Abstract
Purpose
Occult hepatitis B infection (OBI) among HIV positive patients varies widely in different geographic regions. We undertook a study to determine the prevalence of occult hepatitis B infection among HIV infected individuals visiting a health facility in South West Cameroon and characterized occult HBV strains based on sequence analyses.
Methods
Plasma samples (n = 337), which previously tested negative for hepatitis B surface antigen (HBsAg), were screened for antibodies against hepatitis B core (anti-HBc) and surface (anti-HBs) antigens followed by DNA extraction. A 366 bp region covering the overlapping surface/polymerase gene of HBV was then amplified in a nested PCR and the amplicons sequenced using Sanger sequencing. The resulting sequences were then analyzed for genotypes and for escape and drug resistance mutations.
Results
Twenty samples were HBV DNA positive and were classified as OBI giving a prevalence of 5.9%. Out of these, 9 (45%) were anti-HBs positive, while 10 (52.6%) were anti-HBc positive. Additionally, 2 had dual anti-HBs and anti-HBc reactivity, while 6 had no detectable HBV antibodies. Out of the ten samples that were successfully sequenced, nine were classified as genotype E and one as genotype A. Three sequences possessed mutations associated with lamivudine resistance. We detected a number of mutations within the major hydrophilic region of the surface gene where most immune escape mutations occur.
Conclusions
Findings from this study show the presence of hepatitis B in patients without any of the HBV serological markers. Further prospective studies are required to determine the risk factors and markers of OBI.
Similar content being viewed by others
Background
Hepatitis B, a potentially life-threatening liver infection caused by the hepatitis B virus (HBV) is a major global health problem. Of the two billion people infected with the virus, more than 240 million are chronic carriers [1], and more than 686,000 die annually from HBV-related complications, including cirrhosis and hepatocellular carcinoma [2]. A growing body of evidence is emerging showing that the prevalence of HBV is significantly higher amongst HIV-positive individuals, presumably because of the shared transmission risks and risk factors [3, 4]. HIV generally accelerates the natural course of HBV infection and facilitates faster progression of liver disease to cirrhosis and hepatocellular carcinoma (HCC) [5].
Traditionally, HBV is diagnosed by serological techniques to detect antigens or antibodies. The hepatitis B surface antigen (HBsAg) is often used for routine diagnosis since it is considered as the hallmark of infection. During acute infection, antibodies to HBV core antigens (anti-HBc) (initially both IgM and IgG) appear 1–2 weeks after the appearance of HBsAg, while IgG persists during chronic infection. The presence of antibodies to HBsAg (anti-HBs) represents immunity to HBV infection [6].
In the advent of molecular diagnostics, it has been shown that a number of individuals may harbour HBV-DNA at very low levels in their liver and/or serum despite the absence of detectable HBsAg by currently available assays [7]. This is termed occult hepatitis B infection (OBI), characterized by the presence of HBV DNA in the blood and liver in HBsAg-negative individuals, who may or may not have anti-HBc and anti-HBs [6]. There are several mechanisms that have been hypothesized to lead to development of OBI. These include development of HBV S gene mutants that affect the detectability of the virus by conventional HBsAg assays, strong suppression of viral replication and reduced expression of HBsAg, epigenetic mechanisms and co-infection with other viruses [8].
OBI is frequent in persons with HIV, and its prevalence varies considerably in different geographic regions [9]. It has been shown that immunosuppression due to HIV infection could lead to low antibody response to HBsAg and also HBV reactivation [10, 11]. Consequently, among HIV patients who test HBsAg negative, HBV DNA should be determined before starting highly active antiretroviral therapy (HAART) so that anti-HBV antiretrovirals can be included if necessary. The prevalence of OBI however, has been shown to vary in different demographic settings. A recent study in Cameroon reporting on OBI focused on patients from a health centre in the capital Yaoundé [12]. It would be expected therefore that the two regions of Cameroon from where the patients in the current study were derived would provide added information on OBI in this country where both HBV and HIV are endemic.
