Abstract
Background
Telocytes (TCs) are suggested as a new type of interstitial cells with specific telopodes. Our previous study evidenced that TCs differed from fibroblasts and stem cells at the aspect of gene expression profiles. The present study aims to search the characters and patterns of chromosome X genes of TC-specific or TC-dominated gene profiles and fingerprints, investigate the network of principle genes, and explore potential functional association.
Methods
We compared gene expression profiles in chromosome X of pulmonary TCs with mesenchymal stem cells (MSC), fibroblasts (Fb), alveolar type II cells (ATII), airway basal cells (ABC), proximal airway cells (PAC), CD8+ T cells come from bronchial lymph nodes (T-BL), or CD8+ T cells from lungs (T-L) by global analyses, and selected the genes which were consistently up or down regulated (>1 fold) in TCs compared to other cells as TC-specific genes. The functional and characteristic networks were identified and compared by bioinformatics tools.
Results
We selected 31 chromosome X genes as the TC-specific or dominated genes, among which 8 up-regulated (Flna, Msn, Cfp, Col4a5, Mum1l1, Rnf128, Syn1, and Srpx2) and 23 down-regulated (Abcb7, Atf1, Ddx26b, Drp2, Fam122b, Gyk, Irak1, Lamp2, Mecp2, Ndufb11, Ogt, Pdha1, Pola1, Rab9, Rbmx2, Rhox9, Thoc2, Vbp1, Dkc1, Nkrf, Piga, Tmlhe and Tsr2), as compared with other cells.
Conclusions
Our data suggested that gene expressions of chromosome X in TCs are different with those in other cells in the lung tissue. According to the selected TC-specific genes, we infer that pulmonary TCs function as modulators which may enhance cellular growth and migration, resist senescence, protect cells from external stress, regulate immune responses, participate in tissue remodeling and repair, regulate neural function, and promote vessel formation.
Similar content being viewed by others
Background
Telocytes (TCs) as a new type of interstitial cells are characterized with extensive telopodes [1], and found in multiple organs/tissues, including heart [2], trachea and lung [3], digestive tract [4, 5], liver [6], skeletal muscle [7], kidney and urinary tract [8, 9], skin [10], mammary gland [11], and others [12–18]. TCs were suggested to play a key role in supporting and nutrition of associated cells through the interconnection in forms of a complex three-dimensional network within organs/tissues [12, 19–21]. TCs are involved in mechanical support, intercellular signaling, immune surveillance, stem-cell guidance and tissue regeneration [22–24].
The electrophysiological properties of TCs were reported in recent studies [25]. Ion channels on the membrane of TCs are found to be related with the different function of TCs and are involved in multiple biological courses, e.g., SK3 channel modulates the contractility of myometrial [26], large conductance Ca2+-activated K+ current and inwardly rectifying K+ current are expressed on TCs in human heart [27], etc.
Our previous studies initially demonstrated that pulmonary TCs were allocated near the basement membrane of the bronchiolar epithelium, between airway smooth muscle cells, or in pulmonary interstitial space [3].Pulmonary TCs were inferred to play significant roles in lung diseases, e.g. involving in the course of repair after injury, contributing to the development of pulmonary infectious diseases and stimulating the proliferation of fibroblasts during the process of fibrosis, etc. [28]
TCs are mainly identified and defined by the structures of telopodes, podoms and podomers through transmission electron microscopy [4, 22]. CD34/PDGFRα double immunohistochemistry can orientate the diagnosis [29]. The identification course of TCs is relatively complicated and the confusion between TCs and other cells is inevitable. Therefore, there is a need to select several specific biomarkers of TCs to differ between pulmonary TCs and other tissue resident cells, e.g. fibroblasts, stem cells, epithelial cells, and inflammatory cells.
Our previous studies focused on TCs in lung and trachea of the mouse [3] provided genetic evidence that pulmonary TCs differ from stem cells and fibroblasts through comparing the variation of gene expression profiles [30]. Other studies suggested specific microRNA expression signatures as TC-specific biomarkers [31–33]. Our recent studies identified characters and patterns of TC-specific or TC-dominated gene profiles and fingerprints in chromosome 1, 2, 3, 17 and 18, through global comparison between TCs and other cells in the mouse lung tissue [34–36]. The present study aims to search TC-specific or dominated gene profiles and fingerprints of chromosome X, investigate the network of principle genes, and explore potential functional association. We globally compared gene expression profiles of TCs, mesenchymal stem cells (MSCs), fibroblasts (Fbs), alveolar type II cells (ATII), airway basal cells (ABCs), proximal airway cells (PACs), CD8+ T cells from bronchial lymphnodes (T-BL), and CD8+ T cells from lung (T-L), which may interact with TCs in the lung and trachea.
