Abstract
Background
Chemoresistance is one of the major obstacles that lead to poor prognosis in cervical cancer. linc00958 was reported to be an oncogene in cervical cancer. However, its role in mediating chemoresistance remains to be revealed.
Purpose
To explore the regulatory mechanisms of linc00958 in cisplatin-resistant cervical cancer cells and further validate in xenograft mice.
Methods
Online bioinformatic tools were used to conduct the pre-investigation of linc00958/miR-185-5p/RSF-1 and predict the associations between RSF-1 and AKT1/GSK3β/VEGFA in cervical cancer. RT-qPCR measured the RNA expression levels of linc00958/miR-185-5p/RSF-1 in SiHa and SiHa/DDP. Cell survival rates were evaluated by CCK8 methods after cells were exposed to differential concentrations of DDP. Dual-luciferase reporter methods were used to measure luciferase activity. Western blot measured RSF-1 protein and phosphorylated changes of AKT1/GSK3β. Immunofluorescence was employed to observe VEGFA secretion in vitro. Tube formation was applied to evaluate the in-vitro changes of angiogenesis. The SiHa/DDP cells stably transfected with pLKO-sh-NC or pLKO-sh-linc00958 plasmids, were injected into mice, establishing xenograft models. The changes in mice weight and tumor volumes were recorded. H&E staining and Immunohistochemistry (IHC) method was further performed.
Results
linc00958 expression was higher in SiHa/DDP cells. High linc00958 expression was associated with low overall survival. In SiHa/DDP cells linc00958/miR-185-5p/RSF-1 axis inhibited the cellular resistance to cisplatin and suppressed VEGFA and the tube formation through AKT1/GSK3β/VEGFA pathway. The knockdown of linc00958 inhibited RSF-1 and Ki67, curbing tumor growth; it also inhibited VEGFA and CD34, decreasing angiogenesis in mice.
Conclusion
linc00958/miR-185-5p/RSF-1 modulates cisplatin resistance and angiogenesis through AKT1/GSK3β/VEGFA pathway in cervical cancer.
Similar content being viewed by others
Introduction
Cervical cancer (CC) has one of the highest mortality rates in women among all cancers. Currently, resection surgeries, chemotherapy and radiotherapy are available all over the world. However, the overall and disease-free survival rates stay relatively high, which might be correlated with the undesirable outcomes derived from local resection, chemoresistance, radioresistance, tameless metastasis, etc [1, 2]. Cisplatin is a regular alternative for CC patients at locally advanced stage [3], which in practice is also combined with irradiation therapy or other medications [4, 5]. CC patients tend to gain cisplatin resistance, which in turn poses a challenge to the treatment efficacy [6]. As a result, it is of importance to unveil the potential mechanisms of the chemoresistance in CC. Previous researches explained the causes for drug resistance, which include changes in drug flux, detoxification through DNA repair, cell apoptosis, EMT, DNA methylation, gene profile, signaling pathways, etc [7].
During the past decade, studies identified aunderlying functions of molecules in cisplatin-resistance scenario in therapies for nasopharyngeal carcinoma [8], Osteosarcoma [9], lung cancer [10], breast cancer [11],ovarian cancer [12], cervical cancer [13], among which, noncoding RNAs have emerged as part of the new regulators in cisplatin resistance. lncRNA GAS5, was identified to target miR-21 through PTEN/AKT pathway and knockdown of GAS5 or upregulation of miR-21 added to the cellular resistance to cisplatin in SiHa/DDP in vitro [14].lncRNA CASC2/PTEN, competitively binding to miR-21, impacted on the cisplatin resistance in resistant cells via AKT pathway in CC [15]. The RNA interplay loops, defined as competitive endogenous RNAs (ceRNAs), featuring RNAs competing to bind to a certain miRNA [16, 17]. Linc00958 was revealed earlier in cervical cancer as an oncogenic gene, promoting the cell proliferation and invasion [18], and its downregulation sensitized the cells to radiation through miR-5095/RRM2 [19]. In head and neck carcinoma, linc00958 interacts with MYC and mediates cellular resistance to cisplatin and radiation, suggesting that linc00958 might regulate cisplatin resistance in CC [4].
Further, on online bioinformatic tools starBase and Targetscan, linc00958/RSF-1 were predicted to be targeted by miR-185-5p. Though the role of miR-185-5p in CC hasn’t been revealed yet, miR-185-5p was widely considered to increase cisplatin sensitivity in ovarian, lung and oral cancer cells [20,21,22]. Previously, RSF-1 was discovered to be related to paclitaxel sensitivity in CC cell line Hela [23]. In addition, downregulation of lncRNA NEAT1 could inhibit cisplatin resistance by suppressing RSF-1 through competitively binding to let-7-5p in nasopharyngeal carcinoma [24]. On the other hand, Gepia database revealed that in CC tumor tissues, RSF-1 expression is positively correlated with VEGFA, AKT1 and GSK3β expression. AKT/ GSK3β pathway was widely involved in cell survival and cisplatin resistance in cancers [25,26,27,28]. Vascular endothelial growth factor A(VEGFA) is a key factor associated with angiogenesis in tumors29. Taken together, we hypothesized that in CC, linc00958/miR-185-5p/RSF-1 might engage in regulating cisplatin resistance and angiogenesis through AKT1/ GSK3β/ VEGFA pathway. Hence, in this study, we investigated the regulatory effect of linc00958/miR-185-5p/RSF-1 on cisplatin resistance in SiHa/DDP cells and tube formation in vitro; in xenograft mice, we further validated the role of linc00958 in tumor growth and angiogenesis.