Methods
Study design and setting
This was a cross-sectional retrospective study using 337 HBsAg negative plasma samples from 455 HIV positive outpatient clients of the Mutengene Baptist Health Centre located in the South West Region of Cameroon. The health centre offers HIV/AIDS treatment, prevention, medical, spiritual and psychosocial care in the Tiko Health District. The hospital attends to at least 8000 patients per month. Patients come from various towns such as Buea, Limbe, Tiko, Kumba in the South West Region, and Douala, Nkongsamba, in the Littoral Region. Details of the study setting have been reported [13].
Sample collection and processing
All consenting participants completed a questionnaire regarding their socio-demographic characteristics. Five millilitres of venous blood was collected once from each consenting HIV positive adult individual using EDTA vacutainer tubes while two ml of blood was collected from young children (<5 years). Blood was centrifuged for 5 min at 3578×g. Following centrifugation, plasma was aspirated aseptically and aliquoted into sterile labelled cryotubes and stored at −80 °C. These were then transported on dry ice to the HIV/AIDS & Global Health Research laboratories at the University of Venda, South Africa where they were stored at −80 °C until used.
Serological testing
Initially, all samples were screened for HBsAg using a commercially available direct enzyme-linked immunosorbent assay (ELISA) kit (Bioelisa, Biokit, Barcelona, Spain). In this study, samples that were HBsAg negative were further screened for anti-HBc and anti-HBs using another ELISA commercial kit (DRG Instruments GmbH, Germany).
DNA extraction
DNA from HBsAg negative samples was extracted from 100 µl of plasma using the Quick-gDNA mini prep kit (Zymo research, USA) according to the manufacturer’s instructions. The extracted DNA was amplified immediately after extraction or stored at −20 °C awaiting subsequent amplification.
Polymerase chain reaction (PCR) for HBV surface/polymerase gene
A nested PCR was performed in order to amplify the overlapping surface/polymerase gene covering nucleotides 403–768 from the EcoR1 site using primers and protocols described previously [14]. This generates a 366 bp product (Additional file 1: Figure S1). The first round reaction was conducted in a 50 µl volume containing 10 mM Tris-HCI pH 8.3, 50 mM potassium chloride, 0.2 mM dNTP mix, 2.5 mM magnesium chloride, 0.2 ng/µl of each primer, and 2 units of Taq polymerase (Applied Biosystems, PE, Italia). The thermocycling conditions involved 35 cycles of denaturation at 95 °C for 1 min, annealing at 55 °C for 1 min, and extension at 72 °C for 1 min. Five microlitre of the first round PCR product was used as a template for the nested PCR under the same reaction conditions, but performing only 20 cycles. The PCR products were then resolved by 1.5% agarose gel electrophoresis stained with ethidium bromide. PCR amplicons were then purified using the QiAquick PCR Purification Kit (Qiagen, Hilden; Germany) according to the manufacturer’s instructions.
Sequencing and sequence analysis
Purified PCR amplicons were directly sequenced at Inqaba Biotech (Pretoria, South Africa) according to the Sanger protocol. Contiguous nucleotide sequences (contigs) were assembled from resulting forward and reverse reactions using the SeqMan Pro® module of the Lasergene (version 8.1.5) sequence analysis software suite (DNASTAR. Madison, WI.).
The resulting nucleotide sequences were aligned using the Clustal W program implemented in MEGA 6.06 [15]. They were also translated and checked for HBsAg mutations in the S gene and drug resistance associated mutations in the P gene. Phylogenetic reconstruction was carried out using the MrBayes program version 3.1.2 [16]. The Bayesian tree was inferred by running a Markov-chain Monte Carlo (MCMC) algorithm for 1 million generations, sampling at every 100th generation with a burn in setting of 10% of generations. The GTR+G+I model (general time-reversible model with gamma distributed rates of variation among sites and a proportion of invariable sites) was found to be the best-fit model for our dataset. The resulting phylogenetic tree was visualized using FigTree, version 1.3.1 (http://tree.bio.ed.ac.uk/software/figtree/) and used to determine the HBV genotype.