Methods
Isolation and primary culture of TCs from lung tissues
TCs were isolated from mouse lung tissues, cultured in a density of 1 × 105 cells/cm2. TCs usually begin forming telopodes on day 5 and start overlapping and apoptosis on day 10, therefore TCs were harvested on day 5 (TC5) and day 10 (TC10), as described previously [30, 34]. RNAs were isolated and prepared, labeled, and hybridized for DNA microarray (The Mouse 4x44K Gene Expression Array, Agilent, Shanghai, China) with about 39,000+ mouse genes and transcripts represented with public domain annotations, according to the protocol of One-Color Microarray-Based Gene Expression Analysis. The hybridized arrays were washed, fixed and scanned with using the Agilent DNA Microarray Scanner (part number G2505B).
Data collection and mining
We obtained gene expression profiles of pulmonary TCs on days 5 and 10, MSCs, Fbs from our previous study [30], ATII, ABCs, PACs, T-BL, and T-LL from the National Center for Biotechnology Information (NCBI) Gene Expression Omnibus database (GSE6846 [37], GSE27379 [38], GSE28651). The microarray was composed of 45,101 probes. Our first filter eliminated the probe sets without corresponding official symbol, leaving 39, 417 probes and 21,680 genes.
Our earlier study have made the gene expression profile, composed of 23,861 probes, of mouse lung TCs, Fbs and stromal stem/progenitor cells [30]. After eliminating the probes without corresponding official symbol, there are 13,236 probes and 11,532 genes. Only those genes whose expressions were measured in all cells were considered in our analysis. In total, 11,532 genes were analyzed and 335 genes of chromosome X were focused and furthermore analyzed in the present study.
Identification of differentially expressed genes
The present study compared gene expression profiles in chromosome X of pulmonary TCs with MSCs, Fbs, ATIIs, ABCs, PACs, T-BL, or T-L by global analyses, investigated gene expression profiles of chromosome X in different cells to seek for the TC-specific regulated genes and explored potential association of selected genes.
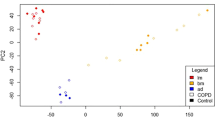
Hierarchical clustering of genes in chromosomes X was performed by TIGR Multi-experiment Viewer (MeV v4.9) (Fig. 1). Gene expression data were normalized and imported into Agilent GeneSpring GX software (version 11.5.1) for further analysis. Up- or down-regulated folds of TCs genes were calculated by comparing with other cells, after the averages of gene expression in cells were obtained from the raw data of multi-databases. The up-regulated folds were defined as the normalized gene expression values of TC5 or TC10 divide those of other cells, while the down-regulated folds were defined as the expression values of other cells devide those of TC5 or TC10. We selected the genes which were consistently up or down regulated (>1 fold) in both TC5 and TC10 as compared to other cells as TC-specific genes (Additional file 1: Table S1).
Hierarchical cluster analysis. Hierarchical cluster analysis of the differentially expressed genes on chromosomes X among telocytes (TCs), mesenchymal stem cells (MSCs), fibroblasts (Fbs), lymphocytes from lungs (T-LL) and from bronchial lymph nodes (T-BL), alveolar type II cells (ATII), proximal airway cells (PAC) and airway basal cells (ABC)
Results
Hierarchical clustering of genes in chromosomes X was performed, as shown in Fig. 1. The result of clustering showed a close relationship of TC5 and TC10, and obvious difference between TCs and other kind of cells.
Gene expression array data showed that 8 chromosome X genes (e.g. Flna, Msn, Cfp, Col4a5, Mum1l1, Rnf128, Syn1, and Srpx2) up-regulated in both TC5 and TC10, as compared with those in other cells (Table 1). Of them, Cfp, Col4a5, Mum1l1, Rnf128, and Syn1 up-regulated 2–5 folds, and Srpx2 more than 5 folds as compared with other cells (Table 1B). 23 genes (e.g. Abcb7, Atf1, Ddx26b, Drp2, Fam122b, Gyk, Irak1, Lamp2, Mecp2, Ndufb11, Ogt, Pdha1, Pola1, Rab9, Rbmx2, Rhox9, Thoc2, Vbp1, Dkc1, Nkrf, Piga, Tmlhe and Tsr2) down-regulated in TCs, of which 5 chromosome X genes (Dkc1, Nkrf, Piga, Tmlhe, and Tsr2) in TCs were more than 2 folds lower than in other cells, as compared with other cells (Table 2).
