Abstract
Background
Exosome-derived microRNAs (exo-miRs) as messengers play important roles, in the cross-talk between genetic [ATP-sensitive potassium channels (KATP) genetic variant rs1799858] and environmental [elevated serum low-density lipoprotein cholesterol (LDL-C) level] factors, but the plasma exo-miRs expression profile and its role in biological processes from genotype to phenotype remain unclear.
Methods
A total of 14 subjects with increased LDL-C serum levels (≥ 1.8 mmol/L) were enrolled in the study. The KATP rs1799858 was genotyped by the Sequenom MassARRAY system. The plasma exo-miRs expression profile was identified by next-generation sequencing.
Results
64 exo-miRs were significantly differentially expressed (DE), among which 44 exo-miRs were up-regulated and 20 exo-miRs were down-regulated in those subjects carrying T-allele (TT + CT) of rs1799858 compared to those carrying CC genotype. The top 20 up-regulated DE-exo-miRs were miR-378 family, miR-320 family, miR-208 family, miR-483-5p, miR-22-3p, miR-490-3p, miR-6515-5p, miR-31-5p, miR-210-3p, miR-17-3p, miR-6807-5p, miR-497-5p, miR-33a-5p, miR-3611 and miR-126-5p. The top 20 down-regulated DE-exo-miRs were let-7 family, miR-221/222 family, miR-619-5p, miR-6780a-5p, miR-641, miR-200a-5p, miR-581, miR-605-3p, miR-548ar-3p, miR-135a-3p, miR-451b, miR-509-3-5p, miR-4664-3p and miR-224-5p. Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) analyses were subsequently implemented to identify the top 10 DE-exo-miRs related specific target genes and signaling pathways. Only 5 DE-exo-miRs were validated by qRT-PCR as follows: miR-31-5p, miR-378d, miR-619-5p, miR-320a-3p and let-7a-5p (all P < 0.05).
Conclusion
These results firstly indicated the plasma exo-miRs expression profile bridging the link between genotype (KATP rs1799858) and phenotype (higher LDL-C serum level), these 5 DE-exo-miRs may be potential target intermediates for molecular intervention points.
Similar content being viewed by others
Background
The low-density lipoprotein cholesterol (LDL-C), as a key atherogenic cholesterol, is an independent risk factor for atherosclerotic cardiovascular diseases (ASCVD), which become a serious public health problem [1]. The increased LDL-C plasma concentration (≥ 1.8 mmol/L) and its related ASCVD is the result of a combination of genetic and environmental factor influences. These environmental factors include lifestyle risk factors (e.g., unhealthy diet, smoking, physical inactivity and obesity, etc.), chemical and physical hazards (e.g., hyperlipidemia, hypertension and hyperglycemia, etc.), and their interactions. Comprehensive management and control of those multiple modifiable environmental risk factors is significant related to lower LDL-C level and lower cardiovascular events risk [2], but those benefits may be dampened by high genetic risk [3]. The genetic susceptibility factor, as an inherent and lifetime risk factor, is powerful independent predictor of high LDL-C level and its related ASCVD. Indeed, the ATP-sensitive potassium channels (KATP) variant rs1799858 was a genetic risk factor for higher LDL-C plasma concentration (≥ 1.8 mmol/L) and its related macro-/micro-vascular arteriosclerotic event risk [4]. However, the mechanism of elevated LDL-C plasma level and its induced ASCVD under specific genetic background of KATP variant rs1799858 remains unclear.
Co-evolution of the genetic and environment factors leads to the development of higher LDL-C serum levels and its related ASCVD. Non-coding RNA, especially the plasma exosome-derived microRNAs (exo-miRs), as the bridge between environmental factors and genetic factors, plays a critical role in this cross-talk process. Exosomes are lipid bilayer extracellular vesicles with a diameter of 30–150 nm secreted by almost all nucleated cells, which mediate cell–cell communication through their components, including microRNAs (miRs), mRNA, DNA, proteins and lipids. The miRs are small and endogenous RNAs (containing about 22–25 nucleotides), which take part in regulating multiple target genes at the post-transcriptional level. Exo-miRs are involved in kinds of physiological or pathological processes in occurrence and development of elevated LDL-C level and its related ASCVD. However, the plasma exo-miRs expression profiles under cross-talk effect between genetic and environment factors remain largely unknown. In this study, using a next-generation sequencing method, we sought to characterize the circulating exo-miRs expression profile in subjects with increased LDL-C level (≥ 1.8 mmol/L) under specific genetic background of KATP polymorphism rs1799858. We then performed Gene Ontology (GO) annotation and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis based on predicted target genes. The top 10 DE-exo-miRs were then confirmed by individual quantitative real-time polymerase chain reaction (qRT-PCR) in 50 increased LDL-C levels (≥ 1.8 mmol/L) subjects with T-allele of rs1799858 and 50 same subjects with counterpart CC genotype. This helped us to facilitate our understanding of the molecular processes from genotype (KATP rs1799858) to phenotype (higher LDL-C serum level).
Methods
Study subjects
A total of 14 subjects with only increased LDL-C serum concentration (≥ 1.8 mmol/L) were recruited into the study from South China. Subjects with other types of dyslipidemia were excluded from the study, including increased levels of triglyceride (TRIG ≥ 1.7 mmol/L), total cholesterol (TC ≥ 5.2 mmol/L) or (and) apolipoprotein B (Apo B ≥ 80 mg/dL), and (or) decreased levels of high-density lipoprotein cholesterol (HDL-C < 1.0 mmol/L) and apolipoprotein AI (Apo AI < 120 mg/dL). All participants with different types of dyslipidemia were newly diagnosed according to guidelines [5]. All subjects combined with smoking, drinking, hypertension (HTN), coronary atherosclerotic heart disease (CAD), type 2 diabetes mellitus (T2D), stroke, abnormal liver function [alanine aminotransferase (ALT) or (and) aspartate aminotransferase (AST) more than 3 times upper limit of normal], abnormal kidney function [estimated glomerular filtration rate (eGFR) less than 90 ml/min·1.73 m2], or (and) any other medical conditions or drugs that may influence blood lipid levels were also excluded from the study. All blood biochemistry analysis was conducted on enrollment to the study by using standard analytical techniques.