Statistical analysis
A database was prepared and processed using SPSS 20 (IBM, Chicago, IL, USA). Fisher’s exact test was used to analyze the socio-demographic and clinical variables associated with OBI. A p value of less than 0.05 was considered statistically significant.
Results
Prevalence of occult HBV infection
HBV DNA was detected in 20 out of the 337 HBsAg negative samples giving an OBI prevalence of 5.9% in this study. OBI was higher among females, the unemployed, patients with low hemoglobin, those with normal white blood cell count and malaria negative individuals. However none of these variables were statistically associated with OBI (Table 1).
Serological profile of occult HBV infection
All the study samples were assayed for anti-HBs and anti-HBc serological markers. Out of the OBI positive samples, 9 (45%) were anti-HBs positive while 10 (52.6%) were anti-HBc positive (Table 2). Additionally, 2 individuals had dual anti-HBs and anti-HBc reactivity while 6 had no detectable HBV antibodies.
Molecular and genetic characterization of occult HBV DNA
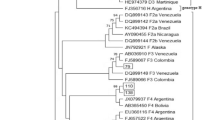
Sequencing of the overlapping surface/polymerase gene was done for 10 out of the 20 OBI positive DNA samples. Phylogenetic analyses indicated that HBV genotype E and A occurred in 9 (90%) and 1 (10%) individuals respectively (Fig. 1).
A bayesian rooted phylogenetic tree constructed using MrBayesver 3.1.2 of the Cameroon OBI sequences from HBV/HIV co-infected individuals. Twenty-eight global sequences obtained from GenBank were included to support tree topology and genotype identification. The ten viruses characterised in this study are named with a prefix BM and highlighted in blue at the taxa
Mutations in the “a” determinant within the major hydrophilic region (MHR) of the S gene may cause a conformational change in HBsAg, leading to undetectability of HBsAg (diagnostic-escape) as well as evading the host’s immune response (immune-escape). The main mutation observed in this region was the G159A mutation observed in eight (80%) of the samples compared to 41.3% among HBsAg positive samples previously observed. Also, nine of the samples possessed leucine at amino acid 127. Examination of the RT domain of the polymerase gene indicated that three of the sequences had mutations associated with drug resistance. These mutations V173L, L180M, T184S and M204V/I are associated with resistance to lamivudine, entecavir and telbivudine (Fig. 2). The three patients harboring these mutations had treatment experience with Lamivudine as part of their antiretroviral therapy.
Discussion
Laboratory detection of Hepatitis B virus infection is crucial for global control and prevention of HBV disease. Among HIV infected individuals under HAART, the increased longevity may facilitate emergence of chronic liver disease which is often a cause of increased morbidity and mortality. A significant proportion of this burden may be attributed to occult hepatitis B virus infection since it has been shown to have hepatopathogenic potential. Furthermore, HAART creates immune reconstitution that can result in immune mediated liver injury and elevation of liver enzymes [17]. This may easily be misclassified as HAART-associated liver toxicity.
In this study, we have found a 5.9% prevalence of OBI among a group of HIV positive individuals in Cameroon and no significant association between the studied demographic parameters and risk factors of the participants and OBI was found.
The prevalence of OBI in this study is similar to the 5.9% found among HIV individuals in Sicily, Italy [18]. However, the observed prevalence is slightly lower than the 6.9 and 9.8% reported in Yaoundé, Cameroon and Khartoum, Sudan respectively [12, 19]. It should be noted however that the prevalence of OBI is dependent on the sensitivity of the DNA assay used, demography and the population studied [7]. Thus, the Sudan study used a more sensitive real time PCR method compared to the conventional PCR method used in this study. We propose that the difference in prevalence with the Yaoundé study may be due to demographics. Among HIV patients, several studies conducted worldwide have reported prevalence of OBI ranging from 0% to more than 90% [20, 21].
The current study did not find any association between OBI and the studied variables. This finding has also been made among Cuban HIV patients [22]. Generally, previous studies have made conflicting findings regarding factors associated with OBI. For example, Stuart et al. [20] have reported a significant association between low CD4 and OBI among HIV patients while other studies have not made this finding [22, 23]. Perhaps this may be due to the different diagnostic algorithms used to define OBI in the various studies.