A set of genes were found specifically up- or down-regulated in pulmonary TCs, as compared with MSCs, Fbs, ATII, ABCs, PACs, T-BL, or T-L, respectively, as listed in Table 3. A set of genes up- or down-regulated more than one fold in TC5 were 178 or 157, 224 or 111, 101 or 234, 99 or 236, 109 or 226, 68 or 267, or 88 or 247 and in TC10 123 or 212, 180 or 155, 75 or 260, 87 or 248, 86 or 249, 51 or 284, or 71 or 264, as compared with MSCs, Fbs, ATII, T-BL, T-L, ABCs, PACs, respectively. Up- or down-regulated genes in both TC5 and TC10 were 119 or 153, 172 or 103, 72 or 231, 79 or 228, 81 or 221, 43 or 259, or 65 or 241, as compared with MSCs, Fbs, ATII, T-BL, T-L, ABCs, PACs, respectively. Details of up- or down- regulated gene variations of chromosome X were listed in Additional file 2: Table S2, including the number and names of up- or down-regulated genes among different cells.
We picked out the top 15 % high-expressed genes in chromosome X of TC10, and compared the distribution of such active gene group with other cells, as shown in Fig. 2. The distribution of the high expressed genes in TC10 was similar to those in TC5, while quite different from MSCs, Fbs, ATII, ABCs, PACs, T-BL or T-L. These high expressed genes are mostly involved in gene transcription, energy metabolism, apoptosis and protein degradation, cell migration and intracellular trafficking, and DNA repair. The relationships, including direct (physical) and indirect (functional) associations, of these genes were analyzed by String Network analysis (http://www.string-db.org). The interaction and potential functional links of those genes are also displayed in Fig. 2.
Expression profiles of the top 15 % up-regulated genes of chomosome X in TCs. Expression profiles of the top 15 % up-regulated genes of chromosome X of TCs isolated and cultured from mouse lungs on days 10 (TC10), as compared with those on days 5 (TC5), MSCs, Fbs, ATII, ABCs, PACs, T-BL, and T-L. The profiles for entire genes are described in Additional file 1. The selected core network and whole mouse network are linked by the documented functional interedractions from various databases (see “Methods”). Genes in each network are indicated in and some of their nearest neighbors are indicated by grey nodes. The top 15 % up-regulated genes within chromosome X of TC10 were selected, and their distribution in each type of cells was compared, and showed the difference between cells
Thirty-one genes were selected as TC-specific or dominated genes, which were up- or down-regulated more than one fold in both TC5 and TC10, as compared with other cells. Among those genes in TCs, 8 were up-regulated, while 23 were down-regulated. Figure 3 demonstrated the distribution of such distinct TC-specific or dominated genes in chromosome X of all cells. The interaction and potential functional links between those genes are also displayed in Fig. 3.
Expression profiles of TC-specific or dominant genes. Expression profiles of the up- or down-regulated genes (more than one fold) in both TC5 and TC10 as compared with MSCs, Fbs, ATII, ABCs, PACs, T-BL and T-L. The profiles for entire genes are described in Additional file 1. The selected core network and whole mouse network are linked by the documented functional interactions from various databases (see “Methods”). Genes in each network are indicated in red and some of their nearest neighbors are indicated by grey nodes. The 8 up-regulated genes in TCs were shown in the upper right corner, while the 23 down-regulated genes in the lower left and their distribution in each type of cells showed in Fig. 2b. These up- or down-regulated genes were considered as TC-specific or TC-dominant genes. The connections between these genes were analyzed and illustrated
Discussion
Chromosome X is one of the two sex-determining chromosomes, which exists in both gender, with one copy in male and two copies in female, spanning about 156 million base pairs and 1805 genes in human cells. Although chromosome X contains only 4 % of all human genes, a large number of disease condition are related with chromosome X, including X-linked diseases and 10 % of diseases with a mendelian pattern of inheritance [39]. There are 20,000–25,000 genes in mouse, and the similarity of genes between human and mouse is about 85 %. Among 2059 genes in chromosome X of the mouse, 335 were measured by bioinformatics tools in the present study. There were about 8 or 23 up- or down-regulated genes of chromosome X in TCs, as compared with stem cells, lung interstitial cells, pneumocytes, airway cells, or lymphocytes (Fig. 4).
