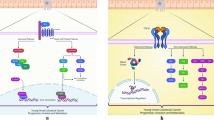
Abstract
Background
Young patients with colorectal cancer (CRC) exhibit poor prognoses compared to older patients due to the difficulty in early diagnosis and treatment. However, the underlying molecular characteristics are still unclear.
Methods
We conducted a comprehensive analysis of 49 CRC patients without hereditary CRC using the whole-exome and RNA sequencing with tumor and matched normal samples. A total of 594 TCGA samples and 4 patient-derived cells were utilized for validation.
Results
Consensus molecular subtype 4 (CMS4) (53.85%) and CMS2 (38.46%) were enriched in the young (≤ 40 years) and old (> 60 years) age groups, respectively. A CMS4-associated gene, platelet-derived growth factor receptor α (PDGFRA), was significantly upregulated in young patients with CRC (FC = 3.21, p = 0.0001) and was negatively correlated with age (p = 0.0001, R = − 0.526). Moreover, PDGFRA showed a positive co-expression with metastasis-related genes in young CRC patients. In vitro validation confirmed that young patient-derived cells (PDCs) showed an enriched expression of PDGFRA compared to old PDCs and a reduced proliferation rate by knockdown of PDGFRA. Furthermore, young CRC patients were more sensitive to regorafenib, a PDGFRA-targeting drug, than old CRC patients.
Conclusions
Our study suggests that CRC in young patients is associated with CMS4 and PDGFRA. In addition, PDGFRA may serve potential of novel therapeutic strategies and represent a predictive biomarker of response to regorafenib for young CRC patients.
Similar content being viewed by others
Background
Colorectal cancer (CRC) is one of the most common cancers and a leading cause of cancer-related deaths worldwide [1, 2]. However, studies on specific therapeutic strategies and appropriate drugs for CRC have not reported successful outcomes [3, 4]. CRC is predominantly a cancer in older adults with a median age at diagnosis of > 60 years [5, 6]. However, population-based studies have reported that CRC incidence has increased in the younger population than in those > 50 years old [5, 6]. Numerous studies have suggested that younger patients have poor prognoses with difficulty in early diagnosis compared to older patients [7,8,9]. In addition to the difficulty in treating young CRC patients, there are few specific treatment strategies for young CRC patients.
According to a study with a selected gene set showing several somatic mutations that tend to be frequent in CRC patients, several early-onset CRCs are caused by inherited genetic mutations [10], except for hereditary cancer syndromes [11]. A gene expression study of young patients without a family history of the disease suggested that upregulated genes, compared with healthy controls, are involved in various biological processes [12]. Based on the insufficient gene set and patient population in previous studies, the knowledge derived so far may be incomplete. Recently, high-throughput genomic and transcriptomic sequencing technologies have attracted attention as tools for detecting a large number of oncogenes [13] and quantifying gene expression levels with a high resolution [14]. Comparative analysis at the genomic and transcriptomic levels will provide greater details and insights into the molecular mechanism of young CRC.
Despite the widespread use of the technique, the CRC classification published so far revealed only similarities [15,16,17,18]. Recently, a more detailed gene expression-based classification of CRC was proposed with four biologically distinct consensus molecular subtypes (CMS) as follows [19]: CMS1 (microsatellite instability (MSI) and immune activation), CMS2 (epithelial type and marked WNT signaling), CMS3 (epithelial type and metabolic dysregulation), and CMS4 (mesenchymal type and activated TGF-β signaling). Additionally, the clinical relevance of CMS4 is associated with poor prognosis, while the others are characterized by a relatively good prognosis [19]. As the CMS allows a more detailed and accurate classification than the existing classification methods, many studies have been recently conducted to diagnose and treat CRC patients[20,21,22,23]. Therefore, the classification of individual CMS groups has great potential to advance precision medicine for CRC patients.
In this study, we conducted a comprehensive analysis of 49 CRC patients to understand the molecular characteristics of young patients through a whole-exome and RNA sequencing with tumor and matched normal samples. To identify young CRC patients’ specific biomarkers, we divided them into three groups: young, middle, and old. A comparison of these three groups could differentiate between young and old age CRC and reveal clearer molecular subtypes and genomic alterations. Our study provides an in-depth understanding of young CRC tumors and novel biomarkers.
Methods
Sample collection
This study was approved by the Institutional Review Board (IRB) of the Samsung Medical Center (IRB Approval No. SMC 2013-11-007-001). Written informed consent was obtained from each patient. The study subjects were 49 patients diagnosed with CRC at the Samsung Medical Center, Seoul, Korea. Tumor and matched adjacent normal tissues were obtained from surgical specimens. All patients were not treated before surgery.
The cancer genome atlas (TCGA) colorectal cancer data sets
The gene expression profile of 594 mRNASeq colorectal adenocarcinoma (COAD) samples from cBioPortal (https://www.cbioportal.org/, TCGA, PanCancer Atlas) was utilized for validation.
Isolation of genomic DNA and RNA
Genomic DNA and RNA in tissues were purified using the AllPrep DNA/RNA Mini Kit (Qiagen, Valencia, CA, USA). Genomic DNA from peripheral blood was extracted using the QIAamp DNA Blood Mini Kit (Qiagen). The genomic DNA concentration and purity were measured using a NanoDrop 8000 UV–Vis Spectrometer (Thermo Scientific Inc., Wilmington, DE, USA) and a Qubit 2.0 Fluorometer (Life Technologies Inc., Grand Island, NY, USA). DNA degradation was estimated by measuring median DNA size and ΔCt values with a 2200 TapeStation Instrument (Agilent Technologies, Santa Clara, CA, USA) and real-time PCR (Agilent Technologies), respectively. For RNA, the concentration and purity were measured with the NanoDrop and Bioanalyzer (Agilent Technologies).