Methods
Ethical statement
This study has been submitted to and approved by Qingdao University Laboratory Animal Welfare Ethics Committee (Approval No. 202208Balb/C12202306025). No human sample was used in this study. All the cell and animal experiments were performed by strictly following the regulations of Qingdao Central Hospital, The Second Affiliated Hospital of Medical College of Qingdao University.
Bioinformatic analysis
Gepia (http://gepia.cancer-pku.cn/) was referred to for lncRNAs with differential expression in cervical cancer, in comparison with normal counterparts. After downloading the gene table, we analyzed on Microsoft Excel software to screen the top differential lncRNAs (Table 1). Furthermore, linc00958 expression was also investigated in various types of cancers on Gepia database. Overall survival analysis was conducted on Gepia to examine the correlation between linc00958 expression and overall survival rate, with 15% as the Cutoff-High and 85% as the Cutoff-Low. The linc00958 expression in cervical cancer tissues at early stage or lymph node-positive stage was analyzed in GEO profile (https://www.ncbi.nlm.nih.gov/). RNAinter database (http://www.rnainter.org/search/) was used to predict that miR-185-5p might target at cisplatin and also RSF-1. linc00958 was predicted as a sponge of miR-185-5p in homo sapiens in starBase (http://starbase.sysu.edu.cn/agoClipRNA.php?source=lncRNA), and miR-185-5p targets were predicted on starBase, PITA (https://genie.weizmann.ac.il/pubs/mir07/mir07_data.html) and RNAInter (http://www.rna-society.org/raid/). In addition, the miR-185-5p was predicted to target cisplatin in RNAInter.
Cell culture
SiHa, SiHa/DDP and HUVECs cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM) with 10% fetal bovine serum (FBS), and 1% penicillin/streptomycin (pen/strep). All cell lines were cultured in a cell incubator with 5% CO2, 99% relative humidity at 37 °C (Thermo Fisher, Shanghai, China).
Transfection
The overexpressed plasmid of linc00958(pc-linc00958) and RSF-1(pc-RSF-1) was constructed using pcDNA3.1 vector (ThermoFisher, Shanghai, China), with the empty vector as pc-NC. The miR-185-5p mimics, miR-185-5p inhibitor and their control mimics NC and inhibitor NC were provided by GenePharma (Suzhou, China). In brief, 6-well plates were used to seed SiHa and SiHa/DDP cells (5 × 105 cells each well). The 2.5 μg of plasmids pc-linc00958, pc-RSF-1, pc-NC, miR-185-5p mimics and mimics NC were mixed with 4 μl Lipo8000 reagent and added into each well respectively (Beyotime, Shanghai, China) and the transfection was performed according to the producer’s suggestions. After 48 h, cells were measured for transfection effect by RT-qPCR method. The knockdown plasmids of linc00958(sh-linc00958 1#, 2#, 3#), VEGFA(sh-VEGFA-1,2,3) and RSF-1(sh-RSF)were constructed using lentivirus interference vector, pLKO.1-EGFP-Puro (Biofeng, Changsha, China), with the empty vector as sh-NC. The mentioned plasmids were constructed by Tianjin Chuanshi Biotech and were sequenced by Shanghai Biotech(China). The lentivirus plasmids were used to infect the cells to downregulate the expression of linc00958 and RSF-1, together with the packaging vectors pLP1, pLP2, pLP VSV-G. The enhanced green fluorescent protein was observed under an inverted fluorescence microscope after 72 h (Nikon, Japan). Puromycin (1 μg/ml) was added to screen the stable transfected cells. The culture was changed once every three days and the concentration was gradually increased to 12 μg/ml.
RT-qPCR
Total RNA from SiHa and SiHa/DDP was extracted using Beyozol reagent (Beyotime). We synthesized cDNA by using RT SuperMix for qPCR (Beyotime). Relative RNA levels were measured on a Roche light cycler 96 by using the SYBR qPCR Master Kit with normalization to GAPDH or U6 (Beyotime). The primer sequences used in this study are listed (Table 2). The relative expression of RNAs was calculated using the comparative Ct method.
Cell counting Kit-8(CCK8) for cytotoxicity
CCK8 was used to examine cell survival after cisplatin treatment (Bioss, Beijing, China). Following manufacturer’s instructions, cells after transfection were seeded in 96-well plates (8000 cells per well) and incubated for 12 h. Then cells were treated with cisplatin (0, 7.5, 15,30,60, 120, 240ug/ml) for 24 h. Thereafter, 10ul CCK8 solution was added into each well. After 2 h-incubation in the cell incubator, OD values were detected on a microplate reader at 450 nm wavelength (Thermo Fisher, Shanghai, China). Each group was repeated for 3 times.
