Abstract
It is widely acknowledged that gastric cancer seriously affects the quality of life and survival of patients. The correlation between the microbiota and gastric cancer has attracted extensive attention in recent years, nonetheless the specific mechanism of its impact on gastric cancer remain largely unclear. Recent studies have shown that in addition to its role in the host’s inflammatory and immune response, the microbiota can also affect the occurrence and development of gastric cancer by affecting the expression of miRNAs. This paper brings together all currently available data on miRNAs, microbiota and gastric cancer, and preliminarily describes the relationship among them.
Similar content being viewed by others
Background
Gastric cancer (GC) is the fifth most common malignant tumor globally, after lung, breast, colorectal and prostate cancers. Even though its incidence rate has declined in recent years, it is still the third leading cause of cancer-related death worldwide [1, 2]. In China, GC ranks third in the incidence and tumor-associated death in the national population-based cancer registry [3]. This finding can be attributed to the fact that most patients with gastric cancer are already at an advanced stage at diagnosis, which is correlated to the high incidence of postoperative local recurrence and distant metastasis and the poor treatment outcomes [4].
The human gastrointestinal microbiota constitutes a complex micro-ecosystem involved in metabolism, promoting the development of the immune system and inhibiting the colonization of pathogens. With the development of sequencing technology, more strains have been found to colonize the stomach. Aviles et al. found decreased numbers of Porphyromonas, Neisseria and Streptococcus and increased Lactic acid bacteria, Pseudomonas aeruginosa and Trichospirillum in gastric cancer by comparing the differences in gastric microbiota of patients with non-atrophic gastritis, intestinal metaplasia and intestinal-type gastric cancer. This finding suggests that these bacteria may play a role in the development of gastric cancer from gastritis [5]. Recent studies on the microbiota in gastric cancer and paracancerous tissues have shown that the mucosa-associated microbiota in tumor tissues and non-tumor tissues have different established microecosystems in terms of composition, structure, interaction network and function. In this regard, it has been shown that Proteobacteria are the main phylum, followed by Firmicutes, Bacteroides, Actinomycetes, Acidobacteria and Fusobacteria in tumor tissues, and acid-producing bacteria are more abundant in non-tumor tissues [6]. Another team analyzed the microbiota of gastric cancer patients’ tumor tissues, adjacent tissues and normal tissues, found significant differences in the microbiota among the three groups. Furthermore, another study demonstrated a higher abundance of Spiricobacter, Halomonas and Shevchella in adjacent tissues, while Streptococcus, Selenomonas, Fusobacterium, Propionibacter and Corynebacterium were more abundant in tumor tissues [7].
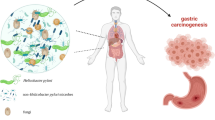
An increasing body of evidence suggests that the microbiota can affect the occurrence and development of tumors in various ways, including chronic inflammation and immune regulation [8,9,10,11,12,13,14,15,16]. In recent years, it has been shown that microRNAs (miRNAs) can influence the survival and composition of gut bacteria [17,18,19]. Based on the existing literature, we hypothesized interactions between the microbiota and the host miRNA affect the occurrence and development of tumors. In this review, we present our current understanding of microbiota-miRNAs interactions in gastric cancer (Fig. 1). After summarizing evidence pointing to their core role, we look forward to the future directions of this rapidly developing field.
Host miRNAs-microbiota interactions in gastric cancer. Microbiota can affect the development of gastric cancer by five ways:①, ②Microbiota directly or through cytotoxic affect host inflammation and immune response;③Microbiota induced methylation of E-cadherin and tumor-suppressor genes;④Microbiota damage host DNA through metabolites;⑤Microbiota can also affect the expression of host miRNAs, which can affect the occurrence and development of gastric cancer. Tumor metabolites can affect microbial colonization, while miRNAs can also affect host metabolism and indirectly affect microbial colonization
Microbiota and gastric cancer
Microbial imbalances affect the occurrence and development of gastric cancer
Many studies have confirmed significant differences in microbial composition between gastric cancer patients and healthy controls, indicating that an imbalanced gastric microbiome is an important cause of gastric cancer [6, 7]. Gastric cancer is driven by inflammation-related factors as many studies have shown that the microbiota can promote the development of gastric cancer by promoting inflammation and regulating immunity.