Whole-exome sequencing
Genomic DNA (1 µg) from each sample was sheared using the Covaris S220 (Covaris, Woburn, MA, USA), and a library was constructed using SureSelect XT Human All Exon V5 and a SureSelect XT Reagent Kit, HSQ (Agilent Technologies). The kit captures 335,756 exons of 21,058 genes, covering ~ 71 Mb of the human genome. After enriched exome libraries were multiplexed, the libraries were sequenced on a HiSeq 2500 platform (Illumina, San Diego, CA, USA). Briefly, a paired-end DNA sequencing library was prepared by gDNA shearing, end-repair, A-tailing, paired-end adaptor ligation, and amplification. After hybridization of the library with bait sequences for 16 h, the captured library was purified and amplified with an index barcode tag, and the library quality and quantity were measured. Sequencing was performed in the 100-bp paired-end mode of the TruSeq Rapid PE Cluster Kit and TruSeq Rapid SBS Kit (Illumina).
Exome-seq data analysis
Raw sequencing reads were aligned to the UCSC hg19 reference genome (downloaded from http://genome.ucsc.edu), using the Burrows–Wheeler Aligner (BWA) [24] version 0.7.5a with default settings. PCR duplicates were marked using Picard-tools-1.93 (http://picard.sourceforge.net/), data cleanup was performed with GATK, and variants were identified with GATK-3.5 [25]. Point mutations were identified using MuTect [26] with paired samples. ANNOVAR [27] was used to annotate variants. Somatic copy number alterations were identified using the Excavator tool [28]. Regions of the genome that were significantly amplified or deleted across samples were detected by the GISTIC tool [29].
RNA sequencing
Library construction for RNA sequencing was carried out with a Truseq RNA Sample Preparation v2 Kit (Illumina). Isolated total RNA was used in a reverse transcription reaction with poly (dT) primers, using SuperScriptTM II Reverse Transcriptase (Invitrogen/Life Technologies). Briefly, an RNA sequencing library was prepared by cDNA amplification, end-repair, 3′ end adenylation, adapter ligation, and amplification. Library quality and quantity were measured using the Bioanalyzer and Qubit. Sequencing of the RNA library was performed on the 100-bp paired-end mode of the TruSeq Rapid PE Cluster Kit and the TruSeq Rapid SBS Kit (Illumina).
RNA-seq data analysis
Reads from the FASTQ files were mapped to the hg19 human reference genome using STAR aligner version 2.5.0a [30] and read counts for each gene were normalized as transcripts per million (TPM) using the RSEM program [31]. Differentially expressed genes (DEGs) were identified using the DESeq R package [32] with a cut-off (|log2 fold-change|> 2 and false discovery rate (FDR) < 0.001) [33]. DEGs were mapped to the pathway using the ReactomeFI Cytoscape [34]. CMS calls in CRC cohorts were predicted based on the gene expression profile using the R package CMSclassifier [19].
Organoid culture
The organoid culture medium was refreshed every 2 days. For the organoid passage, BME was broken up by pipetting, and organoids were collected in a tube. The organoids were centrifuged at 1000 rpm for 3 min, and the medium was removed. Next, 5 mL of Triple Express (Invitrogen) was added, and the organoids were incubated at 37 °C for approximately 5 min. Every minute, a visual check was performed to verify the size of organoids. Care was taken not to treat the organoids too long with the Triple Express. FCS and medium were added, and the cells were spun down at 1500 rpm for 3 min. The pellet was taken up in BME, and the cells were plated in droplets of 5–10 μL each. After allowing the BME to solidify, HICS minus Wnt, both supplemented with 10-µM LY27632, was added to the plates, and organoids were incubated at 37 °C [35].
Cell transfection
For transient transfection assays, PDGFRA siRNAs (#1: 5′ CCUCUAUCCUUCCAAAUGAUU 3′, 5′ UCAUUUGGAAGGAUAGAGGUU 3’, #2: 5′ CCGUCAAAGGAAAGAAGUUUU 3′, 5′ AACUUCUUUCCUUUGACGGUU 3′, Genolution, Seoul, Korea) were transfected into PDCs using Lipofectamine RNAiMAX (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s protocol.
Cell proliferation assay
Cell proliferation was measured in triplicate using a colorimetric assay that determines cellular viability by evaluating the metabolic conversion based on the quantification of the ATP present using CellTiter-Glo (Promega, Madison, WI, USA). The viability of the colon cancer cells was assessed at various times, and assays were performed by adding CellTiter-Glo directly to the culture wells and incubating for 30 min at room temperature. Luminescence was measured using a Mithras microplate reader (Berthold-bio, Bad Wildbad, Germany). Three different experiments were performed for each experimental condition.
Real-time quantitative polymerase chain reaction (RT-qPCR) analysis
Total RNA was extracted from the patient’s tissue (RNAprep Mini kit, Qiagen), and 500 ng RNA was subjected to reverse transcription using a reverse transcription kit (Bioneer). Real-time quantitative PCR amplification was performed with SYBR Green (ABI) in a real-time system (ABI, USA). Human-specific PCR primers (Bioneer) were used to analyze the expression of the following genes: PDGFRA and GAPDH. The mRNA levels of the specific genes were calculated as ΔΔCt and normalized to those of GAPDH.