Genotyping
The extraction of genomic DNA from the whole-blood sample was performed with QIAamp DNA Blood Midi Kit (Qiagen, Dusseldorf, Germany) and stored at -20℃ according to the manufacturer’s protocol. The KATP single nucleotide polymorphism (SNP) rs1799858 were genotyped using MassARRAY platform (Sequenom Co., San Diego, USA) according to previously described methods [4]. The locus-specific primers were designed by Primer 5.0 (Whitehead Institute Cambridge, Massachusetts, USA) according to the gene sequence in GenBank (NC_000011.10:g.17428382C>T) as follow: (1) forward primer (5′–3′): ACGTTGGATGTGAGGCCCCGACAATCCTCC; (2) reverse primer (5′–3′): ACGTTGGATGAGTGGGTCCTCACCTCCAAA; (3) extension primer (5′–3′): GCCACTCAGGGTTGTGAACCGCAA. The accuracy of the genotypes of rs1799858 was determined 100%.
Exosome isolation, exo-miRs sequencing and sequencing data analysis
The test process was carried out according to the following procedure: (1) Sample collection: The whole-blood samples were collected into anticoagulation vacuum tube with EDTA (3 mg/mL) on enrollment, but after a 12-h fasting and a light, low-fat meal the night. Hemolyzed samples were excluded from the experimental workflow. The freshly whole-blood samples were centrifuged within an hour from collection (3000g × 15 min, 4 ℃) to separate plasma. The upper plasma was transferred to a new Eppendorf tube, and then centrifuged (2000g × 20 min, 4 ℃) to remove additional cellular fragments. The cleared supernatant was cautiously transferred to another new Eppendorf tube and stored at − 80 ℃. (2) Isolation exosomes from plasma: Exosomes from the supernatant were isolated with exoEasy Maxi kit (Qiagen, Dusseldorf, Germany) according to the manufacturer’s protocol with modifications described in Stranska et al. [6]. The eluates were collected to low protein binding tubes and stored at − 80 ℃. (3) Extraction RNA from exosomes: Exo-miRs were extracted by HiPure Liquid miRNA Kit/HiPure Serum/Plasma miRNA Kit (Megan, China) according to the manufacturer's instructions. The quantity and integrity of exo-miRs yield was assessed by using the Qubit® 2.0 (Life Technologies, Carlsbad, USA) and Agilent 2200 TapeStation (Agilent Technologies, Carlsbad, USA) separately. (4) Exo-miRs sequencing: exo-miRs sequencing was performed using Illumina platforms (Illumina, Carlsbad, USA) at Ribobio Co. (Guangzhou, China). Briefly, RNAs were successively ligated with 3′- and 5′-RNA adapter. The adapter-ligated RNAs were then submitted to reverse transcription reaction and amplified with a low-cycle. The PCR products were PAGE-size-selected according to manufacturer's protocol of NEBNext® Multiplex Small RNA Library Prep Set for Illumina (New England BioLabs Inc., Massachusetts, USA). The purified exo-miRs library products were assessed using the Agilent 2200 TapeStation, and then sequenced using an Illumina HiSeq2500 with single-end 50 bp. (5) Sequencing data analysis: The clean reads were acquired after quality control and preprocessing of FASTQ. The miRDeep2 was performed to determine known mature exo-miRs based on miRBase21 (https://www.miRBase.org) and predict novel exo-miRs. The expression of exo-miRs was calculated by reads per million (RPM) values. The differential expression of exo-miRs in subjects with different genotypes of rs1799858 was calculated by edgeR algorithm according to the criteria of |log2 (Fold Change)|≥ 1 and P value < 0.05. The online softwares (miRDB, miRTarBase, miRWalk and TargetScan) were performed to predict exo-miRs related targets gene. KOBAS 2.0 software was used to further analysis of GO and KEGG pathway. (6) Validation of top 10 DE-exo-miRs: qRT-PCR array was performed for top 10 DE-exo-miRs in a new verification cohort.
Statistical analysis
All analysis for baseline characteristics was performed with SPSS version 24 (SPSS, Chicago, USA). Categorical variables were presented as frequencies. Continuous variables were presented as mean ± SD. The differences on continuous variables between the two genotypes (CC vs. TT + CT) of rs1799858 in subjects with increased LDL-C serum concentration (≥ 1.8 mmol/L) were assessed by independent-sample t-test while categorical variables by Chi-square test. The DE-exo-miRs between the two different genotypes was also analyzed with edgeR software. Both GO category and KEGG pathway analyses were evaluated by Chi-square test or Fisher’s exact test. The false discovery rate (FDR) was calculated to adjust the P values. If an adjusted P value is less than 0.05, the result is considered as significant.
Results
Clinical baseline characteristics of study subjects
The clinical features among subjects with different genotypes of KATP rs1799858 in this study are shown in Table 1.
DE-exo-miRs between different genotypes of KATP rs1799858 in subjects with elevated LDL-C (≥ 1.8 mmol/L) serum level
The exo-miRs were analyzed with strict data quality control, and a total of 646 exo-miRs were found. In this study a total of 64 exo-miRs were significantly DE between the two genotypes of rs1799858 with filtering out low-expressing exo-miRs (RPM values < 10), as shown in Additional file 1: Figure S1 and Fig. 1. Among the DE-exo-miRs, 44 exo-miRs were up-regulated and 20 exo-miRs were down-regulated in subjects carrying T-allele (TT + CT) of rs1799858 compared to those with CC genotype. The top 20 up-regulated exo-miRs were miR-483-5p, miR-22-3p, miR-490-3p, miR-378g, miR-320e, miR-6515-5p, miR-31-5p, miR-320b, miR-210-3p, miR-17-3p, miR-320d, miR-6807-5p, miR-378b, miR-378a-3p, miR-497-5p, miR-499a-5p, miR-208b-3p, miR-33a-5p, miR-3611 and hsa-miR-126-5p. The top 20 down-regulated exo-miRs were miR-6780a-5p, miR-619-5p, let-7e-5p, let-7i-5p, let-7g-5p, let-7a-5p, let-7f-5p, miR-641, miR-200a-5p, miR-581, miR-222-3p, miR-605-3p, miR-548ar-3p, miR-221-5p, miR-135a-3p, miR-451b, miR-6721-5p, miR-98-5p, miR-4664-3p and miR-224-5p.
GO analysis of Enriched biological processes, cellular component and molecular functions regulated by CTGs of top 10 DE-exo-miRs.
