We prepared viruliferous and nonviruliferous whiteflies as described above and gut total RNA was extracted from pools of approximately 70 guts. Three biological replicates were conducted at the same time. Moreover, 25 whiteflies were collected (24 h after beginning of the gut dissection) from each replication to compare the differences in gene expression between the gut and the whole body. Total RNA was extracted using the TRI-reagent method (Life Technologies, USA). RNA was reverse transcribed using the SYBR® PrimeScript™ RT-PCR Kit II (Takara, Japan). qRT-PCR was performed using PTC-200 Thermocycler (Bio-Rad, USA) with SYBR-Green detection (SYBR® Premix Ex Taq™ II, Takara, Japan). The primer sequences are provided in Additional file 2: Table S2.
References
Scholthof KBG, Adkins S, Czosnek H, Palukaitis P, Jacquot E, Hohn T, et al. Top 10 plant viruses in molecular plant pathology. Mol Plant Pathol. 2011;12:938–54.
Hogenhout SA, Ammar ED, Whitfield AE, Redinbaugh MG. Insect vector interactions with persistently transmitted viruses. Annu Rev Phytopathol. 2008;46:327–59.
Gray S, Gildow FE. Luteovirus-aphid interactions. Annu Rev Phytopathol. 2003;41:539–66.
Ng JCK, Falk BW. Virus-vector interactions mediating nonpersistent and semipersistent transmission of plant viruses. Annu Rev Phytopathol. 2006;44:183–212.
Tomato CH. Yellow leaf curl virus disease: management, molecular biology, breeding for resistance. Netherlands: Springer; 2007.
Czosnek H, Ghanim M. The circulative pathway of begomoviruses in the whitefly vector Bemisia tabaci—insights from studies with Tomato yellow leaf curl virus. Ann Appl Biol. 2002;140:215–31.
Boykin LM, De Barro PJ. A practical guide to identifying members of the Bemisia tabaci species complex: and other morphologically identical species. Front Ecol Evol. 2014;2(45)
Ghanim M. A review of the mechanisms and components that determine the transmission efficiency of Tomato yellow leaf curl virus (Geminiviridae; Begomovirus) by its whitefly vector. Virus Res. 2014;186:47–54.
Rubenstein G, Czosnek H. Long-term association of Tomato yellow leaf curl virus (TYLCV) with its whitefly vector Bemisia tabaci: effect on the insect transmission capacity, longevity and fecundity. J Gen Virol. 1997;78:2683–9.
Luan JB, Li JM, Varela N, Wang YL, Li FF, Bao YY, et al. Global analysis of the transcriptional response of whitefly to Tomato yellow leaf curl China virus reveals the relationship of coevolved adaptations. J Virol. 2011;85:3330–40.
Wang LL, Wang XR, Wei XM, Huang H, Wu JX, Chen XX, et al. The autophagy pathway participates in resistance to Tomato yellow leaf curl virus infection in whiteflies. Autophagy. 2016;12:1560–74.
Hariton Shalev A, Sobol I, Ghanim M, Liu SS, Czosnek H. The whitefly bemisia tabaci knottin-1 gene is implicated in regulating the quantity of Tomato yellow leaf curl virus ingested and transmitted by the insect. Viruses. 2016;8:205.
Rosen R, Kanakala S, Kliot A, Pakkianathan BC, Farich BA, Santana-Magal N, et al. Persistent, circulative transmission of begomoviruses by whitefly vectors. Curr Opin Virol. 2015;15:1–8.
Ohnishi J, Kitamura T, Terami F, Honda KI. A selective barrier in the midgut epithelial cell membrane of the nonvector whitefly Trialeurodes vaporariorum to Tomato yellow leaf curl virus uptake. J Gen Plant Pathol. 2009;75:131–9.
Sylvester ES. Circulative and propagative virus transmission by aphids. Annu Rev Entomol. 1980;25:257–86.