Luciferase assays
The luciferase reporter plasmids constructed by inserting the wild-type(wt) linc00958/RSF-1 segments targeted by miR-185-5p or mutant(mt) counterparts onto pGL3 dual luciferase reporter vector (Promega, Shanghai, China). The plasmids included pGL3-linc00958-wt, pGL3-linc00958-mt, pGL3-RSF-1-wt and pGL3-RSF-1-mt. The indicated luciferase plasmids were transfected into SiHa/DDP cells with the miR-185-5p mimics and mimics NC using Lipo8000 kit (Beyotime). After 48 h, luciferase activity was measured on a multi-functional microplate reader after applying Dual Lumi Luciferase reporter Gene Assay kit (Beyotime). The experiments were performed in triplicate.
Western blot
Protein extracts from cells were prepared using RIPA lysis buffer. Total protein(60 μg) was subjected to SDS-PAGE and transferred to 0.45 μm PVDF membrane (Millipore, USA). Antibodies used in this study were listed and the dilution rates were indicated (Table 3). Membranes were incubated overnight at 4 °C with diluted primary antibody. The membranes were washed using TBST for three times (10 min each time). Then the secondary antibody was used to incubate the membranes for 45 min at room temperature. After washing for another three times, the blotting bands were detected with an ECL kit (Beyotime) on a ECL capture machine (Peiqing, Shanghai, China). The grey values of the bands were analyzed on the analysis software provided by Peiqing.
Immunofluorescence assay (VEGFA)
SiHa/DDP cells were collected in 24-well plates after transfection and washed with PBS. In each well, 200ul paraformaldehyde (4%) was added to fix the cells for 30 min and cold PBS was used to wash the cells for 3 times (5 min each time). Donkey serum (10%) was used to block the cells for 30 min. The primary antibody against VEGFA (bs-4572R, Bioss, Beijing, China) was diluted with 1% donkey serum (1:200) and then added in each well. After overnight incubation at 4℃, the secondary antibody Goat Anti-rabbit IgG H&L/FITC (bs-0295G-FITC, 1:500, Bioss) was added and the cells were incubated in dark for 2 h. Thereafter, cells were washed using PBS for 4 times (5 min each) and DAPI was applied for nucleus staining (Beyotime). The cells were observed under an inverted fluorescence microscope at 100X.
Tube formation
Matrigel (BD, USA)-coated µ-Slide angiogenesis (ibidi, Germany) was used to analyze tube formation of HUVECs. HUVECs (104) were seeded into each well and 50ul supernatant of SiHa/DDP cells was added. At 8 h, images were taken under inverted microscope. The number of mature tubes was counted in from three different fields. The images were taken under microscope at 100X.
Xenograft mice model
The cells stably transfected with pLKO-sh-NC and pLKO-sh-linc00958 (differing from the in-vitro group names, sh-NC and sh-linc00958) were collected and adjusted into 5 × 107 cells/ml using preheated PBS. The cells were centrifuged at 1000 rpm for 10 min and then 80ul serum-free medium was added and 100ul cell suspension was injected subcutaneously into the left back of 12 female Balb/C mice (4 weeks old). After 24 days, the tumors are formed, since which, mice weight was measured and tumor volumes were estimated (volume = length*width2/2) for the following 28 days. On Day 52, mice were killed and tumors were weighed.
H&E staining
The tumor tissues derived from the xenograft mice were fixed using 4% paraformaldehyde and dehydrated and embedded using paraffin. Serial sections were cut into 4um per piece. The slides were dewaxed using xylene first and then ethyl alcohol (100%, 95%, 80%, 70%) was used. Harris hematoxylin and eosin(H&E) was used to stain the sections and neutral gum was used to seal the slides. The sections were observed under microscope.
Immunohistochemistry method
The dewaxed sections were placed into ethyl alcohol (100%, 95%, 85%, 70%, 50% respectively for 40 s each). The 3% H2O2 was used to incubate the slides for 10 min. Antigen retrieval was conducted using sodium citrate buffer (Bioss). Goat serum was used as the blocking solution (Bioss). The primary antibodies against RSF-1, Ki67, CD34 and VEGFA (1:200, Bioss, Table 3) were added to incubate the sections at 37 °C for 1 h and then PBS was used to wash the slides for three times. The secondary Goat-anti-rabbit antibody (bs-40295G-HRP, 1:3000, Bioss) was applied to incubate the sections for 20 min. DAB kit was purchased from Beyotime and applied to stain the sections. The slides were then placed into hematoxylin for 2 min. The neutral gum was used to block the slides. The sections were observed under microscope.
Statistical analysis
All the statistical analysis was performed on Excel (Microsoft, USA) and Graphpad9.0 (Graphpad, USA). Student’s t test was conducted where there were only two groups.