GO analysis was used to identify the biological processes, cellular component and molecular functions for top 10 DE-exo-miRs related CTGs. As shown in Fig. 2, the top 10 DE-exo-miRs related CTGs in subjects carrying T-allele (TT + CT) of rs1799858 were obviously linked to regulation of signaling, apoptotic process, vesicle-mediated transport, homeostatic process, protein complex subunit organization, lipid metabolic process, autophagy, angiogenesis, oxidation–reduction process, response to hypoxia, inflammatory response, and microtubule-based process, relating to cellular components such as vesicle, membrane-bounded organelle (e.g., endoplasmic reticulum, mitochondrion, lysosome), membrane protein complex, microtubule organizing center, transcription factor complex, and transmembrane transporter complex. The molecular functions of the top 10 DE-exo-miRs related target genes were correlated with protein binding, ion binding, DNA binding, enzyme binding, sequence-specific DNA binding, ATP binding, transcription factor activity, kinase activity, lipid binding, oxidoreductase activity and gated channel activity.
KEGG analysis of enrichment pathway regulated by CTGs of top 10 DE-exo-miRs
KEGG analysis was also used to identify the for top 10 DE-exo-miRs related CTGs were evidently enriched in 75 pathways. As shown in Fig. 3 and Additional file 1: Figure S2, the top 30 pathways were involved in environmental information processing (e.g., signaling pathways of PI3K-Akt, MAPK and Ras, etc.), genetic information processing (e.g., protein processing in endoplasmic reticulum), human diseases (e.g., insulin resistance and non-alcoholic fatty liver disease), metabolism (e.g., metabolic pathways), and organismal systems (e.g., signaling pathways of insulin, chemokine, platelet activation and T cell receptor).
KEGG analysis of enrichment pathway regulated by CTGs of top 10 DE-exo-miRs*. *The top signaling pathways were related to metabolism-related diseases or inflammation including those related to human diseases such as insulin resistance, non-alcoholic fatty liver disease, metabolic/insulin/chemokine/T cell receptor signaling pathways and platelet activation; those related to environmental information processing functions including PI3K-Akt/MAPK/Ras/FoxO/Rap1/Hippo/Wnt signaling pathways; and those related to genetic information/cellular processes such as endocytosis and protein processing in endoplasmic reticulum. Our analysis indicated that these pathways may be involved in the molecular processes from genotype (KATP rs1799858) to phenotype (higher LDL-C serum level)
Target interactome of top 10 DE-exo-miRs
There were 1045 CTGs of top 10 DE exo-miRs, and the interactome of these CTGs was determined via STRING online database. As shown in Fig. 4, there were the 10 exo-miRs and 74 CTGs on interaction of exo-miRs/gene and gene/gene by using combined score greater than 0.9 as threshold cutoff. The 10 DE-exo-miRs interacted with target genes, including hypoxia-inducible factor-1α (HIF-1α), nitric oxide synthase (NOS), peroxisome proliferator-activated receptor-α (PPARα), scavenger receptor class B type I (SR-BI), 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase 2 (PFKFB2), lecithin cholesterol acyltransferase (LCAT), peroxisome proliferator-activated receptor-gamma coactivator-1α (PGC-1α), and ATP binding cassette subfamily A member 1 (ABCA1) so on, which resulted in a complex regulatory network affected by obviously and differently regulated exo-miRs in higher serum LDL-C level (≥ 1.8 mmol/L) subjects with T-allele (TT + CT) of rs1799858.
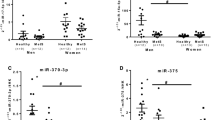
qRT-PCR analysis of top 10 DE-exo-miRs
A total of 50 increased LDL-C levels (≥ 1.8 mmol/L) subjects with T-allele of rs1799858 and 50 same subjects with CC genotype were enrolled to validate the expression of top 10 DE-exo-miRs. Only 5 DE-exo-miRs were successfully validated by qRT-PCR as follows: miR-31-5p (P < 0.001), miR-378d (P = 0.003), miR-619-5p (P = 0.008), miR-320a-3p (P < 0.001) and Let-7a-5p (P < 0.001), as shown in Fig. 5.
Discussion
It's well known that genetic or germline variants (e.g., genotype) have a huge impact on the phenotypic landscape of a population. Complex disease (e.g., cardiovascular disease, cancer, etc.) is defined as a phenotype that is caused by many individual gene events, with a significant contribution from environmental factors. Germline variants influence clinic outcomes of complex disease. Recent studies found that functionally mutations of natural killer cells were positively associated with cancer risk [7, 8]. Similarly, in our previous study found that the KATP SNP rs1799858 was associated with increased risk of elevated LDL-C serum concentration (≥ 1.8 mmol/L) and its related macro-/micro-vascular arteriosclerotic events. However, the underlying mechanism of genetic effects on phenotype mediated by genotype-environment interactions remains elusive. exo-miRs, as the leading factor in inducing genetic susceptibility changes, are messengers in the cross-talk between environmental factors and genetic factors. The exo-miRs expression profile and its role of genetic variants on cellular signaling pathways will facilitate our understanding of the relationships between genotype (e.g., KATP SNP rs1799858) and phenotype (e.g., LDL-C serum concentration ≥ 1.8 mmol/L and its related ASCVD). This is the only study to reveal a distinct exo-miRs expression in subjects with LDL-C serum concentration ≥ 1.8 mmol/L under specific genetic background of KATP polymorphism (rs1799858). (1) We identified the DE-exo-miRs, and respectively screened up-/down-regulating of the top 20 DE-exo-miRs, whose CTGs were identified. (2) GO and KEGG pathway analyses were implemented on those exo-miRs related CTGs. (3) Target interactome network from up-/down-regulating of top 10 DE-exo-miRs was drawn.