TC-specific or dominant genes and their main function. Gene expression array data showed that 8 chromosome X genes up-regulated in both TC5 and TC10, and 23 genes down-regulated, as compared with those in other cells. These genes are selected as TC-dominant or specific genes. The up-regulated genes showed in red color while the down-regulated genes in green color. The up-regulated genes are mostly involved in cytoskeleton and cell migration, and regulation of immune responses. The down-regulated genes participates mainly in gene transcription modulation, energy metastasis, material transportation and so on
Srpx2, Cfp, Col4a5, Mum1l1, Rnf128 and Syn1 over-expressed in chromosome X of TCs, as compared with other cells. Srpx2 (sushi-repeat-containing protein, X-linked 2) gene was the only one of chromosome X that expressed more than 5 folds in pulmonary TCs as compared with all the other cells. The Srpx2-encoded protein was found to be the key factor in the development of speech and language centers in the brain [40], the important mediator in endothelial cell migration and tube formation, and the predictor for tumor growth and metastasis [41–43]. The enrichment of Srpx2 in pulmonary TCs suggests the potential role of TCs in the regulation of neuron function, promotion of vessel formation, and enhance of cellular growth and migration. Those could be performed through the complicated three-dimensional network constructed by the telopodes with other cells. Cfp, Col4a5, Mum1L1, Rnf128 and Syn1 over-expressed 2–5 folds in TCs within the lung tissue. Cfp (complement factor properdin) encodes a plasma glycoprotein that positively regulates the alternative complement pathway of the innate immune system [44]. TCs were found to be associated with the immune cells including lymphocytes [45], basophiles [46], eosinophils and plasma cells [7]. The high-expressed Cfp suggested that TCs might participate in immune responses and play a potential role in the lung infectious diseases. Col4a5 (collagen, type IV, alpha 5) encodes type IV collagen, which is the major structural component of basement membranes and is critical for tissue remodeling and repair [47]. The Col4a5 gene was also found as a TC-dominant gene as compared with Fbs or MSCs in our previous study [30].
Rnf128 (ring finger protein 128, E3 ubiquitin protein ligase) is associated with cellular apoptosis and cytokine regulation, of which over-expression inhibits the production of activation-induced IL2 and IL4 cytokine, and may converse T cells to the anergic phenotype [48, 49]. This implies that TCs may be involved in inflammation, immune surveillance, or tissue repair after injury by regulating immune function and inflammatory cell. Syn1 (synapsin I) encodes neuronal phosphoproteins associated with the cytoplasmic surface of synaptic vesicles, by which TCs may establish synapses as a special type of heterocellular junctions with other neighbouring cells, including lymphocytes, macrophages, or mast cells [46, 50, 51]. Through those connections to immune cells, TCs could integrate signals, and behave as an immune system modulator. Mum1l1 (melanoma associated antigen (mutated) 1-like 1) is a gene related with melanoma or some other tumors, its function in TCs is not quite clear yet.
Genes down-regulated in chromosome X of pulmonary TCs mainly contribute to gene transcription, protein synthesis and energy metabolism. Nkrf (NFKB repressing factor) encodes a transcriptional repressor of nuclear factor kappa B (NF-κB) and is activated for cell survival, proliferation and immune responses. It indicates that TCs per se have relatively high capacities of proliferation and involvements of immune reactions in physiological conditions, although there are needs of direct evidences to show roles and changes of TCs in cancers, infection, autoimmune diseases, or injury [52–55]. Tsr2 [TSR2, 20S rRNA accumulation, homolog (S. cerevisiae)] is another inhibitor of NF-κB, and induces cell apoptosis [56]. Pulmonary TCs may be more involved in activities and processes dominanted by NF-κB such as the production of inflammatory mediators, due to the down-regulation of NF-κB inhibitors in TCs. Dkc1 (dyskeratosis congenita 1, dyskerin) plays an important role in telomerase stabilization and DNA damage response, regulation of a subset of microRNAs, nucleo-cytoplasmic shuttling, and cell adhesion [57]. It suggested that TCs have better abilities to resist senescence and external stresses. Piga (phosphatidylinositol glycan anchor biosynthesis, class A) is a gene related with paroxysmal nocturnal hemoglobinuria and encodes a protein required for synthesis of N-acetylglucosaminyl phosphatidylinositol, the first intermediate in the biosynthetic pathway of a glycolipid on blood cells and to anchor proteins to the cell surface [58]. Tmlhe (trimethyllysine hydroxylase, epsilon) encodes the first enzyme in the carnitine biosynthesis pathway related with the transport of activated fatty acids across the inner mitochondrial membrane [59]. The significance of those two down-regulated genes in TCs still remains unclear. Based on the level of gene expression, the limitation of the study is inevitable, since gene expression level cannot equally reflect the protein level or the function change in the cells. Fold changes of gene expression may not be in direct proportion to the functional significance. More studies will be conducted in the future to detect the relationships among TC-specific genes, TC-specific proteins and the function of TCs.