Previous studies that explored the effect of microbiota on gastric cancer mainly focused on Helicobacter pylori (Hp). Marshall and Warren identified Hp from a gastric biopsy culture in 1984 [20]. Nowadays, it is widely recognized that Hp infection is a high-risk factor for gastric cancer [21]. In this regard, Hp infection can increase secretion of inflammatory cytokines such as interferon-c, TNF-a, IL-1, IL-1B, IL-6, IL-7, IL-8, IL-10 and IL-18, which stimulate a variety of immune cells, including lymphocytes, peripheral monocytes, eosinophils, macrophages, neutrophils, mast cells and dendritic cells [8]. Hp infection also can cause methylations on E-cadherin and tumor-suppressor genes [22]. The cytotoxin-associated gene A (CagA) and vacuolating cytotoxin gene (VacA) are the main virulence factors of Hp. Both CagA and VacA can cause inflammatory reactions and affect the occurrence and development of tumors. CagA can promote the Wnt, MAPK, Erk and NF-κB pathways and inhibit the activity of p53 [23,24,25]. VacA can induce autophagy by upregulating MAPK and ERK signaling, directly activating mitochondria and vascular endothelial growth factors. VacA has also been reported to act on the Wnt pathway and pI3k/Akt signaling pathways [26,27,28,29,30]. Some studies have documented interactions between CagA and VacA, which exhibit antagonistic effects on the NFAT pathway and cell morphology. Importantly, CagA can also inhibit the apoptosis of VacA [31].
In recent years, with the development of sequencing technology, an increasing number of microbes have been discovered in the stomach, such as Firmicutes, Proteobacteria, Bacteroidetes, Fusobacteria and Actinobacteria [32]. Nonetheless, few studies have documented the effect of other microbiota on gastric cancer. Indeed, other bacteria can also affect the occurrence and development of gastric cancer through regulatory pathways, immune cells and their metabolites. For example, Stenoprophomonas and Selenomonas are increased in gastric cancer and positively correlated with BDCA2 + pDCs and Foxp3 + Tregs, while Comannas and Gailla are negatively correlated with BDCA2 + pDCs and Foxp3 + Tregs [33]. Further studies have confirmed that the abundance of Fusobacterium in gastric cancer is associated with a poor prognosis [34,35,36]. Fusobacterium infection has also been associated with tumor-infiltrating lymphocytes and p53 expression [34]. However, the specific mechanisms on gastric cancer progression remain understudied. Interestingly, the levels of P.copri and P. acnes are reportedly increased in gastric cancer patients, affecting the occurrence and development of gastric cancer through the NKG2D system and IL-15 [37]. In contrast, some studies have shown that P.copri levels were decreased in gastric cancer [7]. N-nitroso compounds (NOCs) and lactic acid have been widely reported as the main microbial metabolites associated with gastric cancer. NOCs are produced by many bacteria such as Clostridium, Veillonella, Staphylococcus, and Lactobacillus that are increased in gastric cancer [38]. It is also well established that NOCs are associated with gastric cancer. ROS produced by lactic acid bacteria can damage DNA, promote tumor growth and metastasis, and inhibit tumor apoptosis by promoting NOCs production [33, 39]. Many studies have reported that the abundance of Lactobacillus in patients with gastric cancer is increased, and their lactic acid metabolites can affect the occurrence and development of gastric cancer [40, 41].
Factors affecting microbial colonization
Over the years, an increasing number of studies have emphasized the importance of the balance in the microbiota that colonizes the human body. Microbial imbalance is also an important factor responsible for cancer, described in detail in the previous paragraph. Importantly, microbial colonization of gastric cancer can be affected by the microenvironment (microbial interaction, surgical operation, stage or type of GC and so on) [42,43,44,45,46].
Studies have shown that Hp infection can change the microbial composition of gastric cancer patients [43]. A decreased microbial diversity has been reported in the stomach of HP-positive patients; the main microorganisms were Proteus, followed by Firmicutes, Bacteroidetes and Actinomycetes [47]. Similarly, Anderson et al. found 33 phylotypes in the stomach of HP-positive gastric cancer patients and more than 200 phylotypes in healthy controls [48]. Importantly, genes related to nucleotide transport metabolism and amino acid transport metabolism are significantly enriched in patients’tumor microbiota, and gastric acid secretion increased significantly in the Hp-positive gastric cancer patients [7]. Moreover, HP may lead to a gastric inflammatory environment through CagA and VacA virulence factors, thus affecting the growth of other microorganisms [49]. Leung et al. analyzed the characteristics of individuals with gastric cancer with different histological stages and the gastric microbiota after Hp eradication. Importantly, HP eradication increased the bacterial diversity, and the relative abundance of other bacteria returned to a level similar to healthy controls [50]. Similarly, the gastric microbiota and their metabolites can influence the ability of Hp to colonize the stomach and modulate its pathogenicity and carcinogenic potential [44].