Exosomes are secreted by the nucleated cells in response to surrounding environment changes (e.g., increased LDL-C level). Recent research indicated that subjects carrying T allele (TT + CT) of rs1799858 were associated with elevated risk of higher LDL-C (≥ 1.8 mmol/L) level. It was hypothesized that the exo-miRs expression profile varies by genotypes (CC vs. TT + CT) of rs1799858. By next-generation sequencing, there were 64 significantly DE-exo-miRs in increased LDL-C level subjects carrying T allele (TT + CT) of 1,799,858 compared to those with CC genotype. Among 40 DE-exo-miRs (top 20 up-/down-regulated exo-miRs, respectively; Table 2), mi-31-5p was the highest up-regulated exo-miR (approximately 4.5-fold changes), while miR-6780a-5p was the highest down-regulated exo-miR (approximately 2.1-fold changes). There were also 5 exo-miRs families, including miR-378 (e.g., 378a-3p/b/g), miR-320 (e.g., 320b/d/e), miR-208 (e.g., 208b-3p and 499a-5p), Let-7 (e.g., 7a-5p, 7e-5p, 7f-5p, 7g-5p, 7i-5p and miR-98-5p), and miR221/222 (e.g., miR-221-3p and miR-222-5p), which had highly homologous sequence. miR-378 family, originate from the first intron of the PPARγ coactivator 1 beta gene encoding PGC-1β, is a new emerging miR in oxidation/lipid metabolism [9]. The miR-320 family has intrinsic and conserved function for modulation of glucose metabolism under different pathological conditions [10]. As a member of miR-320 family, miR-320b showed the highest expression level among those exo-miRs. The miR-208 family is almost specifically expressed in heart chamber [11] and closely associated with the development of cardiac diseases (e.g., cardiac fibrosis, myocardial hypertrophy, myocardial infarction, and heart failure, etc.). The expression level of miR-208 family increased approximately by threefold. Different from the three up-regulated exo-miRs families, there were also the other two down-regulated exo-miRs families. The Let-7 family, as one of the first-described miR families involving in endothelial cells (ECs) dysfunction and vascular smooth muscle cells (VSMCs) proliferation in the pathogenesis of atherosclerosis, was the one of two down-regulated Exo-miRs family in this study. In contrast, increasing expression level of let-7 family was a protective effect in regulating inflammation in diabetes-related atherosclerosis [12]. miR-221/222 family also involved in the regulation of atherosclerosis [13], because the increased LDL-C level (especially ox-LDL-C) remarkably inhibited miR-221-3p expression in a concentration-dependent and time-dependent manner [14]. Besides the LDL-C, the miR-221/222 family was also associated with low HDL-C phenotype [15]. Similar to the association of miR-221/222 family with dyslipidemia (LDL-C/HDL-C), this phenomenon was also seen on miR-126-5p [16, 17]. In addition, miR-490-3p [18], miR-320b [19], miR-210-3p [20], miR-17-3p [21] and miR-33a-5p [22] were linked to LDL-C metabolism while miR-22-3p [23], miR-31-5p [24], miR-378b [25], and miR-135a-3p [26] were linked to HDL-C metabolism. These findings suggested that the all the 40 DE-exo-miRs may be association with lipid metabolism, especially on LDL-C.
Participants with T-allele (TT + CT) of rs1799858 were not only associated with increased risk of higher LDL-C level but also with increased risk of atherosclerosis events, including carotid artery stenosis (CAS) ≥ 50% and new-onset/recurrent acute myocardial infarction (AMI). Synchronously, the data reported in this study indicated that those DE-exo-miRs between the two genotypes (CC vs. TT + CT) of rs1799858 were not only involved in lipid metabolism/dyslipidemia as mentioned above but also played a pivotal role in the occurrence and progression of arteriosclerosis [27, 28], such as miR-22-3p, miR-490-3p, miR-210-3p, miR-497-5p, miR-33a-5p, miR-126-5p, miR-451b, miR-320 family (e.g., miR-320b), miR-208 family (e.g., miR-208b-3p), let-7 family (e.g., let-7i-5p, let-7g-5p, let-7a-5p and let-7f-5p) and miR-221/222 family (e.g., miR-222-3p and miR-221-5p), involving in proliferation and migration of VSMCs (e.g., miR-22-3p [29] and miR-490-3p [18]), ECs dysfunction (e.g., miR-22-3p [30], miR-126 [31], miR-221/222 family [13]), plaque angiogenesis (e.g., miR-126 [32]), apoptosis (e.g., miR-210-3p [33], miR-320d [34]) and macrophage lipid deposition (e.g., miR-210-3p [20]). In particular, miR-483-5p [35], miR-31-5p [36], miR-320b [19] and miR-126 [37] were also closely related to the stenosis degree and unstable phenotype of atherosclerotic plaques, suggesting those exo-miRs may be related to the potential risk of acute vascular events. Indeed, in a four-year prospective study on screening potentially important diagnostic and prognostic biomarkers in acute coronary syndrome resulting from CAS, Gacon et al. [38] found that increased miR-208b-3p level were independently associated with AMI risk, which consistent with the HUNT study by Bye et al. [39] who found that let-7g-5p was associated with fatal future AMI in healthy individuals. The miR-483-5p may be linked to in the early phases of AMI [40]. Under specific genetic background of KATP SNP rs1799858, since the accumulation of cardiovascular risk factors (e.g., aging [30], smoking [41],unhealthy diet [26], physical inactivity [42], obesity [43], PM2.5 [44], etc.) to the occurrence of ASCVD events and even death [45], these exo-miRs run through the whole cardiovascular event chain, especially such as miR-483-5p, miR-22-3p, miR-31-5p, miR-126, miR-378 family, miR-320 family, let-7 family and miR-221/222 family. However, it is worth mentioning that there were 5 novel exo-miRs (e.g., miR-6515-5p, miR-6807-5p, miR-3611, miR-641 and miR-605-3p), which has no known association with cardiovascular disease, warrant further investigation.
To further investigate the function of exo-miRs under cross-talk status between higher LDL-C level and different genotypes of rs1799858, GO and KEGG analyses were performed for the 1045 CTGs of top 10 DE-exo-miRs in increased LDL-C ≥ 1.8 mmol/L subjects with T-allele (TT + CT) of rs1799858. GO analyses suggested that enrichment of CTGs played crucial roles in biological processes, cellular component and molecular functions (Fig. 2), consistent with a regulatory role on dyslipidemia and related ASCVD [46] for these exo-miRs in the transcription/translation processes [47]. Many differentially regulated KEGG pathways were identified. The results showed that PI3K-Akt signaling pathway, metabolic pathways, and MAPK signaling pathway were the top 3 differentially regulated pathways (Fig. 3). Importantly, PI3K-Akt pathway, which plays an essential role in cellular physiology by regulating growth factor signals during organismal growth and critical cellular processes (e.g., lipid metabolism, glucose homeostasis, protein synthesis, cell proliferation and survival) in normal physiology and morbid conditions (e.g., obesity and T2D) [48]. MAPK pathway, which is known as an important signal transmitter that transmit signals from receptors on the surface to DNA in the nucleus of the cell, is essential in regulating of lipid homeostasis as well as many other cellular processes (e.g., inflammation, cell differentiation, cell division, cell proliferation, motility, apoptosis and stress response) [49]. Both signaling pathways are not only involved in the regulation of lipid homeostasis but also related to atherosclerosis and ASCVD, manifesting the characteristics of synchronous activation [50]. These top 10 exo-miRs were interacted with target genes, forming a network that was influenced by significantly differently regulated exo-miRs in higher LDL-C (≥ 1.8 mmol/L)level patients with T-allele (TT + CT) of rs1799858 (Fig. 4), but only 5 exo-miRs were further comfirmed based on a new verification cohort (Fig. 5). These findings indicated that these DE-exo-miRs could play an important role in increased LDL-C plasma concentration (≥ 1.8 mmol/L) and its related ASCVD by regulating these two pathways.