Cell lysis and Western blot analysis
To prepare the whole-cell extract, cells were lysed using a Pro-prep buffer (Intron Biotechnology, Seoul, Korea) containing protease inhibitors. A total of 10–40 μg of protein extract was resolved by SDS-PAGE and transferred to polyvinylidene fluoride (PVDF) membranes. The membranes were probed with primary antibodies against PDGFRA (ab65258, Abcam) and β-actin (#3700, Cell Signaling Technology) followed by incubation with secondary antibodies conjugated to horseradish peroxidase (Santa Cruz Biotechnology, CA, USA). β-actin was used as a loading control in the western blot analysis.
High-throughput screening (HTS)
The chip layout was designed for drug screening in a single micropillar chip. In the micropillar chip, ~ 80–100 cells were immociliized with 0.75% alginate. We tested 67 drugs in CRC PDCs. Among them, we revealed regorafenib data in this study. A 50-nL droplet of a 1:1 cell mixture of 1.5% alginate and 950-nL droplet of 3D culture media was dispensed with the ASFA Spotter ST (MBD). After overnight incubation, a 950-nL droplet of the drugs was dispensed with the ASFA Spotter ST and stamped with the micropillar chip containing the cells. The combined chips were incubated for 5 days at 37 °C and 5% CO2 in an incubator for the cell viability assay. After incubation, the micropillar chips were stained with a staining buffer (MBD-STA50, Medical and Bio Device) in a Calcein-AM (Invitrogen, live-cell starting dye) for 1 h in the dark at room temperature. The stained micropillar chips were washed with a staining buffer for 30 min and dried in the dark.
Statistical analysis
Statistical comparisons were performed using GraphPad Prism. Data are expressed as the mean ± standard deviation (SD). P-values < 0.05 were considered statistically significant (*P < 0.05, **P < 0.01, or ***P < 0.001).
Results
CMS classification according to age of CRC patients
To clarify the difference in the molecular characteristics between young and old patients, we divided the samples into three groups according to the age of 49 CRC patients (Table 1). The young (n = 13), middle-aged (n = 23), and old age (n = 13) groups included patients < 40 years old, between 41 and 60 years old, and > 60 years old, respectively. After categorizing the patient groups, CRC patients were classified according to CMS using gene expression profiles [19]. In particular, CMS4 (53.85%) was the most frequent in young CRC, followed by CMS3 (38.46%) and CMS2 (7.69%). CMS1 was absent in the young age group (Fig. 1a). In the middle CRC group, the order of the frequencies was the same as in the young age group, but the frequencies of CMS2 (21.74%) and CMS1 (13.04%) were higher (Fig. 1b). In the old CRC group, the frequency of CMS4 (23.08%) was the lowest, while CMS2 (38.46%) and CMS3 (38.46%) showed the same frequency (Fig. 1c). Thus, CMS4 frequency gradually decreased with age, while CMS2 was more frequent in the old age group (Fig. 1 a-c). Taken together, these results indicate that young CRC patients are closely associated with CMS4, which has been considered as the worst prognosis type [19]. On the other hand, young CRC patients are less related to CMS2, which has a better prognosis than the other CMSs [19].
Differential mRNA expression and gene profiling between young and old CRC
To systematically identify specific gene expression patterns related to young CRC, we detected differentially expressed genes (DEGs) from RNA sequencing data of 49 CRC tissues and their paired adjacent normal tissues. We first identified age-related genes showing a significant difference between normal samples of the young and old age groups (Fig. 2a right panel, log2 fold change (FC) ≥ ± 1, FDR ≤ 0.05). In addition, DEGs in the tumor tissues were obtained by comparing both groups using tumor samples (Fig. 2a left panel, log2 FC ≥ ± 1, FDR ≤ 0.05). Therefore, 155 genes from the DEGs in normal samples and 204 genes in the tumor were obtained. Finally, by eliminating 155 age-related genes in the normal samples, we considered a total of 180 genes as age-related DEGs specific to cancer (Fig. 2b). Among them, 105 genes were significantly upregulated and the other 75 were downregulated (Fig. 2c, log2 fold change ≥ ± 1, p-value ≤ 0.05, FDR ≤ 0.05). To identify cancer-related genes to be considered biomarkers of young CRC patients, we extracted only oncogenes [36] and tumor suppressor genes[37]. A number of oncogenes and tumor suppressor genes were highly upregulated or downregulated in young CRC patients (eight and four upregulated genes, and six and five downregulated genes, respectively) (Tables 2 and 3). Next, we identified the significantly changed pathways (34 upregulated and 26 downregulated) from DEGs using the network-based pathway analysis tool [38] (Additional file 1: Fig. S1). Many CMS4-related pathways [19] were enriched in young CRC patients, including PDGFRA signaling, positive regulation of cell proliferation by VEGF-activated platelet-derived growth factor (PDGF) receptor signaling, cell chemotaxis, regulation of chemotaxis, positive regulation of phospholipase C (PLC) activity, and phosphatidylinositol phosphorylation (Fig. 2d). Notably, these pathways are closely related to metastasis, angiogenesis, proliferation, and chemoresistance, which are related to CMS4 [19] and are associated with PDGFRA.
Differential mRNAs expression and gene profiling between young CRC and old CRC tissue. a Volcano plot of differentially expressed genes. Significant genes are highlighted in red. Dotted lines indicate cutoff values of log2 fold change (FC) ≥ ± 1, FDR ≤ 0.05. b Venn diagram of differentially expressed genes between young and old age groups. The number of genes obtained after a comparison with normal samples (right) and with tumor samples (left). c Heatmap of the expression profile of selected genes. The expression values were normalized by a z-score transformation across samples. d Enriched pathways from significantly changed genes in expression. e Oncoprint of CRC-associated mutations (missense, nonsense, and splicing). f Copy number amplification and deletions that are frequently detected in the samples. The green solid line indicates the q-value cutoff = 0.01
We also identified numerous somatic mutations, including missense, nonsense, and splicing mutations in primary colon tumors of CRC patients (Fig. 2e). The mutation profile showed that a TP53 mutation was more frequent (p = 0.0029) in young CRC (76.9%, 10/13) than in old CRC (23%, 3/13) (Fig. 2e). A comparison of the copy number amplified and deleted genomic regions between both groups (Fig. 2f) showed that the copy number at chromosome 5q22.2 was significantly deleted only in young CRC samples (q-value < 0.01).