Elaine et al. showed that the metabolism and microbial composition of gastric cancer patients were significantly altered after Roux-en-Y gastric bypass (RYGB) surgery [45]. Consistently, Vaidota et al. showed that gastric cancer surgery could affect the composition of gastric microbiota [46]. Liu et al. found that the gastric microenvironment determined the composition and diversity of gastric microbiota rather than the stage or type of GC. The above studies substantiated that the microbiota could affect the occurrence and development of tumors; however, it should be borne in mind that the tumor microenvironment can also regulate the microbial composition.
MiRNAs and gastric cancer
MiRNAs belong to non-coding RNAs that function by regulating downstream target genes. The imbalance of miRNAs is closely related to the occurrence and development of gastric cancer. Besides, the stability of miRNAs enhances their potential as tumor biomarkers. The role of miRNAs during the pathogenesis of gastric cancer and their potential application as biomarkers and therapeutic targets for gastric cancer has attracted much interest.
Clinical application of miRNAs
It has been established that miRNAs can be released from tumor tissue to the serum, plasma, urine, tears, amniotic fluid and gastric juice through exosome particles. Studies have confirmed that circulating miRNAs in the plasma/serum of GC patients are consistent with miRNA expression levels in tissues. Accordingly, they can be used as biomarkers for early diagnosis and recurrence evaluation of gastric cancer [51]. Table 1 lists some circulating miRNAs associated with gastric cancer based on serum/plasma, considered as biomarkers of gastric cancer. Previous studies have shown significant differences in circulating miRNAs between patients with gastric cancer and normal controls.
Recent studies have shown that the serum levels of EV-miR-215-5p in gastric cancer patients were significantly higher than in the healthy subject group, especially in patients with early gastric cancer recurrence. Importantly, serum EV-miR-215-5p levels were significantly correlated with the depth of invasion, TNM stage and lymph node metastasis. Accordingly, serum EV-miR-215-5p has huge prospects as a new biomarker for predicting early gastric cancer recurrence and prognosis of gastric cancer [52].
Wang et al. showed that miR-18a-5p could be a potential therapeutic target for gastric adenocarcinoma [53]. The combination of miR-4257, miR-6785-5p, miR-187-5p and miR-5739 provided a useful and high-precision non-invasive diagnostic means for the detection of early gastric cancer in a large sample size study (n = 1417) [54]. Importantly, Wang et al. showed that miR-214 could be used as a target in patients exhibiting chemotherapy resistance; the injection of exo-anti-214 could reverse the resistance of gastric cancer patients to cisplatin [55]. Overall, the above studies demonstrate that miRNAs have great potential for clinical application when used as biomarkers for diagnosis, prognosis and treatment.
MiRNAs affect the development of gastric cancer
MiRNAs can be used as biomarkers of gastric cancer since they can affect the development of gastric cancer in many ways. It has been found that different miRNAs have different effects on gastric cancer; they can promote or inhibit gastric cancer. miR-96-5p is reportedly overexpressed in gastric cancer cell lines and plasma samples of gastric cancer patients and can promote the proliferation of gastric cancer cells by directly targeting FOXO365. Importantly, it has been shown that miR-93-5p overexpression in gastric cancer cells upregulates the downstream genes CDX2, FoxM1 and CTGF of the Hippo pathway [66]. However, another study has shown that miR-146b-5p is negatively correlated with lymph node metastasis and distant metastasis of gastric cancer and inhibits the malignant development of gastric cancer by targeting TRAF6 [67]. Low expression of miR-339-5p has also been documented to be closely related to metastasis and prognosis of gastric cancer, and miR-339-5p can inhibit the malignant development of GC by negatively regulating ALKBH1 [68]. Table 2 lists the latest studies on the promotion or inhibition of miRNAs on gastric cancer.
Host miRNAs affect cancer metabolism
Changes in host miRNAs can change the metabolism of tumors. Cao et al. showed that the circlmo7-miR-30a-3p-wnt2 axis could provide energy for the growth of GC cells through glutamine metabolism [80]. Enhanced miR-4646-5p can stabilize HIF1A by targeting PHD3 and regulating the expression of ABHD16a and miR-4646-5p itself via a positive feedback mechanism. As a novel phosphatidylserine-specific lipase, ABHD16a participates in lipid metabolism and accumulates lysophosphatidylserine [81]. Other studies have also corroborated that miR-34a has a regulatory effect on lactic acid [82].
Microbiota affect miRNAs expression
An increasing body of evidence suggests that Hp infection can affect host miRNAs expression. For instance, Chang et al. showed that in Hp-positive gastric cancer patients, miR99b-3p, miR-564, and miR-638 were significantly increased, while miR-204-5p, miR-338-5p, miR-375, and miR-548c-3p significantly decreased, compared with Hp-negative patients, [83]. Moreover, miR-18a-3p and miR-4286 have been reported to be induced by Hp infection, while miR-204 was decreased. Previous studies also showed that above miRNAs induced the NF-κB pathway, promoting the growth of gastric cancer [84, 85]. In addition, the expression of many miRNAs is affected by Hp infection, such as miR-146a, miR-1289, miR-223-3p, miR-370, miR-22, miR-133 and so on [86,87,88,89].