Study strengths and limitations
The strength of the study was that this is the first time to characterize the circulating exo-miRs expression profile in biological processes from genotype (KATP variant rs1799858) to phenotype (increased LDL-C serum levels), indicating that the potential role of exo-miRs related epigenetic modification on co-evolution of the genetic and environment factors leads to the development of higher LDL-C serum levels and its related ASCVD. There are some limitations in this study. Firstly, the sample size is small so that large-scale, prospective population-based cohort studies will be conducted to confirm the reported results, such as the relationships between these DE-exo-miRs plasma levels and LDL-C exception level related ASCVD events under specific genetic background of KATP variant rs1799858. Secondly, this was only a preliminary bioinformatics analysis so that the possible miss-distance effect and non-specific effect could not be excluded due to lack of validation at the cellular and molecular levels. Finally, due to the validation of DE-exo-miRs by qRT-PCR at the clinical level is incomplete, it is necessary to carry out further functional verification on those 5 validated DE-exo-miRs and its related pathways at the cellular and molecular levels as follows: to study the function of these exo-miRs at the translational or transcriptional levels based on luciferase system or fluorescent microscopy, to identify the specific nucleotide sequences and exo-miRs binding, and to observe its impact on overall functions of signal pathways after mutation. Therefore, results must be interpreted carefully.
Conclusion
This study firstly indicated that the plasma exo-miRs expression profile bridging the link from genotype (KATP rs1799858) to phenotype (higher LDL-C serum level), and these DE-exo-miRs (especially top 10 DE-exo-miRs) may be potential target intermediates for development of novel diagnosis, prevention, and treatment of LDL-C exception level and its related atherosclerotic vascular disease, warrant further research.
Availability of data and materials
The datasets used and/or analysed during the current study are available from the corresponding author on reasonable request.
Abbreviations
- ABCA1:
-
ATP binding cassette subfamily A member 1
- ABL2:
-
ABL proto-oncogene 2, non-receptor tyrosine kinase
- ACE:
-
Angiotensin converting enzyme
- ACR:
-
Urinary albumin-to-creatinine ratio
- ACTR1A:
-
Actin related protein 1A
- ADAM10:
-
ADAM metallopeptidase domain 10
- AFF4:
-
AF4/FMR2 family member 4
- AGO4:
-
Argonaute RISC component 4
- Alb:
-
Albumin
- ALD:
-
Aldosterone
- ALT:
-
Alanine aminotransferase
- AMER1:
-
APC membrane recruitment protein 1
- AMI:
-
Acute myocardial infarction
- AMER1:
-
APC membrane recruitment protein 1
- Ang I/ II:
-
Angiotensin I/II
- ANKIB1:
-
Ankyrin repeat and IBR domain containing 1
- ANP32E:
-
Acidic nuclear phosphoprotein 32 family member E
- AP1S1:
-
Adaptor related protein complex 1 subunit sigma 1
- Apo AI:
-
Apolipoprotein AI
- Apo B:
-
Apolipoprotein B
- ARGFX:
-
Arginine-fifty homeobox
- ARHGAP12:
-
Rho GTPase activating protein 12
- ARHGEF39:
-
Rho guanine nucleotide exchange factor 39
- ARID1A:
-
AT-rich interaction domain 1A
- ASCVD:
-
Arteriosclerosis cardiovascular disease
- ASNSD1:
-
Asparagine synthetase domain containing 1
- AST:
-
Aspartate aminotransferase
- ATCAY:
-
ATCAY kinesin light chain interacting caytaxin
- ATG9A:
-
Autophagy related 9A
- AURKB:
-
Aurora kinase B
- BACE1:
-
Beta-secretase 1
- BAHD1:
-
Bromo adjacent homology domain containing 1
- BCL7A:
-
BAF chromatin remodeling complex subunit 7A
- BHMT2:
-
Betaine-homocysteine S-methyltransferase 2
- BMI:
-
Body mass index
- BUN:
-
Blood urea nitrogen
- C6orf62:
-
Chromosome 6 open reading frame 62
- C11orf54:
-
Chromosome 11 open reading frame 54
- C15orf40:
-
Chromosome 15 open reading frame 40
- C19orf12:
-
Chromosome 19 open reading frame 12
- CADM1:
-
Cell adhesion molecule 1
- CARD10:
-
Caspase recruitment domain family member 10
- CAS:
-
Carotid artery stenosis
- CASD1:
-
CAS1 domain containing 1
- CAD:
-
Coronary atherosclerotic heart disease
- CCDC125:
-
Coiled-coil domain containing 125
- CCL5:
-
C–C motif chemokine ligand 5
- CCNT1:
-
Cyclin T1
- CDK16:
-
Cyclin dependent kinase 16
- CEP120:
-
Centrosomal protein 120
- CHST6:
-
Carbohydrate sulfotransferase 6
- CPT-1(CPT1A):
-
Carnitine palmitoyltransferase 1
- Cr:
-
Creatinine
- CREB1:
-
CAMP responsive element binding protein 1
- CREG1:
-
Cellular repressor of E1A stimulated genes 1
- CRY2:
-
Cryptochrome circadian regulator 2
- CTC1:
-
CST telomere replication complex component 1
- CTGs:
-
Candidate target genes
- CXCL12:
-
C-X-C motif chemokine ligand 12
- CXorf21:
-
Chromosome X open reading frame 21
- DBP:
-
Diastolic blood pressure
- DE:
-
Differentially expressed
- DLL:
-
Delta-like proteins
- DMD:
-
Dystrophin
- DNAJB9:
-
DnaJ heat shock protein family (Hsp40) member B9
- ECHDC1:
-
Ethylmalonyl-CoA decarboxylase 1
- ECs:
-
Endothelial cells
- eGFR:
-
Estimated glomerular filtration rate
- EIF4A2:
-
Eukaryotic translation initiation factor 4A2
- EPM2AIP1:
-
EPM2A interacting protein 1
- ETS1:
-
ETS proto-oncogene 1, transcription factor
- exo-miRs:
-
Exosome-derived microRNAs
- FAM126B:
-
Family with sequence similarity 126 member B
- FBG:
-
Fasting blood glucose
- FDR:
-
False discovery rate
- FLT-1 (VEGFR1):
-
Fms related receptor tyrosine kinase 1(vascular endothelial growth factor receptor-1)
- GGA3:
-
Golgi associated, gamma adaptin ear containing, ARF binding protein 3
- GLUT(SLC2A1):
-
Glucose transporter 1(solute carrier family 2 member 1)
- GNG5:
-
G protein subunit gamma 5
- GO:
-
Gene Ontology
- HbA1C:
-
Glycosylated hemoglobin
- HDL-C:
-
High-density lipoprotein cholesterol
- HIF-1α:
-
Hypoxia-inducible factor-1α
- HGB:
-
Hemoglobin concentration
- HsCRP:
-
High-sensitivity C-reactive protein
- HTN:
-
Hypertension
- INIP:
-
INTS3 and NABP interacting protein
- K+ :
-
Serum potassium
- KATP:
-
ATP-sensitive potassium channels
- KDR (VEGFR2):
-
Kinase insert domain receptor (vascular endothelial growth factor receptor-2)
- KEGG:
-
Kyoto Encyclopedia of Genes and Genomes
- LCAT:
-
Lecithin cholesterol acyltransferase
- LDL-C:
-
Low-density lipoprotein cholesterol
- LRG-1:
-
Leucine rich alpha-2-glycoprotein 1
- MAU:
-
Microalbumin in urine
- METTL7A:
-
Methyltransferase like 7A
- miRs:
-
MicroRNAs
- Na+ :
-
Serum sodium
- NOS:
-
Nitric oxide synthase
- Notch:
-
Neurogenic locus Notch protein
- NOX:
-
NADPH oxidase
- NRIP1:
-
Nuclear receptor interacting protein 1
- P2hBS:
-
Postprandial blood glucose two hours
- PFKFB2(PFK2):
-
6-Phosphofructo-2-kinase/fructose-2,6-bisphosphatase 2
- PGC-1α:
-
Peroxisome proliferator-activated receptor-gamma coactivator-1α
- PLT:
-
Platelet count
- PPARα:
-
Peroxisome proliferator-activated receptor-α
- qRT-PCR:
-
Quantitative real-time polymerase chain reaction
- RPM:
-
Reads per million
- SBP:
-
Systolic blood pressure
- SNP:
-
Single nucleotide polymorphism
- SNRPB2:
-
Small nuclear ribonucleoprotein polypeptide B2
- SR-BI:
-
Scavenger receptor class B type I
- TC:
-
Total cholesterol
- T2D:
-
Type 2 diabetes mellitus
- TRIG:
-
Triglyceride
- UA:
-
Serum uric acid
- VEGF:
-
Vascular endothelial growth factor
- VKORC1L1:
-
Vitamin K epoxide reductase complex subunit 1 like 1
- VSMCs:
-
Vascular smooth muscle cells
- WBC:
-
White blood cell count
References
Gidding SS, Allen NB. Cholesterol and atherosclerotic cardiovascular disease: a lifelong problem. J Am Heart Assoc. 2019;8:e012924.
Colpani V, Baena CP, Jaspers L, van Dijk GM, Farajzadegan Z, Dhana K, Tielemans MJ, Voortman T, Freak-Poli R, Veloso GGV, et al. Lifestyle factors, cardiovascular disease and all-cause mortality in middle-aged and elderly women: a systematic review and meta-analysis. Eur J Epidemiol. 2018;33:831–45.
Pazoki R, Dehghan A, Evangelou E, Warren H, Gao H, Caulfield M, Elliott P, Tzoulaki I. Genetic predisposition to high blood pressure and lifestyle factors: associations with midlife blood pressure levels and cardiovascular events. Circulation. 2018;137:653–61.
Liu C, Guan T, Lai Y, Zhu J, Kuang J, Shen Y. ATP-sensitive potassium channels gene polymorphism rs1799858 affects the risk of macro-/micro-vascular arteriosclerotic event in patients with increased low-density lipoprotein cholesterol levels. Lipids Health Dis. 2020;19:147.
Mach F, Baigent C, Catapano AL, Koskinas KC, Casula M, Badimon L, Chapman MJ, De Backer GG, Delgado V, Ference BA, et al. 2019 ESC/EAS Guidelines for the management of dyslipidaemias: lipid modification to reduce cardiovascular risk. Eur Heart J. 2020;41:111–88.
Stranska R, Gysbrechts L, Wouters J, Vermeersch P, Bloch K, Dierickx D, Andrei G, Snoeck R. Comparison of membrane affinity-based method with size-exclusion chromatography for isolation of exosome-like vesicles from human plasma. J Transl Med. 2018;16:1.
Milanese JS, Tibiche C, Zou J, Meng Z, Nantel A, Drouin S, Marcotte R, Wang E. Germline variants associated with leukocyte genes predict tumor recurrence in breast cancer patients. NPJ Precis Oncol. 2019;3:28.
Xu X, Li J, Zou J, Feng X, Zhang C, Zheng R, Duanmu W, Saha-Mandal A, Ming Z, Wang E. Association of germline variants in natural killer cells with tumor immune microenvironment subtypes, tumor-infiltrating lymphocytes, immunotherapy response, clinical outcomes, and cancer risk. JAMA Netw Open. 2019;2:e199292.
Machado IF, Teodoro JS, Palmeira CM, Rolo AP. miR-378a: a new emerging microRNA in metabolism. Cell Mol Life Sci. 2019;77:1947–58.
Zhang LL, Ma J, Yang B, Zhao J, Yan BY, Zhang YQ, Li W. Interference with lactate metabolism by mmu-miR-320-3p via negatively regulating GLUT3 signaling in mouse Sertoli cells. Cell Death Dis. 2018;9:964.