High level of PDGFRA expression in young CRC patients
To investigate whether PDGFRA is correlated with the age of CRC patients, its correlation pattern with age was examined. PDGFRA expression was negatively correlated with the age of CRC patients in our cohort (Fig. 3a, Spearman’s correlation coefficient; P = 0.0001, R = − 0.5264). Additionally, TCGA data consistently showed an inverse correlation between its expression and the age of CRC patients (Additional file 2: Fig. S2a, P = 0.0113, R = − 0.1697). Figure 3b shows that the expression of PDGFRA in the young CRC group was significantly higher than that in the old CRC patient group (P = 0.0001). The TCGA data revealed a similar pattern (Additional file 2: Fig. S2b, P = 0.2796), although this data had a limitation due to the small number of young CRC patients. To confirm whether PDGFRA was a young CRC factor rather than a CMS4 factor, its expression was compared between both groups within samples of the CMS4 type. The expression of PDGFRA in the young CRC group was higher than that in the old CRC group, even when considering only the CMS4 type (Fig. 3c, P = 0.0708). There was no significant difference due to the small sample size. To confirm our results, we also used a publicly available gene expression data set (GSE14333) derived from the primary CRC tissue (n = 188). To set similar conditions as our data, we divided it into two groups according to percentile (young CRC group: less than approximately 25th percentile, < 59 years old (n = 49) and old CRC group: greater than approximately 75th percentile, > 76 years old (n = 49)). With the CMS classification according to age, CMS4 significantly increased in the younger group (Fig. 3d, upper panel). The expression of PDGFRA in the younger group was significantly higher than that in the older group (P < 0.001) (Fig. 3d, lower panel).
PDGFRA expression in young and old CRC patients. a Scatter plot of PDGFRA mRNA expression and age. Each point indicates a sample of the patient, distinguished according to CMS types. b Boxplot between PDGFRA expression levels in the young CRC group and old CRC group. c Comparison of PDGFRA expression levels in CMS4 patients. d Pie chart of CMS types for three categories according to age group (upper panel), and boxplot between PDGFRA expression levels in the young CRC group and old CRC group (lower panel)
Co-expression of PDGFRA with a CMS4 related factor
CMS4 is a mesenchymal subtype of CRC and is activated in stromal invasion and angiogenesis [19]. We examined whether PDGFRA was closely associated with epithelial-mesenchymal transition (EMT) markers [39] and angiogenesis markers [40]. Among the EMT markers, six markers, namely Vim, ZEB1, ZEB2, Twist1, Twist2, and MMP2, showed positive co-expression with PDGFRA in our data set (Fig. 4a). Their expression levels were also different than the two groups, showing separate distributions. As with PDGFRA, the samples identified as CMS4 had a high expression. Among the angiogenesis markers, five markers, including FLT1, FLT4, ICAM1, TIE1, and KDR, showed a positive correlation in their gene expression (Fig. 4b). These markers also showed the same pattern as EMT markers. In the TCGA data, these EMT and angiogenesis markers also showed the same results as our sequencing data (Additional file 3: Fig. S3a, b). Each marker positively correlated with PDGFRA and showed a negative correlation with age (Additional file 3: Fig. S3c, d) and a co-expression pattern among each other (Additional file 3: Fig. S3e).
Co-expression of EMT and angiogenesis factor. a Scatter plot of gene expression levels between PDGFRA and EMT-associated signatures. b Scatter plot of gene expression levels between PDGFRA and angiogenesis-associated signatures. Each point represents a sample that is colored according to the CMS type. The comparison of expression levels for the young and old groups is shown by a density plot
PDGFRA expression and PDGFRA-targeted drug response in PDC
To investigate the key roles of PDGFRA in young CRC, we used PDCs derived from young and old age CRC patients that were not treated before surgery. (Table 4). The RT-qPCR analysis showed that PDGFRA expression was higher in young PDCs than in old PDCs (Fig. 5a, Additional file 4: Fig. S4a). Western blot analysis revealed that the expression of PDGFRA was higher in young PDCs than in old PDCs (Fig. 5b). To evaluate the possible roles of PDGFRA in young CRC, we transfected siRNAs in PDCs with two different siRNAs against PDGFRA to compare their proliferation ability using a Cell Titer GLO proliferation assay. Knockdown of PDGFRA in PDCs from young patients significantly reduced viability (Fig. 5c). However, PDCs from old patients did not alter viability compared to the control (Fig. 5d). To determine the PDGFRA-targeted drug for young CRC, we analyzed the correlation between drug sensitivity and gene expression [41]. The Cancer Cell Line Encyclopedia (CCLE) data indicate that sorafenib targeting PDGFRA only had a potential drug response in colorectal cancer (Additional file 4: Fig. S4b). However, our results showed that sorafenib did not show a distinguished drug response according to PDGFRA expression or age in PDCs (Additional file 4: Fig. S4c). Regorafenib is a small molecule multi-kinase inhibitor, including PDGFRA, and was recently approved by the FDA to treat patients with metastatic CRC [42,43,44]. To investigate the treatment strategy for young CRC, we used regorafenib in PDCs. PDCs from young patients were sensitive to regorafenib treatment, whereas PDCs from old patients were relatively resistant to regorafenib (Fig. 5e). When conducting HTS, the young age group was set to younger than 50 years old due to the small number of samples, and the results showed that the IC50 of regorafenib was significantly lower in the young group than in the old group (Additional file 4: Fig. S4d).