Interestingly, after EBV enters the human body, it can cause host genome methylation, abnormal gene expression, disrupt cell signaling pathways, and generate a tumor microenvironment in infected gastric epithelial cells, leading to the occurrence and development of gastric cancer [90]. In addition, latent EBV gene products, such as EBERS, BARF-0, EBNA-1 and LMP2A, are reportedly involved in downregulating the miR-200 family, resulting in decreased expression of E-cadherin or inducing NF-κB/miR-146a/Smad4 pathway in gastric cancer cells. They can also upregulate miR-155-5p expression by NF-κB pathway and inhibit the activation of Smad2 and p-Smad2 [91,92,93].
Perspective
MiRNAs play a key role in the interaction between microbiota and host. The microbiota can affect the occurrence and development of cancer directly or indirectly. Studies have shown that HP infection can increase inflammatory factors such as IL-1, 6 and 7, thus affecting the body's immunity [8]. HP infection can also increase the methylation of E-cad and tumor suppressor genes [22]. Meanwhile, it can also promote Wnt, MAPK, ERK, NF-κB, PI3K pathway and inhibit the p53 pathway by secreting CagA and VacA [21, 25, 87]. At present, a large number of studies have shown that Fusobacteria, Actinobacteria, Proteobacteria and Bacteroidetes are related to gastric cancer, and some strains are significantly related to tumor microenvironment immune factors [32, 34]. Lactic acid and NOCs produced by many strains can also promote tumor development by destroying DNA [33]. Nonetheless, the mechanisms underlying the above findings remain understudied, warranting further studies.
Previous studies have found that miRNAs can be used as potential clinical biomarkers. For example, a significant difference in the expression levels of miR199a-3p, miR-451 and other miRNAs was found between cancer patients and normal controls [56, 57] A significant correlation has been reported between patient prognosis and miRNAs miR-21 and miR-200c, which indicates that miRNAs can be used as prognostic markers [58, 59]. MiRNAs can not only be used as biomarkers of gastric cancer, but also affect Akt and Akt, NF-κB, RAGE, ARF6, TCF4, STAT3 and other signaling pathways promote or inhibit the occurrence and development of gastric cancer [69, 70, 73,74,75,76]. In the meantime, miRNAs can also alter lipid metabolism, energy metabolism and amino acid metabolism, thus indirectly affecting the occurrence and development of cancer [80,81,82].
Some studies have also shown that the microbiota can affect the expression of host miRNAs, and miRNAs can conversely affect microbial colonization by changing the host-related metabolism [89]. However, the specific mechanism of the interaction between host miRNA and colonized microbiota in gastric cancer remains largely unexplored. In future research, an interdisciplinary endeavor is necessary to fill the knowledge gap on miRNA-mediated host-microbiome communication and provide key information for developing new treatment strategies to decrease gastric cancer-associated mortality rates.
Availability of data and materials
Not applicable.
References
Bray F, Ferlay J, Soerjomataram I, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394–424.
Ferlay J, Colombet M, Soerjomataram I, et al. Estimating the global cancer incidence and mortality in 2018: GLOBOCAN sources and methods. Int J Cancer. 2019;144(8):1941–53.
McGuire S. World Cancer Report 2014. Geneva, Switzerland: World Health Organization, International Agency for Research on Cancer, WHO Press, 2015. Adv Nutr. 2016;7(2):418–9.
Siegel RL, Miller KD, Jemal A. Cancer statistics, 2020. CA Cancer J Clin. 2020;70(1):7–30.
Aviles-Jimenez F, Vazquez-Jimenez F, Medrano-Guzman R, et al. Stomach microbiota composition varies between patients with non-atrophic gastritis and patients with intestinal type of gastric cancer. Sci Rep. 2014;4:4202.
Chen XH, Wang A, Chu AN, et al. Mucosa-associated microbiota in gastric cancer tissues compared with non-cancer tissues. Front Microbiol. 2019;10:1261.
Liu X, Shao L, Liu X, et al. Alterations of gastric mucosal microbiota across different stomach microhabitats in a cohort of 276 patients with gastric cancer. EBioMedicine. 2019;40:336–48.
Meng C, Bai C, Brown TD, et al. Human gut microbiota and gastrointestinal cancer. Genomics Proteomics Bioinformatics. 2018;16(1):33–49.
Hattori N, Ushijima T. Epigenetic impact of infection on carcinogenesis: mechanisms and applications. Genome Med. 2016;8(1):10.