Conclusions
In conclusion, the present study initially compared genetic variations of chromosome X of pulmonary TCs with MSCs, Fbs, ATIIs, ABCs, PACs, or bronchial and lung lymphocytes by global analyses. Our data demonstrated that 8 or 23 genes of TC chromosome X up- or down-regulated, respectively, indicating that biological functions of TCs may be mainly fulfilled through the network constructed by the telopodes with connecting cells and act as an integrated modulator to regulate neural function and immune responses, promote vessel formation and cellular growth and migration, and participate in tissue remodeling and repair. TCs may have the potential function to support cell survival, anti-senescence, and protection from stresses. Pulmonary TCs with stronger NF-kB-dominant activities may play the important and critical roles in pathogeneses of lung diseases, although further studies on those chromosome X genes of pulmonary TC are needed.
Abbreviations
- TC:
-
telocyte
- TC10:
-
telocytes isolated and cultured from mouse lungs on days 10
- TC5:
-
telocytes isolated and cultured from mouse lungs on days 5
- MSC:
-
mesenchymal stem cell
- Fb:
-
fibroblast
- ATII:
-
alveolar type II cell
- ABC:
-
airway basal cell
- PAC:
-
proximal airway cell
- T-BL:
-
CD8+ T cells come from bronchial lymph node
- T-L:
-
CD8+ T cells from lungs
References
Popescu L, Faussone-Pellegrini MS. TELOCYTES–a case of serendipity: the winding way from Interstitial Cells of Cajal (ICC), via Interstitial Cajal-Like Cells (ICLC) to TELOCYTES. J Cell Mol Med. 2010;14:729–40.
Gherghiceanu M, Popescu L. Cardiomyocyte precursors and telocytes in epicardial stem cell niche: electron microscope images. J Cell Mol Med. 2010;14:871–7.
Zheng Y, Li H, Manole CG, Sun A, Ge J, Wang X. Telocytes in trachea and lungs. J Cell Mol Med. 2011;15:2262–8. doi:10.1111/j.1582-4934.2011.01404.x.
Cantarero Carmona I, Luesma Bartolomé MJ, Junquera Escribano C. Identification of telocytes in the lamina propria of rat duodenum: transmission electron microscopy. J Cell Mol Med. 2011;15:26–30. doi:10.1111/j.1582-4934.2010.01207.x.
Rusu MC, Nicolescu MI, Jianu AM, Lighezan R, Manoiu VS, Paduraru D. Esophageal telocytes and hybrid morphologies. Cell Biol Int. 2012;36:1079–88. doi:10.1042/cbi20120007.
Xiao J, Wang F, Liu Z, Yang C. Telocytes in liver: electron microscopic and immunofluorescent evidence. J Cell Mol Med. 2013;17:1537–42. doi:10.1111/jcmm.12195.
Popescu LM, Manole E, Serboiu CS, Manole CG, Suciu LC, Gherghiceanu M, et al. Identification of telocytes in skeletal muscle interstitium: implication for muscle regeneration. J Cell Mol Med. 2011;15:1379–92. doi:10.1111/j.1582-4934.2011.01330.x.
Gevaert T, De Vos R, Van Der Aa F, Joniau S, van den Oord J, Roskams T, et al. Identification of telocytes in the upper lamina propria of the human urinary tract. J Cell Mol Med. 2012;16:2085–93. doi:10.1111/j.1582-4934.2011.01504.x.
Qi G, Lin M, Xu M, Manole CG, Wang X, Zhu T. Telocytes in the human kidney cortex. J Cell Mol Med. 2012;16:3116–22. doi:10.1111/j.1582-4934.2012.01582.x.
Rusu MC, Mirancea N, Manoiu VS, Valcu M, Nicolescu MI, Paduraru D. Skin telocytes. Ann Anat. 2012;194:359–67. doi:10.1016/j.aanat.2011.11.007.