Kakimoto Y, Tanaka M, Kamiguchi H, Hayashi H, Ochiai E, Osawa M. MicroRNA deep sequencing reveals chamber-specific miR-208 family expression patterns in the human heart. Int J Cardiol. 2016;211:43–8.
Brennan E, Wang B, McClelland A, Mohan M, Marai M, Beuscart O, Derouiche S, Gray S, Pickering R, Tikellis C, et al. Protective effect of let-7 miRNA family in regulating inflammation in diabetes-associated atherosclerosis. Diabetes. 2017;66:2266–77.
Xue Y, Wei Z, Ding H, Wang Q, Zhou Z, Zheng S, Zhang Y, Hou D, Liu Y, Zen K, et al. MicroRNA-19b/221/222 induces endothelial cell dysfunction via suppression of PGC-1alpha in the progression of atherosclerosis. Atherosclerosis. 2015;241:671–81.
Zhuang X, Li R, Maimaitijiang A, Liu R, Yan F, Hu H, Gao X, Shi H. miR-221-3p inhibits oxidized low-density lipoprotein induced oxidative stress and apoptosis via targeting a disintegrin and metalloprotease-22. J Cell Biochem. 2019;120:6304–14.
Zhou Y, Liu M, Li J, Wu B, Tian W, Shi L, Zhang J, Sun Z. The inverted pattern of circulating miR-221-3p and miR-222-3p associated with isolated low HDL-C phenotype. Lipids Health Dis. 2018;17:188.
Ben-Aicha S, Escate R, Casani L, Padro T, Pena E, Arderiu G, Mendieta G, Badimon L, Vilahur G. High-density lipoprotein remodelled in hypercholesterolaemic blood induce epigenetically driven down-regulation of endothelial HIF-1alpha expression in a preclinical animal model. Cardiovasc Res 2020;116:1288–99.
Chen Z, Pan X, Sheng Z, Yan G, Chen L, Ma G. Baicalin suppresses the proliferation and migration of Ox-LDL-VSMCs in atherosclerosis through upregulating miR-126-5p. Biol Pharm Bull. 2019;42:1517–23.
Sun Y, Chen D, Cao L, Zhang R, Zhou J, Chen H, Li Y, Li M, Cao J, Wang Z. MiR-490-3p modulates the proliferation of vascular smooth muscle cells induced by ox-LDL through targeting PAPP-A. Cardiovasc Res. 2013;100:272–9.
Zhang R, Qin Y, Zhu G, Li Y, Xue J. Low serum miR-320b expression as a novel indicator of carotid atherosclerosis. J Clin Neurosci. 2016;33:252–8.
Qiao XR, Wang L, Liu M, Tian Y, Chen T. MiR-210-3p attenuates lipid accumulation and inflammation in atherosclerosis by repressing IGF2. Biosci Biotechnol Biochem. 2020;84:321–9.
Zambrano T, Hirata RDC, Hirata MH, Cerda A, Salazar LA. Statins differentially modulate microRNAs expression in peripheral cells of hyperlipidemic subjects: a pilot study. Eur J Pharm Sci. 2018;117:55–61.
Mao M, Lei H, Liu Q, Chen Y, Zhao L, Li Q, Luo S, Zuo Z, He Q, Huang W, et al. Effects of miR-33a-5P on ABCA1/G1-mediated cholesterol efflux under inflammatory stress in THP-1 macrophages. PLoS ONE. 2014;9:e109722.
Desgagne V, Guerin R, Guay SP, Boyer M, Hutchins E, Picard S, Marechal A, Corbin F, Keuren-Jensen KV, Arsenault BJ, Bouchard L. Human high-density lipoprotein microtranscriptome is unique and suggests an extended role in lipid metabolism. Epigenomics. 2019;11:917–34.
Desgagne V, Guerin R, Guay SP, Corbin F, Couture P, Lamarche B, Bouchard L. Changes in high-density lipoprotein-carried miRNA contribution to the plasmatic pool after consumption of dietary trans fat in healthy men. Epigenomics. 2017;9:669–88.
Lu YL, Jing W, Feng LS, Zhang L, Xu JF, You TJ, Zhao J. Effects of hypoxic exercise training on microRNA expression and lipid metabolism in obese rat livers. J Zhejiang Univ Sci B. 2014;15:820–9.
Desgagne V, Guay SP, Guerin R, Corbin F, Couture P, Lamarche B, Bouchard L. Variations in HDL-carried miR-223 and miR-135a concentrations after consumption of dietary trans fat are associated with changes in blood lipid and inflammatory markers in healthy men—an exploratory study. Epigenetics. 2016;11:438–48.
Vegter EL, Ovchinnikova ES, van Veldhuisen DJ, Jaarsma T, Berezikov E, van der Meer P, Voors AA. Low circulating microRNA levels in heart failure patients are associated with atherosclerotic disease and cardiovascular-related rehospitalizations. Clin Res Cardiol. 2017;106:598–609.
Huesca-Gomez C, Torres-Paz YE, Martinez-Alvarado R, Fuentevilla-Alvarez G, Del Valle-Mondragon L, Torres-Tamayo M, Soto ME, Gamboa R. Association between the transporters ABCA1/G1 and the expression of miR-33a/144 and the carotid intima media thickness in patients with arterial hypertension. Mol Biol Rep. 2020;47:1321–9.
Huang SC, Wang M, Wu WB, Wang R, Cui J, Li W, Li ZL, Li W, Wang SM. Mir-22-3p inhibits arterial smooth muscle cell proliferation and migration and neointimal hyperplasia by targeting HMGB1 in arteriosclerosis obliterans. Cell Physiol Biochem. 2017;42:2492–506.
Takeda E, Suzuki Y, Sato Y. Age-associated downregulation of vasohibin-1 in vascular endothelial cells. Aging Cell. 2016;15:885–92.
Schober A, Nazari-Jahantigh M, Wei Y, Bidzhekov K, Gremse F, Grommes J, Megens RT, Heyll K, Noels H, Hristov M, et al. MicroRNA-126-5p promotes endothelial proliferation and limits atherosclerosis by suppressing Dlk1. Nat Med. 2014;20:368–76.
Sezer Zhmurov C, Timirci-Kahraman O, Amadou FZ, Fazliogullari O, Basaran C, Catal T, Zeybek U, Bermek H. Expression of Egfl7 and miRNA-126-5p in symptomatic carotid artery disease. Genet Test Mol Biomark. 2016;20:125–9.