Slightly more than half (52.6%) of the OBI positive individuals were anti-HBc positive, while 45% were anti-HBs positive. Our findings on the anti-HBc status is consistent with several studies that indicate the role of the anti-HBc profile as the most common serological surrogate of OBI although higher prevalence of anti-HBc alone does not necessarily reflect significantly higher frequency of OBI [24, 25]. Dual anti-HBc and anti-HBs reactivity was observed in 2 (20%) of the samples. Additionally, we identified six OBI positive individuals with no serologic evidence of infection (negative for HBsAg, anti-HBc, and anti-HBs). This is an indication that occult HBV can occur either in patients with serological evidence of past “apparently resolved” HBV infection, or also in individuals with no evident history of exposure to HBV [25]. Indeed this finding has epidemiological implications in that the burden of HBV in Cameroon, often determined by serological assays, is significantly underestimated. This ‘silent’ infection also has clinical implications since it leads to an increased risk of hepatocellular carcinoma [26].
The ten HBV strains sequenced in this study showed the circulation of both genotype E and A with genotype E predominating. Our findings are in agreement with previous studies in Cameroon that have documented the circulation of these two genotypes [12, 27]. Drug resistant patterns indicate that three of the strains had mutations associated with resistance to lamivudine, telbivudine and entecavir. Further, that the individuals harboring viruses with these mutations were all lamivudine experienced. Importantly though, none of the viruses harbored mutations associated with tenofovir and adefovir resistance. This finding is in agreement with the recommendation to include tenofovir as part of HAART among HIV/HBV co-infected patients [28]. Regarding mutations in the “a” determinant of the S gene, majority of the viruses had a G159A mutation. In addition, all the viruses had 127Lin this region. The 127L mutation has previously been associated with OBI [29], while the role of G159A in OBI is unclear.
The findings in this study should be interpreted in the light of several limitations. First, the HBV viral loads which are also often used as a marker of OBI were not available. Secondly, the study utilized a qualitative and not quantitative ELISA format, thus antibody titres were not determined. Thirdly, owing to resource constraints only 50% of the OBI positive samples were sequenced and lastly, the lack of liver enzyme levels precludes our ability to correlate the results reported here with clinical outcomes.
Conclusion
In conclusion, we report a 5.9% prevalence of OBI among HIV infected individuals in Cameroon and the dominance of HBV genotype E. We also show the occurrence of OBI in the absence of any serologic evidence of infection; and the presence of mutations associated with lamivudine drug resistance in the study cohort. Further prospective studies in a larger cohort are needed to determine the clinical implications of OBI in this region.
References
WHO. Hepatitis B fact sheet. 2016. http://www.who.int/mediacentre/factsheets/fs204/en/. Accessed 20 Sept 2016.
Global, regional, and national age–sex specific all-cause and cause-specific mortality for 240 causes of death, 1990–2013: a systematic analysis for the global burden of disease study 2013. Lancet. 2014;385(9963):117–171. doi:10.1016/S0140-6736(14)61682-2.
Diop-Ndiaye H, Toure-Kane C, Etard JF, Lo G, Diaw P, Ngom-Gueye NF, et al. Hepatitis B, C seroprevalence and delta viruses in HIV-1 Senegalese patients at HAART initiation (retrospective study). J Med Virol. 2008;80(8):1332–6. doi:10.1002/jmv.21236.
Alter MJ. Epidemiology of viral hepatitis and HIV co-infection. J Hepatol. 2006;44(1 Suppl):S6–9. doi:10.1016/j.jhep.2005.11.004.
Vallet-Pichard A, Pol S. Natural history and predictors of severity of chronic hepatitis C virus (HCV) and human immunodeficiency virus (HIV) co-infection. J Hepatol. 2006;44(1 Suppl):S28–34. doi:10.1016/j.jhep.2005.11.008.
Trepo C, Chan HL, Lok A. Hepatitis B virus infection. Lancet. 2014;384(9959):2053–63. doi:10.1016/S0140-6736(14)60220-8.