Ammar ED, Gomez-Luengo RG, Gordon DT, Hogenhout SA. Characterization of Maize Iranian mosaic virus and comparison with Hawaiian and other isolates of Maize mosaic virus (Rhabdoviridae). J Phytopathol. 2005;153:129–36.
Ammar ED, Hogenhout SA. A neurotropic route for Maize mosaic virus (Rhabdoviridae) in its planthopper vector Peregrinus maidis. Virus Res. 2008;131:77–85.
Ghanim M, Brumin M, Popovski S. A simple, rapid and inexpensive method for localization of Tomato yellow leaf curl virus and Potato leafroll virus in plant and insect vectors. J Virol Methods. 2009;159:311–4.
Pakkianathan BC, Kontsedalov S, Lebedev G, Mahadav A, Zeidan M, Czosnek H, et al. Replication of Tomato yellow leaf curl virus in its whitefly vector, Bemisia tabaci. J Virol. 2015;89:9791–803.
Wang L, Feng Z, Wang X, Wang X, Zhang X. DEGseq: an R package for identifying differentially expressed genes from RNA-seq data. Bioinformatics. 2010;26:136–8.
Gitler AD, Lu MM, Epstein JA. PlexinD1 and semaphorin signaling are required in endothelial cells for cardiovascular development. Dev Cell. 2004;7:107–16.
Klaus JP, Eisenhauer P, Russo J, Mason AB, Do D, King B, et al. The intracellular cargo receptor ERGIC-53 is required for the production of infectious arenavirus, coronavirus, and filovirus particles. Cell Host Microbe. 2013;14:522–34.
Scarselli E, Ansuini H, Cerino R, Roccasecca MR, Acali S, Filocamo G, et al. The human scavenger receptor class B type I is a novel candidate receptor for the hepatitis C virus. EMBO J. 2002;21:5017–25.
Yamayoshi S, Yamashita Y, Li J, Hanagata N, Minowa T, Takemura T, et al. Scavenger receptor B2 is a cellular receptor for enterovirus 71. Nature Med. 2009;15:798–801.
Jones PM, George AM. The ABC transporter structure and mechanism: perspectives on recent research. Cellular Molr Life Sci. 2004;61:682–99.
Cole SP, Bhardwaj G, Gerlach JH, Mackie JE, Grant CE, Almquist KC, et al. Overexpression of a transporter gene in a multidrug-resistant human lung cancer cell line. Science. 1992;258:1650–4.
Hinoshita E, Taguchi K, Inokuchi A, Uchiumi T, Kinukawa N, Shimada M, et al. Decreased expression of an ATP-binding cassette transporter, MRP2, in human livers with hepatitis C virus infection. J Hepatol. 2001;35:765–73.
Emmett SR, Dove B, Mahoney L, Wurm T, Hiscox JA. The cell cycle and virus infection. Methods Mol Biol. 2005;296:197–218.
Ascencio-Ibáñez JT, Sozzani R, Lee TJ, Chu TM, Wolfinger RD, Cella R, et al. Global analysis of Arabidopsis gene expression uncovers a complex array of changes impacting pathogen response and cell cycle during geminivirus infection. Plant Physiol. 2008;148:436–54.
Zhang Q, Sakamoto K, Wagner KU. D-type cyclins are important downstream effectors of cytokine signaling that regulate the proliferation of normal and neoplastic mammary epithelial cells. Mol Cell Endocrinol. 2014;382:583–92.
Song CL, Ren JH, Ran LK, Li YG, Li XS, Chen X, et al. Cyclin D2 plays a regulatory role in HBV replication. Virology. 2014;462:149–57.
Chaurushiya MS, Weitzman MD. Viral manipulation of DNA repair and cell cycle checkpoints. DNA Repair. 2009;8:1166–76.
Huang N, Wu W, Yang K, Passarelli AL, Rohrmann GF, Clem RJ. Baculovirus infection induces a DNA damage response that is required for efficient viral replication. J Virol. 2011;85:12547–56.