Mou Y, Wang Y, Li J, Lu S, Duan C, Du Z, et al. Immunohistochemical characterization and functional identification of mammary gland telocytes in the self-assembly of reconstituted breast cancer tissue in vitro. J Cell Mol Med. 2013;17:65–75. doi:10.1111/j.1582-4934.2012.01646.x.
Nicolescu MI, Popescu LM. Telocytes in the interstitium of human exocrine pancreas: ultrastructural evidence. Pancreas. 2012;41:949–56. doi:10.1097/MPA.0b013e31823fbded.
Popescu BO, Gherghiceanu M, Kostin S, Ceafalan L, Popescu LM. Telocytes in meninges and choroid plexus. Neurosci Lett. 2012;516:265–9. doi:10.1016/j.neulet.2012.04.006.
Hatta K, Huang ML, Weisel RD, Li RK. Culture of rat endometrial telocytes. J Cell Mol Med. 2012;16:1392–6. doi:10.1111/j.1582-4934.2012.01583.x.
Suciu L, Popescu LM, Gherghiceanu M, Regalia T, Nicolescu MI, Hinescu ME, et al. Telocytes in human term placenta: morphology and phenotype. Cells Tissues Organs. 2010;192:325–39. doi:10.1159/000319467.
Nicolescu MI, Bucur A, Dinca O, Rusu MC, Popescu LM. Telocytes in parotid glands. Anat Rec Hoboken. 2012;295:378–85. doi:10.1002/ar.21540.
Corradi LS, Jesus MM, Fochi RA, Vilamaior PS, Justulin LA Jr, Goes RM, et al. Structural and ultrastructural evidence for telocytes in prostate stroma. J Cell Mol Med. 2013;17:398–406. doi:10.1111/jcmm.12021.
Luesma MJ, Gherghiceanu M, Popescu LM. Telocytes and stem cells in limbus and uvea of mouse eye. J Cell Mol Med. 2013;17:1016–24. doi:10.1111/jcmm.12111.
Gherghiceanu M, Popescu LM. Cardiac telocytes—their junctions and functional implications. Cell Tissue Res. 2012;348:265–79.
Creţoiu SM, Creţoiu D, Popescu LM. Human myometrium—the ultrastructural 3D network of telocytes. J Cell Mol Med. 2012;16:2844–9.
Popescu L, Gherghiceanu M, Cretoiu D, Radu E. The connective connection: interstitial cells of Cajal (ICC) and ICC-like cells establish synapses with immunoreactive cells: electron microscope study in sity. J Cell Mol Med. 2005;9:714–30.
Popescu LM, Gherghiceanu M, Suciu LC, Manole CG, Hinescu ME. Telocytes and putative stem cells in the lungs: electron microscopy, electron tomography and laser scanning microscopy. Cell Tissue Res. 2011;345:391–403.
Hinescu ME, Gherghiceanu M, Suciu L, Popescu LM. Telocytes in pleura: two- and three-dimensional imaging by transmission electron microscopy. Cell Tissue Res. 2011;343:389–97. doi:10.1007/s00441-010-1095-0.
Vannucchi MG, Bani D, Faussone-Pellegrini MS. Telocytes contribute as cell progenitors and differentiation inductors in tissue regeneration. Curr Stem cell Res Ther 2015 [Epub ahead of print].
Cretoiu SM, Popescu LM. Telocytes revisited. Biomol Concept. 2014;5:353–69.
Rosenbaum ST, Svalø J, Nielsen K, Larsen T, Jørgensen JC, Bouchelouche P. Immunolocalization and expression of small-conductance calcium-activated potassium channels in human myometrium. J Cell Mol Med. 2012;16:3001–8.
Sheng J, Shim W, Lu J, Lim SY, Ong BH, Lim TS, et al. Electrophysiology of human cardiac atrial and ventricular telocytes. J Cell Mol Med. 2014;18:355–62.
Zheng Y, Bai C, Wang XD. Potential significance of telocytes in the pathogenesis of lung diseases. Expert Rev Respir Med. 2012;6(1):45–9. doi:10.1586/ers.11.91.
Vannucchi MG, Traini C, Manetti M, Ibba-Manneschi L, Faussone-Pellegrini MS. Telocytes express PDGFRα in the human gastrointestinal tract. J Cell Mol Med. 2013;17:1099–108.
Zheng Y, Zhang M, Qian M, Wang L, Cismasiu VB, Bai C, et al. Genetic comparison of mouse lung telocytes with mesenchymal stem cells and fibroblasts. J Cell Mol Med. 2013;17:567–77. doi:10.1111/jcmm.12052.