Chemin I, Trepo C. Clinical impact of occult HBV infections. J Clin Virol. 2005;34(Suppl 1):S15–21. doi:10.1016/S1386-6532(05)80005-8.
Sood AK, Pangotra C, Manrai M. Prevalence of occult hepatitis B infection in patients visiting tertiary care hospital. Med J Armed Forces India. 2016;72(2):140–4. doi:10.1016/j.mjafi.2016.02.006.
Ramezani A, Banifazl M, Eslamifar A, Sofian M, Aghakhani A. Occult hepatitis B infection in different high risk patients. Hepat Mon. 2012;12(7):467–8. doi:10.5812/hepatmon.7094.
Burnett RJ, François G, Kew MC, Leroux-Roels G, Meheus A, Hoosen AA, et al. Hepatitis B virus and human immunodeficiency virus co-infection in sub-Saharan Africa: a call for further investigation. Liver Int. 2005;25(2):201–13. doi:10.1111/j.1478-3231.2005.01054.x.
Sagnelli C, Macera M, Pisaturo M, Zampino R, Coppola M, Sagnelli E. Occult HBV infection in the oncohematological setting. Infection. 2016. doi:10.1007/s15010-016-0891-1.
Salpini R, Forkam J, Ceccareilli L, Santoro MM, Nanfack A, Sosso SM, et al. High burden of HBV-infection and atypical HBV strains among HIV-infected Cameroonians. Curr HIV Res. 2016;14(2):165–71.
Magoro T, Gachara G, Mavhandu L, Lum E, Kimbi H, Ndip R. Serologic and genetic characterization of hepatitis B virus in HIV infected individuals from the South and Littoral Regions of Cameroon. Virol J. 2016;13:178. doi:10.1186/s12985-016-0636-x.
Marrone A, Zampino R, Karayannis P, Cirillo G, Cesaro G, Guerrera B, et al. Clinical reactivation during lamivudine treatment correlates with mutations in the precore/core promoter and polymerase regions of hepatitis B virus in patients with anti-hepatitis B e-positive chronic hepatitis. Aliment Pharmacol Ther. 2005;22(8):707–14. doi:10.1111/j.1365-2036.2005.02653.x.
Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 2013;30(12):2725–9. doi:10.1093/molbev/mst197.
Ronquist F, Huelsenbeck JP. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 2003;19(12):1572–4.
Shire NJ, Rouster SD, Stanford SD, Blackard JT, Martin CM, Fichtenbaum CJ, et al. The prevalence and significance of occult hepatitis B virus in a prospective cohort of HIV-infected patients. JAIDS J Acquir Immune Defic Syndr. 2007;44(3):309–14. doi:10.1097/QAI.0b013e31802e29a9.
Tramuto F, Maida CM, Colomba GME, Di Carlo P, Vitale F. Prevalence of occult hepatitis B virus infection in a cohort of HIV-positive patients resident in Sicily, Italy. Biomed Res Int. 2013;2013:859583. doi:10.1155/2013/859583.
Yousif M, Mudawi H, Hussein W, Mukhtar M, Nemeri O, Glebe D, et al. Genotyping and virological characteristics of hepatitis B virus in HIV-infected individuals in Sudan. Int J Infect Dis. 2014;29:125–32. doi:10.1016/j.ijid.2014.07.002.
Cohen Stuart JW, Velema M, Schuurman R, Boucher CA, Hoepelman AI. Occult hepatitis B in persons infected with HIV is associated with low CD4 counts and resolves during antiretroviral therapy. J Med Virol. 2009;81(3):441–5. doi:10.1002/jmv.21422.
Gupta S, Singh S. Occult hepatitis B virus infection in ART-naive HIV-infected patients seen at a tertiary care centre in north India. BMC Infect Dis. 2010;10:53. doi:10.1186/1471-2334-10-53.
Marite B, Montalvo MC, Rodriguez Lde L, Sariego S, Verdasquera D, Vincent M, et al. Occult hepatitis B in Cuban HIV patients. MEDICC Rev. 2011;13(2):32–7.