PDGFRA expression and PDGFRA-targeted drug response in PDC. a Boxplot between PDGFRA expression levels (RT-qPCR) in young and old PDCs. b Western blot of PDGFRA in young and old PDCs. c Viability by knockdown of PDGFRA in PDCs from young patients. d Viability by knockdown of PDGFRA in PDCs from old patients. e Sensitivity to regorafenib, an anti-cancer drug targeting PDGFRA, for PDCs. All patients used for PDC did not have any treatment before surgery
Discussion
We first classified the CMS for young CRC patients and suggested a biomarker according to CMS. Moreover, we utilized PDCs to explore the function of the biomarker in young CRC patients. Previous studies on young CRC patients have focused only on a few cancer gene mutations and cancer-related gene expression or case reports, which were analyzed by categorizing patients into only two groups of young and old [45,46,47,48,49]. We divided the patients into young, middle-aged, and old age groups and focused on the CMS that gradually changed with age, as observed in a large amount of sequencing data of CRC patients. We found that the proportion of CMS4 gradually increased from the old group to the young group in the CRC patients. It is worth noting that our results demonstrated interesting findings that have not been previously reported in other studies on the CMS status of young CRC patients. From transcriptomic data, we found that PDGFRA, which is associated with CMS4, was significantly upregulated in young CRC patients than in old CRC patients. The protein level of PDGFRA was also higher in young CRC patients using PDCs. Additionally, when PDGFRA was downregulated by siRNA, the proliferation rate was significantly lower in young PDCs than in old PDCs. These results indicated that PDGFRA could possibly be a biomarker for young CRC patients.
We found that young CRC specific pathways include PDGFRA signaling, positive regulation of cell proliferation by VEGF-activated PDGF receptor signaling, cell chemotaxis, regulation of chemotaxis, positive regulation of PLC activity, and phosphatidylinositol phosphorylation. They are closely related to metastasis, angiogenesis, proliferation, and chemoresistance, which reflect the characteristics of the CMS4 type [19]. PDGFRA plays a major role in these pathways and is expressed in mesenchymal cells and epithelial cancer cells undergoing EMT [50]. PDGF, a ligand of PDGFRA, promotes cancer angiogenesis in the cancer stroma [51] and has positive regulation of cell proliferation and angiogenesis by VEGF-activated PDGFRA [52]. The chemotaxis of cancer cells plays an important role in cancer progression, metastasis, and drug resistance [53, 54]. PDGFRA promotes chemotaxis of cancer cells [53, 55, 56]. Activation of the PLC pathway increases cisplatin resistance and promotes cancer growth and metastasis [57,58,59]. PDGFRA induces activation of the PLC pathway and interacts with it [60,61,62,63]. These results suggest that PDGFRA may play a pivotal role in CMS4-associated pathways.
DNA sequencing data showed that TP53 mutations were frequently detected in the young CRC group than the old CRC group. A recent study reported that the TP53 mutation was also more frequent in young-onset CRC [64]. This was the same condition as the young CRC group that is < 40 years old [64]. Although both studies were consistent, the reason for a large number of TP53 mutations in young CRC patients remains unknown. In addition, the copy number at chromosome 5q22.2 was deleted only in the young CRC group. A high deletion rate in chromosome 5q22.2 may explain poor clinical outcomes in young CRC patients, which is because the region contains several tumor suppressor genes, such as APC and MCC [65,66,67]. Based on these results, more functional studies are required further study in TP53 mutation and deletion of chromosome 5q22.2 for young CRC patients.
Our data showed that regorafenib was more sensitive to PDCs of young CRC in which PDGFRA was upregulated. Regorafenib is a multi-kinase inhibitor [68, 69] that has been mainly indicated worldwide for patients with metastatic CRC [43, 69]. However, our data showed that regorafenib might be a potential candidate for treating young CRC patients. Therefore, regorafenib could be used for young CRC patients and for metastatic CRC.
Conclusions
In conclusion, the CMS classification for young CRC patients may better inform clinicians of the prognosis and potential of novel therapeutic strategies. Our study suggests that CRC in young patients is associated with CMS4, PDGFRA, and CMS4-related genes. Our data will provide a greater understanding of young CRC patients and the prognostic value of biomarkers.
Availability of data and materials
The data supporting the finding of this study are available within the article are available from the corresponding author upon request.
Abbreviations
- CRC:
-
Colorectal cancer
- CMS:
-
Consensus molecular subtypes
- MSI:
-
Microsatellite instability
- COAD:
-
Colorectal adenocarcinoma
- DEGs:
-
Differentially expressed genes
- TPM:
-
Transcripts per million
- FC:
-
Fold change
- EMT:
-
Epithelial–mesenchymal transition
- CCLE:
-
The Cancer Cell Line Encyclopedia
References
Hubbard JM, Grothey A. Colorectal cancer in 2014: progress in defining first-line and maintenance therapies. Nat Rev Clin Oncol. 2015;12(2):73–4.
Lee YS, Kim SY, Song SJ, Hong HK, Lee Y, Oh BY, et al. Crosstalk between CCL7 and CCR3 promotes metastasis of colon cancer cells via ERK-JNK signaling pathways. Oncotarget. 2016;7(24):36842–53.