One-way ANOVA followed by Tukey’s correction was applied when multiple comparisons were made between groups. Two-way ANOVA and Sidak’s multiple comparison test were performed in cell survival analysis. P < 0.05 was considered significant.
Results
Downregulation of linc00958 decreased cisplatin resistance in cervical cancer cells
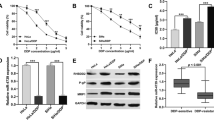
Based on Gepia database, linc00958 is among the top upregulated lncRNAs in CC tissues in comparison with the normal counterparts (Fig. 1A&D) and it is also aberrantly upregulated in other cancers, including head and neck squamous cancer, lung cancer, ovarian cancer, etc. (Fig. 1B). Based on GEO profile, linc00958 expression was not associated with the lymph node metastasis in CC (Fig. 1C). Furthermore, low linc00958 was correlated with high overall survival rates among CC patients (Fig. 1E).
Downregulation of linc00958 decreased cisplatin resistance in cervical cancer cells. A Gepia database provided the differentially expressed genes (DEG) in cervical cancer tissues. We screened the top DEGs in Excel and generated the bar graph. B Gepia online tool showed the expression of linc00958 in cancers. C GEO profile was used to analyze whether linc00958 was associated with the lymph nodes in CC patients. D-E Differential expression of linc00958 in CC; Overall survival curve in correlation to linc00958 in CC (Gepia). F-H CCK8 was used to evaluate the cisplatin resistance in SiHa, SiHa/DDP before and after the knockdown of linc00958
In this study, RT-qPCR results showed that linc00958 expression was higher in the cisplatin-resistant cells SiHa/DDP compared to parental SiHa cells (Suppl 1A). After validation of transfection efficiency, by RT-qPCR methods, (Suppl B-F), CCK8 results showed that SiHa/DDP was more reluctant to respond to increasing concentrations of cisplatin with half maximal inhibitory concentration (IC50) as 30ug/ml (Fig. 1F). The knockdown of linc00958 in SiHa cells enhanced the cellular sensitivity to cisplatin (Fig. 1G). Further, knockdown of linc00958 in SiHa/DDP cells reduced the IC50 of cisplatin, suggesting that the downregulation of linc00958 was correlated with inhibition of cisplatin resistance (Fig. 1H).
linc00958 mediated the cisplatin resistance via miR-185-5p/RSF-1 axis in cervical cancer cells
RNAInter predicted that miR-185-5p might be related to cisplatin and RSF-1 (Fig. 2A-B). StarBase and PITA databases were applied to further predict the potential binding sites between miR-185-5p and linc00958/RSF-1 (Fig. 2C-D). Luciferase reporter assays in SiHa/DDP cells after co-transfection validated that miR-185-5p targeted RSF-1 and linc00958 sponged miR-185-5p (Fig. 2E-F). RT-qPCR results also revealed that miR-185-5p expression was lower in cisplatin-resistant cell line (Fig. 2G). Furthermore, upregulation of linc00958 could inhibit miR-185-5p in cells (Fig. 2H). On the other hand, RSF-1 mRNA expression was significantly higher in SiHa/DDP cells compared to SiHa (Fig. 2I). Gepia online database showed that linc00958 and RSF-1 were positively correlated in CC tissues (Fig. 2J); In SiHa/DDP cells, knockdown of linc00958 could decrease RSF-1 and linc00958 upregulation could enhance RSF-1 in mRNA and protein levels (Fig. 2K-L). Upregulation of miR-185-5p could decrease mRNA and protein levels of RSF-1 (Fig. 2M-N). Combined together, linc00958 could upregulate RSF-1 and serve as a sponge of miR-185-5p. Furthermore, the upregulation of miR-185-5p or RSF-1 downregulation inhibited the cisplatin resistance in SiHa/DDP cells, suggesting that linc00958 mediated the cisplatin resistance via miR-185-5p/RSF-1 axis in cervical cancer cells (Fig. 2O-P).