Putoczki TL, Thiem S, Loving A, et al. Interleukin-11 is the dominant IL-6 family cytokine during gastrointestinal tumorigenesis and can be targeted therapeutically. Cancer Cell. 2013;24(2):257–71.
Gensollen T, Iyer SS, Kasper DL, et al. How colonization by microbiota in early life shapes the immune system. Science. 2016;352(6285):539–44.
Palm NW, de Zoete MR, Flavell RA. Immune-microbiota interactions in health and disease. Clin Immunol. 2015;159(2):122–7.
Brown DG, Rao S, Weir TL, et al. Metabolomics and metabolic pathway networks from human colorectal cancers, adjacent mucosa, and stool. Cancer Metab. 2016;4:11.
Carino A, Graziosi L, D’Amore C, et al. The bile acid receptor GPBAR1 (TGR5) is expressed in human gastric cancers and promotes epithelial-mesenchymal transition in gastric cancer cell lines. Oncotarget. 2016;7(38):61021–35.
Louis P, Hold GL, Flint HJ. The gut microbiota, bacterial metabolites and colorectal cancer. Nat Rev Microbiol. 2014;12(10):661–72.
Smith PM, Howitt MR, Panikov N, et al. The microbial metabolites, short-chain fatty acids, regulate colonic Treg cell homeostasis. Science. 2013;341(6145):569–73.
Yuan C, Burns MB, Subramanian S, et al. Interaction between host microRNAs and the gut microbiota in colorectal cancer. mSystems. 2018. https://doi.org/10.1128/mSystems.00205-17.
Liu S, da Cunha AP, Rezende RM, et al. The host shapes the gut microbiota via fecal microRNA. Cell Host Microbe. 2016;19(1):32–43.
Teng Y, Ren Y, Sayed M, et al. Plant-derived exosomal microRNAs shape the gut microbiota. Cell Host Microbe. 2018;24(5):637-652.e8.
Marshall BJ, Warren JR. Unidentified curved bacilli in the stomach of patients with gastritis and peptic ulceration. Lancet. 1984;1(8390):1311–5.
Nejati S, Karkhah A, Darvish H, et al. Influence of Helicobacter pylori virulence factors CagA and VacA on pathogenesis of gastrointestinal disorders. Microb Pathog. 2018;117:43–8.
Sitaraman R. Helicobacter pylori DNA methyltransferases and the epigenetic field effect in cancerization. Front Microbiol. 2014;5:115.
Murata-Kamiya N, Kurashima Y, Teishikata Y, et al. Helicobacter pylori CagA interacts with E-cadherin and deregulates the beta-catenin signal that promotes intestinal transdifferentiation in gastric epithelial cells. Oncogene. 2007;26(32):4617–26.
Saadat I, Higashi H, Obuse C, et al. Helicobacter pylori CagA targets PAR1/MARK kinase to disrupt epithelial cell polarity. Nature. 2007;447(7142):330–3.
Yong X, Tang B, Li BS, et al. Helicobacter pylori virulence factor CagA promotes tumorigenesis of gastric cancer via multiple signaling pathways. Cell Commun Signal. 2015;13:30.
Mashima H, Suzuki J, Hirayama T, et al. Involvement of vesicle-associated membrane protein 7 in human gastric epithelial cell vacuolation induced by Helicobacter pylori-produced VacA. Infect Immun. 2008;76(6):2296–303.
Galmiche A, Rassow J. Targeting of Helicobacter pylori VacA to mitochondria. Gut Microbes. 2010;1(6):392–5.
Ki MR, Lee HR, Goo MJ, et al. Differential regulation of ERK1/2 and p38 MAP kinases in VacA-induced apoptosis of gastric epithelial cells. Am J Physiol Gastrointest Liver Physiol. 2008;294(3):G635–47.
Caputo R, Tuccillo C, Manzo BA, et al. Helicobacter pylori VacA toxin up-regulates vascular endothelial growth factor expression in MKN 28 gastric cells through an epidermal growth factor receptor-, cyclooxygenase-2-dependent mechanism. Clin Cancer Res. 2003;9(6):2015–21.
Liu N, Zhou N, Chai N, et al. Helicobacter pylori promotes angiogenesis depending on Wnt/beta-catenin-mediated vascular endothelial growth factor via the cyclooxygenase-2 pathway in gastric cancer. BMC Cancer. 2016;16:321.
Yokoyama K, Higashi H, Ishikawa S, et al. Functional antagonism between Helicobacter pylori CagA and vacuolating toxin VacA in control of the NFAT signaling pathway in gastric epithelial cells. Proc Natl Acad Sci USA. 2005;102(27):9661–6.