Siegel R, Desantis C, Jemal A. Colorectal cancer statistics, 2014. CA Cancer J Clin. 2014;64(2):104–17.
Hong HK, Pyo DH, Kim TW, Yun NH, Lee YS, Song SJ, et al. Efficient primary culture model of patientderived tumor cells from colorectal cancer using a Rho-associated protein kinase inhibitor and feeder cells. Oncol Rep. 2019;42(5):2029–38.
Chew MH, Koh PK, Ng KH, Eu KW. Improved survival in an Asian cohort of young colorectal cancer patients: an analysis of 523 patients from a single institution. Int J Colorectal Dis. 2009;24(9):1075–83.
O’Connell JB, Maggard MA, Liu JH, Etzioni DA, Livingston EH, Ko CY. Rates of colon and rectal cancers are increasing in young adults. Am Surg. 2003;69(10):866–72.
Chan KK, Dassanayake B, Deen R, Wickramarachchi RE, Kumarage SK, Samita S, et al. Young patients with colorectal cancer have poor survival in the first twenty months after operation and predictable survival in the medium and long-term: analysis of survival and prognostic markers. World J Surg Oncol. 2010;8:82.
Cusack JC, Giacco GG, Cleary K, Davidson BS, Izzo F, Skibber J, et al. Survival factors in 186 patients younger than 40 years old with colorectal adenocarcinoma. J Am Coll Surg. 1996;183(2):105–12.
Endreseth BH, Romundstad P, Myrvold HE, Hestvik UE, Bjerkeset T, Wibe A, et al. Rectal cancer in the young patient. Dis Colon Rectum. 2006;49(7):993–1001.
Pearlman R, Frankel WL, Swanson B, Zhao W, Yilmaz A, Miller K, et al. Prevalence and spectrum of germline cancer susceptibility gene mutations among patients with early-onset colorectal cancer. JAMA Oncol. 2017;3(4):464–71.
Berg M, Danielsen SA, Ahlquist T, Merok MA, Agesen TH, Vatn MH, et al. DNA sequence profiles of the colorectal cancer critical gene set KRAS-BRAF-PIK3CA-PTEN-TP53 related to age at disease onset. PLoS One. 2010;5(11):e13978.
Hong Y, Ho KS, Eu KW, Cheah PY. A susceptibility gene set for early onset colorectal cancer that integrates diverse signaling pathways: implication for tumorigenesis. Clin Cancer Res. 2007;13(4):1107–14.
Berger MF, Mardis ER. The emerging clinical relevance of genomics in cancer medicine. Nat Rev Clin Oncol. 2018;15(6):353–65.
Ozsolak F, Milos PM. RNA sequencing: advances, challenges and opportunities. Nat Rev Genet. 2011;12(2):87–98.
Cancer Genome Atlas Network. Comprehensive molecular characterization of human colon and rectal cancer. Nature. 2012;487(7407):330–7.
Roepman P, Schlicker A, Tabernero J, Majewski I, Tian S, Moreno V, et al. Colorectal cancer intrinsic subtypes predict chemotherapy benefit, deficient mismatch repair and epithelial-to-mesenchymal transition. Int J Cancer. 2014;134(3):552–62.
Budinska E, Popovici V, Tejpar S, D’Ario G, Lapique N, Sikora KO, et al. Gene expression patterns unveil a new level of molecular heterogeneity in colorectal cancer. J Pathol. 2013;231(1):63–76.
Sadanandam A, Lyssiotis CA, Homicsko K, Collisson EA, Gibb WJ, Wullschleger S, et al. A colorectal cancer classification system that associates cellular phenotype and responses to therapy. Nat Med. 2013;19(5):619–25.
Guinney J, Dienstmann R, Wang X, de Reynies A, Schlicker A, Soneson C, et al. The consensus molecular subtypes of colorectal cancer. Nat Med. 2015;21(11):1350–6.
Thanki K, Nicholls ME, Gajjar A, Senagore AJ, Qiu S, Szabo C, et al. Consensus molecular subtypes of colorectal cancer and their clinical implications. Int Biol Biomed J. 2017;3(3):105–11.
Sveen A, Bruun J, Eide PW, Eilertsen IA, Ramirez L, Murumagi A, et al. Colorectal cancer consensus molecular subtypes translated to preclinical models uncover potentially targetable cancer cell dependencies. Clin Cancer Res. 2018;24(4):794–806.
Linnekamp JF, Hooff SRV, Prasetyanti PR, Kandimalla R, Buikhuisen JY, Fessler E, et al. Consensus molecular subtypes of colorectal cancer are recapitulated in in vitro and in vivo models. Cell Death Differ. 2018;25(3):616–33.
Becht E, de Reynies A, Giraldo NA, Pilati C, Buttard B, Lacroix L, et al. Immune and stromal classification of colorectal cancer is associated with molecular subtypes and relevant for precision immunotherapy. Clin Cancer Res. 2016;22(16):4057–66.
Li H, Durbin R. Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics. 2009;25(14):1754–60.
DePristo MA, Banks E, Poplin R, Garimella KV, Maguire JR, Hartl C, et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet. 2011;43(5):491–8.
Cibulskis K, Lawrence MS, Carter SL, Sivachenko A, Jaffe D, Sougnez C, et al. Sensitive detection of somatic point mutations in impure and heterogeneous cancer samples. Nat Biotechnol. 2013;31(3):213–9.
Wang K, Li M, Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010;38(16):e164.
Magi A, Tattini L, Cifola I, D’Aurizio R, Benelli M, Mangano E, et al. EXCAVATOR: detecting copy number variants from whole-exome sequencing data. Genome Biol. 2013;14(10):R120.