The interactions between linc00958/miR-185-5p/RSF-1 in cervical cancer cells. A-B RNAInter was applied to predict cisplatin targets and also miR-185-5p and RSF-1. C-D Starbase database provided the putative binding sites between miR-185-5p and RSF-1/linc00958. E–F Luciferase reporter gene assays. G-I&K-L RT-qPCR measured RNA levels of miR-185-5p/RSF-1 in SiHa and SiHa/DDP cells, in SiHa/DDP cells with loss or gain of linc00958. Western blot measured RSF-1 protein expression. J Spearman’s correlation analysis between linc00958 and RSF-1 in CC tissues on Gepia. M–N Impact of miR-185-5p upregulation on RSF-1. O-P CCK8 evaluated the cisplatin resistance in SiHa/DDP cells with loss of RSF-1 or gain of miR-185-5p. ****P < 0.0001, ***P = < 0.0002, **P < 0.002, *P < 0.03
AKT1/GSK3β pathway were modulated by linc00958/miR-185-5p/RSF-1 axis in cervical cancer
RSF1 was correlated with AKT1 and GSK3β in CC tissues according to Spearman’s correlation analysis on Gepia (Fig. 3A-B). RT-qPCR methods validated that in SiHa/DDP cells, upregulation of linc00958 could enhance AKT1 mRNA expression but downregulation of linc00958 didn’t impact on AKT1 mRNA expression significantly (Fig. 3C), while linc00958 regulation didn’t mediate the mRNA of GSK3β in cells (Fig. 3D). However, western blot results supported that phosphorylated AKT1/AKT (ser473) and phosphorylated GSK3β were promoted by linc00958 upregulation and inhibited by downregulation of linc00958 in SiHa/DDP cells (Fig. 3E-H). The efficiency of the downregulated plasmids for RSF-1 were validated using RT-qPCR and western blot (Supp 1F-G). miR-185-5p upregulation or knockdown of RSF-1 inactivated AKT1/GSK3β pathway (Fig. 3I-P). In addition, western blot results confirmed that miR-185-5p inhibitor could upregulate RSF-1 protein and activate AKT1/GSK3β pathway while the knockdown of RSF-1 could partly reverse this (Suppl 2A-G).
linc00958/miR-185-5p/RSF-1 modulates the AKT1/GSK3β pathway and VEGFA. A-B Spearman’s correlation analysis between RSF-1 and AKT1/GSK3β (Gepia). C-P Cells were transfected to regulate the expression of linc00958/miR-185-5p/RSF-1. RT-qPCR measured the mRNA expression of AKT1 and GSK3β while western blot detected the protein levels of phospho-AKT1(ser473), phosphor-AKT (ser473), AKT, phosphor-GSK3β and GSK3β. ****P < 0.0001, ***P = < 0.0002, **P < 0.002, *P < 0.03
Linc00958/miR-185-5p/RSF-1 axis modulated tube formation through VEGFA pathway
VEGFA expression was closely correlated with RSF-1 in CC tissues (Fig. 4A). High VEGFA expression was associated with low overall survival rates among CC patients (Fig. 4B). In SiHa/DDP cells, downregulation of RSF-1 could inhibit VEGFA mRNA expression (Fig. 4C). Furthermore, immunofluorescence results showed that VEGFA secretion was inhibited by knockdown of linc00958 and RSF-1 or the overexpression of miR-185-5p in SiHa/DDP cells (Fig. 4D-F). The loss of linc00958/RSF-1 or gain of miR-185-5p in supernatant inhibited the tube formation of HUVECs, blocking angiogenesis in vitro (Fig. 4G-L). The loss of miR-185-5p in supernatant promoted VEGFA secretion and the tube formation while the knockdown of RSF-1 could reverse these (Suppl 2H-J). Furthermore, DDP significantly inhibited VEGFA secretion in SiHa/DDP cells and suppressed the tube formation but miR-185-5p downregulation could reverse the effect of DDP (Suppl 3A-C). After transfection of sh-VEGFA (-1,2,3) in SiHa/DDP cells, it was confirmed that the SiHa/DDP cells in sh-VEGFA-2 group presented the best knockdown effect of VEGFA (Suppl 3D-E). The knockdown of VEGFA in supernatant led to significant inhibition in tube formation (Suppl 3F-G). However, in the cells with VEGFA blocked, neither knockdown of linc00958 nor upregulation of miR-185-5p could further inhibit the tube formation, suggesting that linc00958/miR-185-5p regulated the tube formation through VEGFA pathway (Suppl 3F-G).
linc00958/miR-185-5p/RSF-1 axis modulated tube formation through VEGFA pathway. A Spearman’s correlation analysis between RSF-1 and VEGFA in CC(Gepia). B Overall survival in association with VEGFA mRNA expression in CC patients (Gepia). C-E Immunofluorescence assays to observe VEGFA secretion in SiHa cells. F-L Tube formation assays. **P < 0.002
Loss of linc00958 inhibited the tumor growth and angiogenesis in xenograft model
In vivo, tumor growth was inhibited by loss of linc00958 in mice (Fig. 5A-D). H&E staining showed that downregulation of linc00958 was associated with less severe pathology (Fig. 5E). IHC showed that RSF-1 and the tumor growth biomarker Ki67, were inhibited in the group where linc00958 was knocked down, revealing that linc00958 downregulation in mice could inhibit RSF-1 and Ki67, suppressing the tumor growth (Fig. 5F-G). IHC results also validated that linc00958 downregulation could inhibit the tumor microvessel density marker CD34 and VEGFA in tumors, supporting that the knockdown of linc00958 inhibited the angiogenesis in xenograft model (Fig. 5H-I).