Ma H, Chen P, Sang C, Huang D, Geng Q, Wang L. Modulation of apoptosis-related microRNAs following myocardial infarction in fat-1 transgenic mice vs wild-type mice. J Cell Mol Med. 2018;22:5698–707.
Shen H, Lu S, Dong L, Xue Y, Yao C, Tong C, Wang C, Shu X. hsa-miR-320d and hsa-miR-582, miRNA biomarkers of aortic dissection, regulate apoptosis of vascular smooth muscle cells. J Cardiovasc Pharmacol. 2018;71:275–82.
Li S, Lee C, Song J, Lu C, Liu J, Cui Y, Liang H, Cao C, Zhang F, Chen H. Circulating microRNAs as potential biomarkers for coronary plaque rupture. Oncotarget. 2017;8:48145–56.
Infante T, Forte E, Punzo B, Cademartiri F, Cavaliere C, Soricelli A, Salvatore M, Napoli C. Correlation of circulating miR-765, miR-93-5p, and miR-433-3p to obstructive coronary heart disease evaluated by cardiac computed tomography. Am J Cardiol. 2019;124:176–82.
Leistner DM, Boeckel JN, Reis SM, Thome CE, De Rosa R, Keller T, Palapies L, Fichtlscherer S, Dimmeler S, Zeiher AM. Transcoronary gradients of vascular miRNAs and coronary atherosclerotic plaque characteristics. Eur Heart J. 2016;37:1738–49.
Gacon J, Badacz R, Stepien E, Karch I, Enguita FJ, Zmudka K, Przewlocki T, Kablak-Ziembicka A. Diagnostic and prognostic micro-RNAs in ischaemic stroke due to carotid artery stenosis and in acute coronary syndrome: a four-year prospective study. Kardiol Pol. 2018;76:362–9.
Bye A, Rosjo H, Nauman J, Silva GJ, Follestad T, Omland T, Wisloff U. Circulating microRNAs predict future fatal myocardial infarction in healthy individuals—The HUNT study. J Mol Cell Cardiol. 2016;97:162–8.
Li L, Li S, Wu M, Chi C, Hu D, Cui Y, Song J, Lee C, Chen H. Early diagnostic value of circulating microRNAs in patients with suspected acute myocardial infarction. J Cell Physiol. 2019;234:13649–58.
Sundar IK, Li D, Rahman I. Small RNA-sequence analysis of plasma-derived extracellular vesicle miRNAs in smokers and patients with chronic obstructive pulmonary disease as circulating biomarkers. J Extracell Vesicles. 2019;8:1684816.
Barber JL, Zellars KN, Barringhaus KG, Bouchard C, Spinale FG, Sarzynski MA. The effects of regular exercise on circulating cardiovascular-related microRNAs. Sci Rep. 2019;9:7527.
Iacomino G, Russo P, Stillitano I, Lauria F, Marena P, Ahrens W, De Luca P, Siani A. Circulating microRNAs are deregulated in overweight/obese children: preliminary results of the I.Family study. Genes Nutr. 2016;11:7.
Rodosthenous RS, Coull BA, Lu Q, Vokonas PS, Schwartz JD, Baccarelli AA. Ambient particulate matter and microRNAs in extracellular vesicles: a pilot study of older individuals. Part Fibre Toxicol. 2016;13:13.
Yin Z, Guo Y, Zhang J, Zhang Q, Li L, Wang S, Wang C, He Y, Zhu S, Li C, et al. Association between an indel polymorphism in the 3’UTR of COL1A2 and the risk of sudden cardiac death in Chinese populations. Leg Med. 2017;28:22–6.
Xu J, Chen Z, Wang Y, Wang X, Chen L, Yuan T, Tang X, Lu Y, Chen H, Chen M, et al. Several circulating miRNAs related to hyperlipidemia and atherosclerotic cardiovascular diseases. Lipids Health Dis. 2019;18:104.
Liu X, Fortin K, Mourelatos Z. MicroRNAs: biogenesis and molecular functions. Brain Pathol. 2008;18:113–21.
Huang X, Liu G, Guo J, Su Z. The PI3K/AKT pathway in obesity and type 2 diabetes. Int J Biol Sci. 2018;14:1483–96.
Nunez LR, Jesch SA, Gaspar ML, Almaguer C, Villa-Garcia M, Ruiz-Noriega M, Patton-Vogt J, Henry SA. Cell wall integrity MAPK pathway is essential for lipid homeostasis. J Biol Chem. 2008;283:34204–17.
Morello F, Perino A, Hirsch E. Phosphoinositide 3-kinase signalling in the vascular system. Cardiovasc Res. 2009;82:261–71.
Acknowledgements
The authors wish to thank all the study participants from the South China Cardiovascular Related Disease Cohort (SCCDC, since July 2010), research staff and students who participated in this work.
Funding
This study was funded by the Guangzhou Science and Technology Project of China (201804010214), the Guangdong Natural Science Foundation of China (S2011040004458), the National Natural Science Foundation of China (81100235), xinxin-Merck Cardiovascular Scientific Research Fund (2017-CCA-xinxin merck fund-012) and the Guangdong Science and Technology Planning Project of China (2014A020212372).
Author information
Authors and Affiliations
Contributions
CL, literature search, study format, writing protocol, processing data, data interpretation, analysing data and revising manuscript; YXL, literature search, recruiting patients, data interpretation, qRT-PCR, and writing manuscript; SSY, sample collection and genotyping; JFZ and YS, recruiting patients, following up patients and collecting data. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
This study was approved by the Ethics Committee of Guangzhou First People’s Hospital, South China University of Technology (K-2017-043-02).
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Additional file 1: Figure S1.
Volcano map of DE-exo-miRs between different genotypes of KATP rs1799858 in subjects with elevated LDL-C (≥1.8 mmol/L) serum level. Figure S2. Bubble map for KEGG analysis of enrichment pathway regulated by CTGs of top 10 DE-exo-miRs.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Liu, C., Lai, Y., Ying, S. et al. Plasma exosome-derived microRNAs expression profiling and bioinformatics analysis under cross-talk between increased low-density lipoprotein cholesterol level and ATP-sensitive potassium channels variant rs1799858. J Transl Med 18, 459 (2020). https://doi.org/10.1186/s12967-020-02639-8
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12967-020-02639-8