Mermel CH, Schumacher SE, Hill B, Meyerson ML, Beroukhim R, Getz G. GISTIC2.0 facilitates sensitive and confident localization of the targets of focal somatic copy-number alteration in human cancers. Genome Biol. 2011;12(4):R41.
Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29(1):15–21.
Li B, Dewey CN. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics. 2011;12:323.
Anders S, Huber W. Differential expression analysis for sequence count data. Genome Biol. 2010;11(10):R106.
Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26(1):139–40.
Wu G, Dawson E, Duong A, Haw R, Stein L. ReactomeFIViz: a Cytoscape app for pathway and network-based data analysis. F1000Res. 2014;3:146.
van de Wetering M, Francies HE, Francis JM, Bounova G, Iorio F, Pronk A, et al. Prospective derivation of a living organoid biobank of colorectal cancer patients. Cell. 2015;161(4):933–45.
Liu Y, Sun J, Zhao M. ONGene: a literature-based database for human oncogenes. J Genet Genomics. 2017;44(2):119–21.
Zhao M, Kim P, Mitra R, Zhao J, Zhao Z. TSGene 2.0: an updated literature-based knowledgebase for tumor suppressor genes. Nucleic Acids Res. 2016;44(D1):D1023-31.
Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13(11):2498–504.
Vu T, Datta PK. Regulation of EMT in colorectal cancer: a culprit in metastasis. Cancers (Basel). 2017. https://doi.org/10.3390/cancers9120171.
Mousa L, Salem ME, Mikhail S. Biomarkers of angiogenesis in colorectal cancer. Biomark Cancer. 2015;7(Suppl 1):13–9.
Qin Y, Conley AP, Grimm EA, Roszik J. A tool for discovering drug sensitivity and gene expression associations in cancer cells. PLoS ONE. 2017;12(4):e0176763.
Wilhelm SM, Dumas J, Adnane L, Lynch M, Carter CA, Schutz G, et al. Regorafenib (BAY 73–4506): a new oral multikinase inhibitor of angiogenic, stromal and oncogenic receptor tyrosine kinases with potent preclinical antitumor activity. Int J Cancer. 2011;129(1):245–55.
Miura K, Satoh M, Kinouchi M, Yamamoto K, Hasegawa Y, Philchenkov A, et al. The preclinical development of regorafenib for the treatment of colorectal cancer. Expert Opin Drug Discov. 2014;9(9):1087–101.
Schroeder B, Li Z, Cranmer LD, Jones RL, Pollack SM. Targeting gastrointestinal stromal tumors: the role of regorafenib. Onco Targets Ther. 2016;9:3009–16.
Ballester V, Rashtak S, Boardman L. Clinical and molecular features of young-onset colorectal cancer. World J Gastroenterol. 2016;22(5):1736–44.
Venugopal A, Stoffel EM. Colorectal cancer in young adults. Curr Treat Options Gastroenterol. 2019;17(1):89–98.
Zhu B, Wang Y, Wang X, Wu S, Zhou L, Gong X, et al. Evaluation of the correlation of MACC1, CD44, Twist1, and KiSS-1 in the metastasis and prognosis for colon carcinoma. Diagn Pathol. 2018;13(1):45.
Vatandoust S, Price TJ, Ullah S, Roy AC, Beeke C, Young JP, et al. Metastatic colorectal cancer in young adults: a study from the South Australian Population-based registry. Clin Colorectal Cancer. 2016;15(1):32–6.
Wang MJ, Ping J, Li Y, Adell G, Arbman G, Nodin B, et al. The prognostic factors and multiple biomarkers in young patients with colorectal cancer. Sci Rep. 2015;5:10645.
Heldin CH, Lennartsson J. Structural and functional properties of platelet-derived growth factor and stem cell factor receptors. Cold Spring Harb Perspect Biol. 2013;5(8):a009100.
Jechlinger M, Sommer A, Moriggl R, Seither P, Kraut N, Capodiecci P, et al. Autocrine PDGFR signaling promotes mammary cancer metastasis. J Clin Invest. 2006;116(6):1561–70.
Ball SG, Shuttleworth CA, Kielty CM. Vascular endothelial growth factor can signal through platelet-derived growth factor receptors. J Cell Biol. 2007;177(3):489–500.
Roussos ET, Condeelis JS, Patsialou A. Chemotaxis in cancer. Nat Rev Cancer. 2011;11(8):573–87.
Chen MC, Baskaran R, Lee NH, Hsu HH, Ho TJ, Tu CC, et al. CXCL2/CXCR2 axis induces cancer stem cell characteristics in CPT-11-resistant LoVo colon cancer cells via Galphai-2 and Galphaq/11. J Cell Physiol. 2019;234(7):11822–34.
Yamada K, Hamashima T, Ishii Y, Yamamoto S, Okuno N, Yoshida N, et al. Different PDGF receptor dimers drive distinct migration modes of the mouse skin fibroblast. Cell Physiol Biochem. 2018;51(3):1461–79.
Watts TL, Cui R, Szaniszlo P, Resto VA, Powell DW, Pinchuk IV. PDGF-AA mediates mesenchymal stromal cell chemotaxis to the head and neck squamous cell carcinoma tumor microenvironment. J Transl Med. 2016;14(1):337.
Dai L, Chen X, Lu X, Wang F, Zhan Y, Song G, et al. Phosphoinositide-specific phospholipase Cgamma1 inhibition induces autophagy in human colon cancer and hepatocellular carcinoma cells. Sci Rep. 2017;7(1):13912.