Cismasiu VB, Radu E, Popescu LM. miR-193 expression differentiates telocytes from other stromal cells. J Cell Mol Med. 2011;15:1071–4. doi:10.1111/j.1582-4934.2011.01325.x.
Gherghiceanu M, Popescu LM. Cardiac telocytes—their junctions and functional implications. Cell Tissue Res. 2012;348:265–79. doi:10.1007/s00441-012-1333-8.
Manole CG, Cismasiu V, Gherghiceanu M, Popescu LM. Experimental acute myocardial infarction: telocytes involvement in neo-angiogenesis. J Cell Mol Med. 2011;15:2284–96. doi:10.1111/j.1582-4934.2011.01449.x.
Sun X, Zheng M, Zhang M, Qian M, Zheng Y, Li M, et al. Differences in the expression of chromosome 1 genes between lung telocytes and other cells: mesenchymal stem cells, fibroblasts, alveolar type II cells, airway epithelial cells and lymphocytes. J Cell Mol Med. 2014;18:801–10.
Zheng M, Sun X, Zhang M, Qian M, Zheng Y, Li M, et al. Variations of chromosomes 2 and 3 gene expression profiles among pulmonary telocytes, pneumocytes, airway cells, mesenchymal stem cells and lymphocytes. J Cell Mol Med. 2014;18:2044–60.
Wang J, Ye L, Jin M, Wang X. Global analyses of Chromosome 17 and 18 genes of lung telocytes compared with mesenchymal stem cells, fibroblasts, alveolar type II cells, airway epithelial cells, and lymphocytes. Biol Direct. 2015;10:9.
Xu Y, Ikegami M, Wang Y, Matsuzaki Y, Whitsett JA. Gene expression and biological processes influenced by deletion of Stat3 in pulmonary type II epithelial cells. BMC Genom. 2007;8:455. doi:10.1186/1471-2164-8-455.
Chen W, Ware LB. Prognostic factors in the acute respiratory distress syndrome. Clin Transl Med. 2015;4(1):65. doi:10.1186/s40169-015-0065-2.
Ross MT, Grafham DV, Coffey AJ, Scherer S, McLay K, Muzny D, et al. The DNA sequence of the human X chromosome. Nature. 2005;434:325–37.
Sia GM, Clem RL, Huganir RL. The human language-associated gene SRPX2 regulates synapse formation and vocalization in mice. Science. 2013;342:987–91. doi:10.1126/science.1245079.
Yamada T, Oshima T, Yoshihara K, Sato T, Nozaki A, Shiozawa M, et al. Impact of overexpression of Sushi repeat-containing protein X-linked 2 gene on outcomes of gastric cancer. J Surg Oncol. 2014;109:836–40. doi:10.1002/jso.23602.
Tanaka K, Arao T, Tamura D, Aomatsu K, Furuta K, Matsumoto K, et al. SRPX2 is a novel chondroitin sulfate proteoglycan that is overexpressed in gastrointestinal cancer. PLoS One. 2012;7:e27922. doi:10.1371/journal.pone.0027922.
Tanaka K, Arao T, Maegawa M, Matsumoto K, Kaneda H, Kudo K, et al. SRPX2 is overexpressed in gastric cancer and promotes cellular migration and adhesion. Int J Cancer. 2009;124:1072–80. doi:10.1002/ijc.24065.
Saggu G, Cortes C, Emch HN, Ramirez G, Worth RG, Ferreira VP. Identification of a novel mode of complement activation on stimulated platelets mediated by properdin and C3(H2O). J Immunol. 2013;190:6457–67. doi:10.4049/jimmunol.1300610.
Wang XY, Berezin I, Mikkelsen HB, Der T, Bercik P, Collins SM, et al. Pathology of interstitial cells of Cajal in relation to inflammation revealed by ultrastructure but not immunohistochemistry. Am J Pathol. 2002;160:1529–40.
Popescu LM, Gherghiceanu M, Cretoiu D, Radu E. The connective connection: interstitial cells of Cajal (ICC) and ICC-like cells establish synapses with immunoreactive cells. Electron microscope study in situ. J Cell Mol Med. 2005;9:714–30.
Massella L, Muda AO, Faraggiana T, Bette C, Renieri A, Rizzoni G. Epidermal basement membrane alpha 5(IV) expression in females with Alport syndrome and severity of renal disease. Kidney Int. 2003;64:1787–91. doi:10.1046/j.1523-1755.2003.00251.x.