Ferreira RM, Pereira-Marques J, Pinto-Ribeiro I, et al. Gastric microbial community profiling reveals a dysbiotic cancer-associated microbiota. Gut. 2018;67(2):226–36.
Ling Z, Shao L, Liu X, et al. Regulatory T cells and plasmacytoid dendritic cells within the tumor microenvironment in gastric cancer are correlated with gastric microbiota dysbiosis: a preliminary study. Front Immunol. 2019;10:533.
Nie S, Wang A, Yuan Y. Comparison of clinicopathological parameters, prognosis, micro-ecological environment and metabolic function of gastric cancer with or without Fusobacterium sp. Infection. J Cancer. 2021;12(4):1023–32.
Hsieh YY, Tung SY, Pan HY, et al. Increased abundance of clostridium and fusobacterium in gastric microbiota of patients with gastric cancer in Taiwan. Sci Rep. 2018;8(1):158.
Boehm ET, Thon C, Kupcinskas J, et al. Fusobacterium nucleatum is associated with worse prognosis in Lauren’s diffuse type gastric cancer patients. Sci Rep. 2020;10(1):16240.
Gunathilake MN, Lee J, Choi IJ, et al. Association between the relative abundance of gastric microbiota and the risk of gastric cancer: a case-control study. Sci Rep. 2019;9(1):13589.
Zhang S, Shi D, Li M, et al. The relationship between gastric microbiota and gastric disease. Scand J Gastroenterol. 2019;54(4):391–6.
Jones RM, Mercante JW, Neish AS. Reactive oxygen production induced by the gut microbiota: pharmacotherapeutic implications. Curr Med Chem. 2012;19(10):1519–29.
Vinasco K, Mitchell HM, Kaakoush NO, et al. Microbial carcinogenesis: lactic acid bacteria in gastric cancer. Biochim Biophys Acta Rev Cancer. 2019;1872(2): 188309.
Castaño-Rodríguez N, Goh KL, Fock KM, et al. Dysbiosis of the microbiome in gastric carcinogenesis. Sci Rep. 2017;7(1):15957.
Sheflin AM, Whitney AK, Weir TL. Cancer-promoting effects of microbial dysbiosis. Curr Oncol Rep. 2014;16(10):406.
Yang J, Zhou X, Liu X, et al. Role of the gastric microbiome in gastric cancer: from carcinogenesis to treatment. Front Microbiol. 2021;12: 641322.
Espinoza J, Matsumoto A, Tanaka H, et al. Gastric microbiota: an emerging player in Helicobacter pylori-induced gastric malignancies. Cancer Lett. 2018;414:147–52.
Li JV, Ashrafian H, Sarafian M, et al. Roux-en-Y gastric bypass-induced bacterial perturbation contributes to altered host-bacterial co-metabolic phenotype. Microbiome. 2021;9(1):139.
Maksimaityte V, Bausys A, Kryzauskas M, et al. Gastrectomy impact on the gut microbiome in patients with gastric cancer: a comprehensive review. World J Gastrointest Surg. 2021;13(7):678–88.
Parsons BN, Ijaz UZ, D’Amore R, et al. Comparison of the human gastric microbiota in hypochlorhydric states arising as a result of Helicobacter pylori-induced atrophic gastritis, autoimmune atrophic gastritis and proton pump inhibitor use. PLoS Pathog. 2017;13(11):e1006653.
Andersson AF, Lindberg M, Jakobsson H, et al. Comparative analysis of human gut microbiota by barcoded pyrosequencing. PLoS ONE. 2008;3(7): e2836.
Zhang RG, Duan GC, Fan QT, et al. Role of Helicobacter pylori infection in pathogenesis of gastric carcinoma. World J Gastrointest Pathophysiol. 2016;7(1):97–107.
Li TH, Qin Y, Sham PC, et al. Alterations in gastric microbiota after H. Pylori eradication and in different histological stages of gastric carcinogenesis. Sci Rep. 2017;7:44935.
Mitchell PS, Parkin RK, Kroh EM, et al. Circulating microRNAs as stable blood-based markers for cancer detection. Proc Natl Acad Sci USA. 2008;105(30):10513–8.
Chen R, Yang M, Huang W, et al. Cascades between miRNAs, lncRNAs and the NF-κB signaling pathway in gastric cancer (review). Exp Ther Med. 2021;22(1):769.
Wang L, Zhang M, Wang J, et al. Diagnostic and therapeutic potencies of miR-18a-5p in mixed-type gastric adenocarcinoma. J Cell Biochem. 2021. https://doi.org/10.1002/jcb.29927.
Abe S, Matsuzaki J, Sudo K, et al. A novel combination of serum microRNAs for the detection of early gastric cancer. Gastric Cancer. 2021. https://doi.org/10.1007/s10120-021-01161-0.