Cho HJ, Baek KE, Nam IK, Park SM, Kim IK, Park SH, et al. PLCgamma is required for RhoGDI2-mediated cisplatin resistance in gastric cancer. Biochem Biophys Res Commun. 2011;414(3):575–80.
Zhang B, Wang F, Dai L, Cai H, Zhan Y, Gang S, et al. Lentivirus-mediated PLCgamma1 gene short-hairpin RNA suppresses tumor growth and metastasis of human gastric adenocarcinoma. Oncotarget. 2016;7(7):8043–54.
Li X, Ba Q, Liu Y, Yue Q, Chen P, Li J, et al. Dihydroartemisinin selectively inhibits PDGFRalpha-positive ovarian cancer growth and metastasis through inducing degradation of PDGFRalpha protein. Cell Discov. 2017;3:17042.
Heldin CH. Targeting the PDGF signaling pathway in tumor treatment. Cell Commun Signal. 2013;11:97.
Ip CKM, Ng PKS, Jeong KJ, Shao SH, Ju Z, Leonard PG, et al. Neomorphic PDGFRA extracellular domain driver mutations are resistant to PDGFRA targeted therapies. Nat Commun. 2018;9(1):4583.
Hofer MD, Fecko A, Shen R, Setlur SR, Pienta KG, Tomlins SA, et al. Expression of the platelet-derived growth factor receptor in prostate cancer and treatment implications with tyrosine kinase inhibitors. Neoplasia. 2004;6(5):503–12.
Yurgelun MB, Masciari S, Joshi VA, Mercado RC, Lindor NM, Gallinger S, et al. Germline TP53 mutations in patients with early-onset colorectal cancer in the colon cancer family registry. JAMA Oncol. 2015;1(2):214–21.
Staub E, Grone J, Mennerich D, Ropcke S, Klamann I, Hinzmann B, et al. A genome-wide map of aberrantly expressed chromosomal islands in colorectal cancer. Mol Cancer. 2006;5:37.
Srivastava S, Verma M, Henson DE. Biomarkers for early detection of colon cancer. Clin Cancer Res. 2001;7(5):1118–26.
Al-Sohaily S, Biankin A, Leong R, Kohonen-Corish M, Warusavitarne J. Molecular pathways in colorectal cancer. J Gastroenterol Hepatol. 2012;27(9):1423–31.
Ettrich TJ, Seufferlein T. Regorafenib. Recent Results Cancer Res. 2018;211:45–56.
Dhillon S. Regorafenib: a review in metastatic colorectal cancer. Drugs. 2018;78(11):1133–44.
Acknowledgements
This work was supported by a National Research Foundation of Korea (NRF) Grant funded by the Korean Government (NRF-2017H1A2A1045221-Fostering Core Leaders of the Future Basic Science Program/Global Ph.D. Fellowship Program).
The biospecimens for this study were provided by Samsung Medical Center BioBank (20140002).
Funding
This research was supported by a grant of the Korea Health Technology R&D Project through the Korea Health Industry Development Institute (KHIDI), funded by the Ministry of Health & Welfare, Republic of Korea (Grant Number: HI15C1593).
This research was supported by the Samsung Medical Center and by the National Research Foundation of Korea (NRF), with a grant funded by the Korean government (MSIT) (NRF-2019R1F1A1061365).
Author information
Authors and Affiliations
Contributions
TWK and YBC conceived the study design. TWK, HKH, CL, SK, and JGJ conducted the experiments. WYL, SYY, HCK, JWH, YAP, and YBC provided patients sample. TWK wrote the initial manuscript drafts. JGJ, WYP and YBC performed critical editing. All authors have read and approved the final manuscript.
Corresponding authors
Ethics declarations
Ethics approval and consent to participate
Ethics approval for human use was obtained from the Samsung Medical Center on Institutional Review Board (Approval Number: SMC 2013-11-007-001).
Consent for publication
Not applicable.
Competing interest
The authors declare no potential conflicts of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Additional file 1: Fig. S1.
Significantly changed pathways from DEGs. a Biological pathways from upregulated genes in young CRC patients. b Biological pathways from downregulated genes in young CRC patients.
Additional file 2: Fig. S2.
PDGFRA expression between young and old CRC patients in TCGA dataset. a Scatter plot of PDGFRA mRNA expression and age. b Boxplot between PDGFRA expression levels in the young CRC group and in the old CRC group.
Additional file 3: Fig. S3.
Co-expression of EMT and angiogenesis factor in TCGA. a Scatter plot of gene expression levels between PDGFRA and EMT-associated signatures. b Scatter plot of gene expression levels between PDGFRA and angiogenesis-associated signatures. c Scatter plot of mRNA expression of EMT markers and age. d Scatter plot of mRNA expression of angiogenesis markers and age. e Heatmap of co-expression patterns between EMT and angiogenesis markers.
Additional file 4: Fig. S4.
PDGFRA expression and PDGFRA-targeted drug response in PDCs. a Boxplot of PDGFRA expression levels (RT-qPCR) for PDC samples (derived from young CRC: #1 and #2; old CRC: #3 and #4). b Prediction of drug sensitivity from gene expression data based on CCLE cell lines. c Viability by knockdown of PDGFRA in PDCs from young (#1 and #2) and old (#3 and #4) patients. d Comparison of sensitivity to regorafenib, an anti-cancer drug targeting PDGFRA between old and young PDCs.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Kim, T.W., Hong, H.K., Lee, C. et al. The role of PDGFRA as a therapeutic target in young colorectal cancer patients. J Transl Med 19, 446 (2021). https://doi.org/10.1186/s12967-021-03088-7
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12967-021-03088-7