Su LL, Iwai H, Lin JT, Fathman CG. The transmembrane E3 ligase GRAIL ubiquitinates and degrades CD83 on CD4 T cells. J Immunol. 2009;183:438–44. doi:10.4049/jimmunol.0900204.
MacKenzie DA, Schartner J, Lin J, Timmel A, Jennens-Clough M, Fathman CG, et al. GRAIL is up-regulated in CD4 + CD25 + T regulatory cells and is sufficient for conversion of T cells to a regulatory phenotype. J Biol Chem. 2007;282:9696–702. doi:10.1074/jbc.M604192200.
Rusu MC, Jianu AM, Mirancea N, Didilescu AC, Manoiu VS, Paduraru D. Tracheal telocytes. J Cell Mol Med. 2012;16:401–5. doi:10.1111/j.1582-4934.2011.01465.x.
Chen X, Zheng Y, Manole CG, Wang X, Wang Q. Telocytes in human oesophagus. J Cell Mol Med. 2013;17:1506–12. doi:10.1111/jcmm.12149.
King PT. Inflammation in chronic obstructive pulmonary disease and its role in cardiovascular disease and lung cancer. Clin Transl Med. 2015;4:26.
Ho SC, Lee KY, Chan YF, Kuo LW, Ito K, Adcock IM, et al. Neutrophil elastase represses IL-8/CXCL8 synthesis in human airway smooth muscle cells through induction of NF-kappa B repressing factor. J Immunol. 2009;183:411–20. doi:10.4049/jimmunol.0803729.
Swindell WR, Sarkar MK, Stuart PE, Voorhees JJ, Elder JT, Johnston A, Gudjonsson JE. Psoriasis drug development and GWAS interpretation through in silico analysis of transcription factor bindingsites. Clin Transl Med. 2015;4:13. doi:10.1186/s40169-015-0054-5.
López E, Muñoz S, Pascual J, Madero L. Relevant phosphoproteomic and mass spectrometry: approaches useful in clinical research. Clin Transl Med. 2012;1:2.
Graves CA, Abboodi FF, Tomar S, Wells J, Pirisi L. The translational significance of epithelial-mesenchymal transition in head and neck cancer. Clin Transl Med. 2014;3:39.
Gu BW, Bessler M, Mason PJ. A pathogenic dyskerin mutation impairs proliferation and activates a DNA damage response independent of telomere length in mice. Proc Natl Acad Sci U S A. 2008;105:10173–8. doi:10.1073/pnas.0803559105.
Johnston JJ, Gropman AL, Sapp JC, Teer JK, Martin JM, Liu CF, et al. The phenotype of a germline mutation in PIGA: the gene somatically mutated in paroxysmal nocturnal hemoglobinuria. Am J Hum Genet. 2012;90:295–300. doi:10.1016/j.ajhg.2011.11.031.
Nava C, Lamari F, Heron D, Mignot C, Rastetter A, Keren B, et al. Analysis of the chromosome X exome in patients with autism spectrum disorders identified novel candidate genes, including TMLHE. Transl Psychiatry. 2012;2:e179. doi:10.1038/tp.2012.102.
Authors' contributions
YCZ planned and performed studies, analyzed data and wrote the manuscript; and XDW made study plans, overviewed data, and prepare the manuscript. MHZ, DLS, and LY analysed data and performed data mining. All authors read and approved the final manuscript.
Acknowledgements
The work was supported by Shanghai Leading Academic Discipline Project (B115), Zhongshan Distinguished Professor Grant (XDW), The National Nature Science Foundation of China (91230204, 81270099, 81320108001, 81270131), The Shanghai Committee of Science and Technology (12JC1402200, 12431900207, 11410708600).
Compliance with ethical guidelines
Competing interests The authors declare that they have no competing interests.
Author information
Authors and Affiliations
Corresponding author
Additional files
12967_2015_669_MOESM1_ESM.xlsx
Additional file 1. Data profiles for all genes in chromosome X.
12967_2015_669_MOESM2_ESM.xlsx
Additional file 2. Details of up-regulated or down-regulated gene expression variations in chromosome X.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Zhu, Y., Zheng, M., Song, D. et al. Global comparison of chromosome X genes of pulmonary telocytes with mesenchymal stem cells, fibroblasts, alveolar type II cells, airway epithelial cells, and lymphocytes. J Transl Med 13, 318 (2015). https://doi.org/10.1186/s12967-015-0669-8
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12967-015-0669-8