Marathi UK, Dahlen M, Sunnerhagen P, Romero AV, Ramagli LS, Siciliano MJ, et al. RAD1, a human structural homolog of the Schizosaccharomyces pombe RAD1Cell cycle checkpoint gene. Genomics. 1998;54:344–7.
Brooks WS, Banerjee S, Crawford DF. G2E3 is a nucleo-cytoplasmic shuttling protein with DNA damage responsive localization. Exp Cell Res. 2007;313:665–76.
Schmidt F, Karnitz LM, Dobbelstein M. G2E3 attenuating replicative stress. Aging (Albany NY). 2015;7:527.
Lai J, Chen H, Teng K, Zhao Q, Zhang Z, Li Y, et al. RKP, a RING finger E3 ligase induced by BSCTV C4 protein, affects geminivirus infection by regulation of the plant cell cycle. Plant J. 2009;57:905–17.
Zasloff M. Antimicrobial peptides of multicellular organisms. Nature. 2002;415:389–95.
Jiravanichpaisal P, Lee BL, Söderhäll K. Cell-mediated immunity in arthropods: hematopoiesis, coagulation, melanization and opsonization. Immunobiology. 2006;211:213–36.
Wang Y, Willott E, Kanost MR. Organization and expression of the hemolin gene, a member of the immunoglobulin superfamily in an insect, Manduca sexta. Insect Mol Biol. 1995;4:113–23.
Franc NC, Heitzler P, White K. Requirement for croquemort in phagocytosis of apoptotic cells in Drosophila. Science. 1999;284:1991–4.
Kocks C, Cho JH, Nehme N, Ulvila J, Pearson AM, Meister M, et al. Eater, a transmembrane protein mediating phagocytosis of bacterial pathogens in Drosophila. Cell. 2005;123:335–46.
De Gregorio E, Spellman PT, Rubin GM, Lemaitre B. Genome-wide analysis of the Drosophila immune response by using oligonucleotide microarrays. Proc Natl Acad Sci U S A. 2001;98:12590–5.
Hoffmann JA, Reichhart JM. Drosophila innate immunity: an evolutionary perspective. Nat Immunol. 2002;3:121–6.
Vojtek AB, Der CJ. Increasing complexity of the Ras signaling pathway. J Biol Chem. 1998;273:19925–8.
Soldatos AN, Metheniti A, Mamali I, Lambropoulou M, Marmaras VJ. Distinct LPS-induced signals regulate LPS uptake and morphological changes in medfly hemocytes. Insect Biochem Molec. 2003;33:1075–84.
Rusten TE, Lindmo K, Juhász G, Sass M, Seglen PO, Brech A, et al. Programmed autophagy in the Drosophila fat body is induced by ecdysone through regulation of the PI3K pathway. Dev Cell. 2004;7(17)
Berry DL, Baehrecke EH. Growth arrest and autophagy are required for salivary gland cell degradation in Drosophila. Cell. 2007;131:1137–48.
Mamali I, Tatari MN, Micheva I, Lampropoulou M, Marmaras VJ. Apoptosis in medfly hemocytes is regulated during pupariation through FAK, Src, ERK, PI-3K p85a, and Akt survival signaling. J Cell Biochem. 2007;101:331–47.
De Winter P, Rayne RC, Coast GM. The effects of intracellular signalling pathway inhibitors on phagocytosis by haemocytes of Manduca sexta. J Insect Physiol. 2007;53:975–82.
Lamprou I, Mamali I, Dallas K, Fertakis V, Lampropoulou M, Marmaras VJ. Distinct signalling pathways promote phagocytosis of bacteria, latex beads and lipopolysaccharide in medfly haemocytes. Immunology. 2007;121:314–27.
Surachetpong W, Singh N, Cheung KW, Luckhart S. MAPK ERK signaling regulates the TGF-β1-dependent mosquito response to Plasmodium falciparum. PLoS Pathog. 2009;5:e1000366.