Wang X, Zhang H, Bai M, et al. Exosomes serve as nanoparticles to deliver anti-miR-214 to reverse chemoresistance to cisplatin in gastric cancer. Mol Ther. 2018;26(3):774–83.
Li C, Li JF, Cai Q, et al. miRNA-199a-3p in plasma as a potential diagnostic biomarker for gastric cancer. Ann Surg Oncol. 2013;20(Suppl 3):S397-405.
Konishi H, Ichikawa D, Komatsu S, et al. Detection of gastric cancer-associated microRNAs on microRNA microarray comparing pre- and post-operative plasma. Br J Cancer. 2012;106(4):740–7.
Wu J, Li G, Wang Z, et al. Circulating microRNA-21 is a potential diagnostic biomarker in gastric cancer. Dis Markers. 2015;2015: 435656.
Valladares-Ayerbes M, Reboredo M, Medina-Villaamil V, et al. Circulating miR-200c as a diagnostic and prognostic biomarker for gastric cancer. J Transl Med. 2012;10:186.
Song MY, Pan KF, Su HJ, et al. Identification of serum microRNAs as novel non-invasive biomarkers for early detection of gastric cancer. PLoS ONE. 2012;7(3): e33608.
Liu X, Kwong A, Sihoe A, et al. Plasma miR-940 may serve as a novel biomarker for gastric cancer. Tumour Biol. 2016;37(3):3589–97.
Jiang X, Jiang M, Xu M, et al. Identification of diagnostic utility and molecular mechanisms of circulating miR-551b-5p in gastric cancer. Pathol Res Pract. 2019;215(5):900–4.
Wang N, Wang L, Yang Y, et al. A serum exosomal microRNA panel as a potential biomarker test for gastric cancer. Biochem Biophys Res Commun. 2017;493(3):1322–8.
Ranjbar R, Hesari A, Ghasemi F, et al. Expression of microRNAs and IRAK1 pathway genes are altered in gastric cancer patients with Helicobacter pylori infection. J Cell Biochem. 2018;119(9):7570–6.
He X, Zou K. MiRNA-96-5p contributed to the proliferation of gastric cancer cells by targeting FOXO3. J Biochem. 2020;167(1):101–8.
Meng H, Li YY, Han D, et al. MiRNA-93-5p promotes the biological progression of gastric cancer cells via Hippo signaling pathway. Eur Rev Med Pharmacol Sci. 2019;23(11):4763–9.
Ding JN, Zang YF, Ding YL. MiRNA-146b-5p inhibits the malignant progression of gastric cancer by targeting TRAF6. Eur Rev Med Pharmacol Sci. 2020;24(17):8837–44.
Wang C, Huang Y, Zhang J, et al. MiRNA-339–5p suppresses the malignant development of gastric cancer via targeting ALKBH1. Exp Mol Pathol. 2020;115: 104449.
Xiao F, Cheng Z, Wang P, et al. MicroRNA-28-5p inhibits the migration and invasion of gastric cancer cells by suppressing AKT phosphorylation. Oncol Lett. 2018;15(6):9777–85.
Ye T, Yang M, Huang D, et al. MicroRNA-7 as a potential therapeutic target for aberrant NF-κB-driven distant metastasis of gastric cancer. J Exp Clin Cancer Res. 2019;38(1):55.
Lin J, Liu Z, Liao S, et al. Elevated microRNA-7 inhibits proliferation and tumor angiogenesis and promotes apoptosis of gastric cancer cells via repression of Raf-1. Cell Cycle. 2020;19(19):2496–508.
Lu Q, Chen Y, Sun D, et al. MicroRNA-181a functions as an oncogene in gastric cancer by targeting Caprin-1. Front Pharmacol. 2018;9:1565.
Xu XC, Zhang WB, Li CX, et al. Up-regulation of MiR-1915 inhibits proliferation, invasion, and migration of Helicobacter pylori-infected gastric cancer cells via targeting RAGE. Yonsei Med J. 2019;60(1):38–47.
Qiu Y, Yuan Y, Luo P. MiR-1299 functions as a tumor suppressor to inhibit the proliferation and metastasis of gastric cancer by targeting ARF6. Genes Genomics. 2021. https://doi.org/10.1007/s13258-021-01124-w.
He MQ, Wan JF, Zeng HF, et al. miR-133a-5p suppresses gastric cancer through TCF4 down-regulation. J Gastrointest Oncol. 2021;12(3):1007–19.
Li Z, Fan H, Chen W, et al. MicroRNA-653–5p promotes gastric cancer proliferation and metastasis by targeting the SOCS6-STAT3 pathway. Front Mol Biosci. 2021;8: 655580.