linc00958 downregulation inhibited the tumor growth and angiogenesis in xenograft model. A Images of xenograft mice and derived tumors. B Tumor weight. C-D Weight curve of mice and tumor volume since the first day when tumors could be detected. E H&E staining. F-I IHC measured the changes of RSF-1, Ki67, VEGFA and CD34. Scale: 100um. ****P < 0.0001.*P < 0.03
Discussion
Drug resistance is one of the major concerns leading to bad prognosis in cancer patients [30, 31]. Hence, it is of significance to reveal molecular mechanisms involved. This study focused on the investigation into linc00958 in mediating cisplatin resistance in cervical cancer. linc00958 was validated to upregulate RSF-1 as ceRNA of miR-185-5p in SiHa/DDP cells. Further, linc00958/RSF-1 downregulation or miR-185-5p upregulation could alleviate the cisplatin resistance and tube formation in cervical cancer cells via AKT1/GSK3β/VEGFA pathway. In cisplatin-resistant CC cell-graft mice models, stable knockdown of linc00958 inhibited the tumor growth and angiogenesis through decreasing RSF-1.
Previously, linc00958 downregulation was confirmed to decrease chemo- and radio-resistance in head and neck squamous cell carcinoma in vitro [4]. This study confirmed in CC cell and mice models that linc00958 could inhibit cisplatin resistance. In gastric cancer, miR-185-5p was predicted in the regulation of cisplatin and fluorouracil resistance among gastric cancer patients [32]. However, no former study has been focused on miR-185-5p in cervical cancer yet. In this study, we validated in CC that miR-185-5p upregulation inhibited the cisplatin resistance in SiHa/DDP cells. RSF-1, remodeling and spacing factor 1, engaged in DNA damage repair and cell apoptosis, was reported to reduce cisplatin resistance in malignant melanoma [33] and nasopharyngeal carcinoma [24]. Furthermore, two of our previous in-vitro studies discovered that the RSF-1 downregulation enhanced cell sensitivity to paclitaxel [23] and increased radiosensitivity [34] in CC. In this research, we further discovered that in vitro, depletion of RSF-1 in SiHa/DDP cells could attenuate the cisplatin resistance and tube formation; in xenograft mice, RSF-1 was decreased by the knockdown of linc00958 and associated with the inhibition of tumor growth and angiogenesis.
Interestingly, through online bioinformatics, we noted that RSF-1 was correlated with AKT1, GSK3β and VEGFA expression in CC tissues. Phosphorylation of AKT1 at ser473 is closely involved in cell proliferation, apoptosis and tumor growth [35,36,37]. The AKT inhibitors including capivasertib, uprosertib and ipatasertib could inhibit the cancer cell survival and drug resistance [38]. Previous research reported that lncRNAs could regulate AKT pathway in cancers [39]. In CC, lncRNA RP1-93H18.6 suppressed the tumorigenesis by inactivating AKT pathway [40]. In this study, we validated that linc00958 could sponge miR-185-5p and upregulate RSF-1 as a ceRNA and linc00958/miR-185-5p/RSF-1 axis could regulate the phosphorylation of AKT1(ser473) and GSK3β, thereby mediating the cell survival and cisplatin resistance in vitro.
VEGF family are essential for blood vessel formation, among which, the VEGFA is a key factor involved with tumor angiogenesis [41]. In CC patients after radiochemotherapy, high VEGFA serum level is correlated with higher rate of early recurrence [42]. In CC patients at advanced stage, the canonical humanized VEGFA monoclonal antibody, bevacizumab, combined with conventional chemotherapy (cisplatin or paclitaxel), could prolong overall survival for 3.7 months [43]. It is of significance to find new molecular mechanisms that regulate VEGFA secretion, which are potential to intervene the CC progression [44]. Previously, depletion of lncRNA HAND2-AS1 could inhibit CC progression by inhibiting VEGFA secretion [45]. Nuclear factor 90(NF90) facilitated angiogenesis in CC by mediating VEGFA [46]. In this study, we revealed that depletion of linc00958/miR-185-5p/RSF-1could inhibit the VEGFA secretion and tube formation in cisplatin-resistant SiHa/DDP cells. In CC mice with knockdown of linc00958, the tumor microvessel density biomarker CD34 and VEGFA were significantly inhibited, revealing that knockdown of linc00958 could attenuate tumor angiogenesis.
Conclusion
Taken together, this study reported a new regulatory axis linc00958/miR-185-5p/RSF-1 in cisplatin resistance and tube formation in CC via AKT1/GSK3β/VEGFA pathway and knockdown of linc00958 could also inhibit the tumor growth and angiogenesis.
Availability of data and materials
The analyzed data sets generated during the present study are available from the corresponding author on reasonable request.
Abbreviations
- CC:
-
Cervical cancer
- ceRNAs:
-
Competitive endogenous RNAs
- DMEM:
-
Dulbecco’s modified Eagle’s medium
- FBS:
-
Fetal bovine serum
- pen/strep:
-
Penicillin/streptomycin
- VEGF:
-
Vascular endothelial growth factor
References
Liontos M, Kyriazoglou A, Dimitriadis I, Dimopoulos MA, Bamias A. Systemic therapy in cervical cancer: 30 years in review. Crit Rev Oncol Hematol. 2019;137:9–17.