Fabris P, Biasin MR, Giordani MT, Berardo L, Menini V, Carlotto A, et al. Impact of occult HBV infection in HIV/HCV co-infected patients: HBV-DNA detection in liver specimens and in serum samples. Curr HIV Res. 2008;6(2):173–9.
Mphahlele MJ, Lukhwareni A, Burnett RJ, Moropeng LM, Ngobeni JM. High risk of occult hepatitis B virus infection in HIV-positive patients from South Africa. J Clin Virol. 2006;35(1):14–20. doi:10.1016/j.jcv.2005.04.003.
Firnhaber C, Chen CY, Evans D, Maskew M, Schulz D, Reyneke A, et al. Prevalence of hepatitis B virus (HBV) co-infection in HBV serologically-negative South African HIV patients and retrospective evaluation of the clinical course of mono- and co-infection. Int J Infect Dis. 2012;16(4):e268–72. doi:10.1016/j.ijid.2011.12.007.
Chen JD, Yang HI, Iloeje UH, You SL, Lu SN, Wang LY, et al. Carriers of inactive hepatitis B virus are still at risk for hepatocellular carcinoma and liver-related death. Gastroenterology. 2010;138(5):1747–54. doi:10.1053/j.gastro.2010.01.042.
Kouanfack C, Aghokeng AF, Mondain AM, Bourgeois A, Kenfack A, Mpoudi-Ngole E, et al. Lamivudine-resistant HBV infection in HIV-positive patients receiving antiretroviral therapy in a public routine clinic in Cameroon. Antivir Ther. 2012;17(2):321–6. doi:10.3851/IMP1911.
Soriano V, Puoti M, Bonacini M, Brook G, Cargnel A, Rockstroh J, et al. Care of patients with chronic hepatitis B and HIV co-infection: recommendations from an HIV-HBV International Panel. AIDS. 2005;19(3):221–40.
Svicher V, Cento V, Bernassola M, Neumann-Fraune M, Van Hemert F, Chen M, et al. Novel HBsAg markers tightly correlate with occult HBV infection and strongly affect HBsAg detection. Antiviral Res. 2012;93(1):86–93. doi:10.1016/j.antiviral.2011.10.022.
Authors’ contributions
PB, HKK and RNN designed the study, EL recruited study subjects, collected samples and demographic data, TM and LM performed the laboratory analyses, GG analyzed the data and prepared the manuscript with contributions from all the authors. All authors read and approved the manuscript.
Acknowledgements
The authors would like to thank the study subjects for their participation.
Competing interests
The authors declare that they have no competing interests.
Availability of data and materials
Please contact author for data requests.
Compliance with ethical standards
This study protocol was approved by the institutional review board of the Cameroon Baptist Health Board, Cameroon (IRB2012-01) and the Research Ethics Committee of the University of Venda, South Africa (SMNS/14/MBY/21/2110). Signed informed consent was obtained from all study participants or their legal guardians prior to sample collection.
Funding
The study was not specifically funded. TM was supported by the National Research Foundation. PB is supported by the National Research Foundation and Medical Research Council of South Africa. GG is supported by award number D43 TW009359 from the Fogarty International Center, National Institutes of Health. None of the supporting funding bodies played a role in the execution of the study, data analysis, or writing of the manuscript. The information presented here is solely the responsibility of the authors.
Author information
Authors and Affiliations
Corresponding author
Additional file
12981_2017_136_MOESM1_ESM.docx
Additional file 1: Figure S1. A representation gel showing amplified HBV DNA from HBsAg negative plasma. Lane L is a 100 bp molecular weight marker. Expected DNA band sizes of 366 bp were readily detected for samples on lanes 2, 5, 7, 8, 10–14. Lane P is a positive control obtained from the PCR optimization process and confirmed by sequencing. Lane N is a negative control (sterilized distilled water used as template).
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Gachara, G., Magoro, T., Mavhandu, L. et al. Characterization of occult hepatitis B virus infection among HIV positive patients in Cameroon. AIDS Res Ther 14, 11 (2017). https://doi.org/10.1186/s12981-017-0136-0
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12981-017-0136-0