Huangfu L, He Q, Han J, et al. MicroRNA-135b/CAMK2D axis contribute to malignant progression of gastric cancer through EMT process remodeling. Int J Biol Sci. 2021;17(8):1940–52.
Zhou K, Cao D, Wang Y, et al. Hsa-miR-30a-3p attenuates gastric adenocarcinoma proliferation and metastasis via APBB2. Aging (Albany NY). 2021;13(12):16763–72.
Luo D, Fan H, Ma X, et al. miR-1301–3p promotes cell proliferation and facilitates cell cycle progression via targeting SIRT1 in gastric cancer. Front Oncol. 2021;11: 664242.
Cao J, Zhang X, Xu P, et al. Circular RNA circLMO7 acts as a microRNA-30a-3p sponge to promote gastric cancer progression via the WNT2/β-catenin pathway. J Exp Clin Cancer Res. 2021;40(1):6.
Yang L, Hou Y, Du YE, et al. Mirtronic miR-4646–5p promotes gastric cancer metastasis by regulating ABHD16A and metabolite lysophosphatidylserines. Cell Death Differ. 2021. https://doi.org/10.1038/s41418-021-00779-y.
Ping W, Senyan H, Li G, et al. Increased lactate in gastric cancer tumor-infiltrating lymphocytes is related to impaired T cell function due to miR-34a deregulated lactate dehydrogenase A. Cell Physiol Biochem. 2018;49(2):828–36.
Chang H, Kim N, Park JH, et al. Different microRNA expression levels in gastric cancer depending on Helicobacter pylori infection. Gut Liver. 2015;9(2):188–96.
Tsai CC, Chen TY, Tsai KJ, et al. NF-κB/miR-18a-3p and miR-4286/BZRAP1 axis may mediate carcinogenesis in Helicobacter pylori-associated gastric cancer. Biomed Pharmacother. 2020;132: 110869.
Chen P, Guo H, Wu X, et al. Epigenetic silencing of microRNA-204 by Helicobacter pylori augments the NF-κB signaling pathway in gastric cancer development and progression. Carcinogenesis. 2020;41(4):430–41.
Zhang YM, Noto JM, Hammond CE, et al. Helicobacter pylori-induced posttranscriptional regulation of H-K-ATPase α-subunit gene expression by miRNA. Am J Physiol Gastrointest Liver Physiol. 2014;306(7):G606–13.
Yang F, Xu Y, Liu C, et al. NF-κB/miR-223-3p/ARID1A axis is involved in Helicobacter pylori CagA-induced gastric carcinogenesis and progression. Cell Death Dis. 2018;9(1):12.
Saito Y, Suzuki H, Tsugawa H, et al. Dysfunctional gastric emptying with down-regulation of muscle-specific microRNAs in Helicobacter pylori-infected mice. Gastroenterology. 2011;140(1):189–98.
Dastmalchi N, Safaralizadeh R, Banan Khojasteh SM. The correlation between microRNAs and Helicobacter pylori in gastric cancer. Pathog Dis. 2019. https://doi.org/10.1093/femspd/ftz039.
Sun K, Jia K, Lv H, et al. EBV-positive gastric cancer: current knowledge and future perspectives. Front Oncol. 2020;10: 583463.
Shinozaki A, Sakatani T, Ushiku T, et al. Downregulation of microRNA-200 in EBV-associated gastric carcinoma. Cancer Res. 2010;70(11):4719–27.
Kim DH, Chang MS, Yoon CJ, et al. Epstein-Barr virus BARF1-induced NFκB/miR-146a/SMAD4 alterations in stomach cancer cells. Oncotarget. 2016;7(50):82213–27.
Shi Q, Zhang Y, Liu W, et al. Latent membrane protein 2A inhibits expression level of Smad2 through regulating miR-155-5p in EBV-associated gastric cancer cell lines. J Med Virol. 2020;92(1):96–106.
Acknowledgements
Not applicable.
Funding
This study was supported by the project of the Regional Diagnosis and Treatment Center of the Health Planning Committee (No. JBZX-201903) and Zhejiang Science and Technology Planning Project (No.2021C03119).
Author information
Authors and Affiliations
Contributions
YY participated in the conception and design of the manuscript. CXL and HYY participated in drafting the article and preparing the figure. LW, LJ and CCZ participated in preparing the tables. TLS and YXF have all reviewed. All authors have read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors have declared that no competing interest exists.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Yang, Y., Huang, Y., Lin, W. et al. Host miRNAs-microbiota interactions in gastric cancer. J Transl Med 20, 52 (2022). https://doi.org/10.1186/s12967-022-03264-3
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12967-022-03264-3