Masadah R, Rauf S, Pratama MY, Tiribelli C, Pascut D. The role of microRNAs in the cisplatin- and radio-resistance of cervical cancer. Cancers 2021; 13.
Markman M. Advances in cervical cancer pharmacotherapies. Expert Rev Clin Pharmacol. 2014;7:219–23.
Huang S, Zhan Z, Li L, Guo H, Yao Y, Feng M, Deng J, Xiong J. LINC00958-MYC positive feedback loop modulates resistance of head and neck squamous cell carcinoma cells to chemo- and radiotherapy in vitro. Onco Targets Ther. 2019;12:5989–6000.
Chen X, Zou H, Li H, Lin R, Su M, Zhang W, Zhou Y, Zhang P, Hou M, Deng X, Zou C. Weekly Versus Triweekly Cisplatin-Based Chemotherapy Concurrent With Radiotherapy in the Treatment of Cervical Cancer: A Meta-Analysis. International journal of gynecological cancer : official journal of the International Gynecological Cancer Society. 2017;27:344–9.
Bi L, Ma F, Tian R, Zhou Y, Lan W, Song Q, Cheng X. AJUBA increases the cisplatin resistance through hippo pathway in cervical cancer. Gene. 2018;644:148–54.
Zhu H, Luo H, Zhang W, Shen Z, Hu X, Zhu X. Molecular mechanisms of cisplatin resistance in cervical cancer. Drug Des Dev Ther. 2016;10:1885–95.
Cui Z, Pu T, Zhang Y, Wang J, Zhao Y. Long non-coding RNA LINC00346 contributes to cisplatin resistance in nasopharyngeal carcinoma by repressing miR-342-5p. Open Biol. 2020;10: 190286.
Ferretti VA, León IE. Long Non-coding RNAs in Cisplatin Resistance in Osteosarcoma. Curr Treat Options Oncol. 2021;22:41.
Zhang J, Xu C, Gao Y, Wang Y, Ding Z, Zhang Y, Shen W, Zheng Y, Wan Y. A novel long non-coding RNA, MSTRG.51053.2 regulates cisplatin resistance by sponging the miR-432–5p in non-small cell lung cancer cells. Front Oncol. 2020;10:215.
Campos-Parra AD, López-Urrutia E, Orozco Moreno LT, López-Camarillo C, Meza-Menchaca T, Figueroa González G, Bustamante Montes LP, Pérez-Plasencia C. Long non-coding RNAs as new master regulators of resistance to systemic treatments in breast cancer. Int J Mol Sci 2018; 19.
Long X, Song K, Hu H, Tian Q, Wang W, Dong Q, Yin X, Di W. Long non-coding RNA GAS5 inhibits DDP-resistance and tumor progression of epithelial ovarian cancer via GAS5-E2F4-PARP1-MAPK axis. J Exp Clin Cancer Res. 2019;38:345.
Shi F, Su J, Liu Z, Wang J, Wang T. miR-144 reverses cisplatin resistance in cervical cancer via targeting LHX2. J Cell Biochem. 2019;120:15018–26.
Wen Q, Liu Y, Lyu H, Xu X, Wu Q, Liu N, Yin Q, Li J, Sheng X. Long noncoding RNA GAS5, which acts as a tumor suppressor via microRNA 21, regulates cisplatin resistance expression in cervical cancer. Int J Gynecol Cancer. 2017;27:1096–108.
Feng Y, Zou W, Hu C, Li G, Zhou S, He Y, Ma F, Deng C, Sun L. Modulation of CASC2/miR-21/PTEN pathway sensitizes cervical cancer to cisplatin. Arch Biochem Biophys. 2017;623–624:20–30.
Tay Y, Rinn J, Pandolfi PP. The multilayered complexity of ceRNA crosstalk and competition. Nature. 2014;505:344–52.
Yang XZ, Cheng TT, He QJ, Lei ZY, Chi J, Tang Z, Liao QX, Zhang H, Zeng LS, Cui SZ. LINC01133 as ceRNA inhibits gastric cancer progression by sponging miR-106a-3p to regulate APC expression and the Wnt/β-catenin pathway. Mol Cancer. 2018;17:126.
Wang L, Zhong Y, Yang B, Zhu Y, Zhu X, Xia Z, Xu J, Xu L. LINC00958 facilitates cervical cancer cell proliferation and metastasis by sponging miR-625-5p to upregulate LRRC8E expression. J Cell Biochem. 2020;121:2500–9.
Zhao H, Zheng GH, Li GC, Xin L, Wang YS, Chen Y, Zheng XM. Long noncoding RNA LINC00958 regulates cell sensitivity to radiotherapy through RRM2 by binding to microRNA-5095 in cervical cancer. J Cell Physiol. 2019;234:23349–59.
Pei K, Zhu JJ, Wang CE, Xie QL, Guo JY. MicroRNA-185-5p modulates chemosensitivity of human non-small cell lung cancer to cisplatin via targeting ABCC1. Eur Rev Med Pharmacol Sci. 2016;20:4697–704.