Katsuma S, Mita K, Shimada T. ERK-and JNK-dependent signaling pathways contribute to Bombyx mori nucleopolyhedrovirus infection. J Virol. 2007;81:13700–9.
McClure SJ, Robinson PJ. Dynamin, endocytosis and intracellular signalling (review). Mol Membr Biol. 1996;13:189–215.
Nakano MY, Boucke K, Suomalainen M, Stidwill RP, Greber UF. The first step of adenovirus type 2 disassembly occurs at the cell surface, independently of endocytosis and escape to the cytosol. J Virol. 2000;74:7085–95.
Sieczkarski SB, Brown HA, Whittaker GR. Role of protein kinase C βII in influenza virus entry via late endosomes. J Virol. 2003;77:460–9.
Chu JJH, Leong PWH, Ng ML. Analysis of the endocytic pathway mediating the infectious entry of mosquito-borne flavivirus West Nile into Aedes albopictus Mosquito (C6/36) cells. Virology. 2006;349:463–75.
Minden A, Lin A, Claret FX, Abo A, Karin M. Selective activation of the JNK signaling cascadeand c-Jun transcriptional activity by the small GTPases Rac and Cdc42Hs. Cell. 1995;81:1147–57.
Waskiewicz AJ, Flynn A, Proud CG, Cooper JA. Mitogen-activated protein kinases activate the serine/threonine kinases Mnk1 and Mnk2. EMBO J. 1997;16:1909–20.
Mamali I, Kapodistria K, Lampropoulou M, Marmaras VJ. Elk-1 is a novel protein-binding partner for FAK, regulating phagocytosis in medfly hemocytes. J Cell Biochem. 2008;103:1895–911.
Mizutani T, Kobayashi M, Eshita Y, Shirato K, Kimura T, Ako Y, et al. Involvement of the JNK-like protein of the Aedes albopictus Mosquito cell line, C6/36, in phagocytosis, endocytosis and infection of West Nile virus. Insect Mol Boil. 2003;12:491–9.
Zhou X, Xie Y, Tao X, Zhang Z, Li Z, Fauquet CM. Characterization of DNAβ associated with begomoviruses in China and evidence for co-evolution with their cognate viral DNA-AFN1. J Gen Virol. 2003;84:237–47.
Su YL, Li JM, Li M, Luan JB, Ye XD, Wang XW, et al. Transcriptomic analysis of the salivary glands of an invasive whitefly. PLoS One. 2012;7:e39303.
Grabherr MG, Haas BJ, Yassour M, Levin JZ, Thompson DA, Amit I, et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat Biotechnol. 2011;29:644–52.
Acknowledgements
Not applicable.
Funding
Financial support for this study was provided by the National Natural Science Foundation of China (31390421), the National Key Research and Development Program (2016YFC1200601) and China Agriculture Research System (CARS-23-D07).
Availability of data and materials
The datasets generated and analysed during the current study are available in the NCBI repository, https://www.ncbi.nlm.nih.gov/bioproject/PRJNA407873/.
Author information
Authors and Affiliations
Contributions
LG, YQL, SSL and XWW designed the whole study. LG, LXQ and RXS performed the experiment. LG analyzed the data. LG and XWW wrote the manuscript and prepared all figures. All authors reviewed the manuscript. All Authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Additional files
Additional file 1: Table S1.
DEGs involved in Lysosome. Displayed are the fold changes of DEGs involved in Lysosome of viruliferous guts in comparison to nonviruliferous guts. aThe function of the homologous gene. bFC, fold change (log2 ratio) of gene expression. (XLSX 8 kb)
Additional file 2: Table S2.
Sequences of primers used in the study. Displayed are the sequence of the primers used for qRT-PCR. Primers were synthesized by GenScript (Nanjing, China). (XLSX 9 kb)
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Geng, L., Qian, LX., Shao, RX. et al. Transcriptome profiling of whitefly guts in response to Tomato yellow leaf curl virus infection. Virol J 15, 14 (2018). https://doi.org/10.1186/s12985-018-0926-6
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12985-018-0926-6