Xiang Y, Ma N, Wang D, Zhang Y, Zhou J, Wu G, Zhao R, Huang H, Wang X, Qiao Y, Li F, Han D, Wang L, Zhang G, Gao X. MiR-152 and miR-185 co-contribute to ovarian cancer cells cisplatin sensitivity by targeting DNMT1 directly: a novel epigenetic therapy independent of decitabine. Oncogene. 2014;33:378–86.
Ratovitski EA. Phospho-ΔNp63α regulates AQP3, ALOX12B, CASP14 and CLDN1 expression through transcription and microRNA modulation. FEBS Lett. 2013;587:3581–6.
Wang X, Sheu JJ, Lai MT, Yin-Yi Chang C, Sheng X, Wei L, Gao Y, Wang X, Liu N, Xie W, Chen CM, Ding WY, Sun L. RSF-1 overexpression determines cancer progression and drug resistance in cervical cancer. Biomedicine. 2018;8:4.
Liu F, Tai Y, Ma J. LncRNA NEAT1/let-7a-5p axis regulates the cisplatin resistance in nasopharyngeal carcinoma by targeting Rsf-1 and modulating the Ras-MAPK pathway. Cancer Biol Ther. 2018;19:534–42.
Mohapatra P, Shriwas O, Mohanty S, Ghosh A, Smita S, Kaushik SR, Arya R, Rath R, Das Majumdar SK, Muduly DK, Raghav SK, Nanda RK, Dash R. CMTM6 drives cisplatin resistance by regulating Wnt signaling through the ENO-1/AKT/GSK3β axis. JCI Insight 2021; 6.
Chen L, Wang J, Cai X, Chen S, Zhang J, Li B, Chen W, Guo X, Luo H, Chen J. Cyclometalated Ru(II)-isoquinoline complexes overcome cisplatin resistance of A549/DDP cells by downregulation of Nrf2 via Akt/GSK-3β/Fyn pathway. Bioorg Chem. 2022;119: 105516.
Nath N, Rana A, Nagini S, Mishra R: Glycogen synthase kinase-3β inactivation promotes cervical cancer progression, invasion, and drug resistance. Biotechnology and applied biochemistry 2021.
el Arafa SA, Zhu Q, Barakat BM, Wani G, Zhao Q, El-Mahdy MA, Wani AA. Tangeretin sensitizes cisplatin-resistant human ovarian cancer cells through downregulation of phosphoinositide 3-kinase/Akt signaling pathway. Can Res. 2009;69:8910–7.
Siveen KS, Prabhu K, Krishnankutty R, Kuttikrishnan S, Tsakou M, Alali FQ, Dermime S, Mohammad RM, Uddin S. Vascular Endothelial Growth Factor (VEGF) Signaling in Tumour Vascularization: Potential and Challenges. Curr Vasc Pharmacol. 2017;15:339–51.
Wu Q, Yang Z, Nie Y, Shi Y, Fan D. Multi-drug resistance in cancer chemotherapeutics: mechanisms and lab approaches. Cancer Lett. 2014;347:159–66.
Li B, Jiang J, Assaraf YG, Xiao H, Chen ZS, Huang C. Surmounting cancer drug resistance: New insights from the perspective of N(6)-methyladenosine RNA modification. Drug resistance updates : reviews and commentaries in antimicrobial and anticancer chemotherapy. 2020;53: 100720.
Sun J, Zhao J, Yang Z, Zhou Z, Lu P. Identification of gene signatures and potential therapeutic targets for acquired chemotherapy resistance in gastric cancer patients. Journal of gastrointestinal oncology. 2021;12:407–22.
He J, Fu L, Li Q. Rsf-1 regulates malignant melanoma cell viability and chemoresistance via NF-κB/Bcl-2 signaling. Mol Med Rep. 2019;20:3487–98.
Tian J, Kong E, Wang X, Xie Z, Chang CY, Sheu JJ, Hao Q, Sun L. RSF-1 siRNA Enhances Tumor Radiosensitivity in Cervical Cancer via Enhanced DNA Damage, Cell Cycle Redistribution, and Promotion of Apoptosis. Onco Targets Ther. 2020;13:3061–71.
Liu J, Liu Y, Zhang J, Liu D, Bao Y, Chen T, Tang T, Lin J, Luo Y, Jin Y, Zhang J. Indole hydrazide compound ZJQ-24 inhibits angiogenesis and induces apoptosis cell death through abrogation of AKT/mTOR pathway in hepatocellular carcinoma. Cell Death Dis. 2020;11:926.
Diao W, Guo Q, Zhu C, Song Y, Feng H, Cao Y, Du M, Chen H. USP18 promotes cell proliferation and suppressed apoptosis in cervical cancer cells via activating AKT signaling pathway. BMC Cancer. 2020;20:741.
Revathidevi S, Munirajan AK. Akt in cancer: Mediator and more. Semin Cancer Biol. 2019;59:80–91.
Jabbarzadeh Kaboli P, Salimian F, Aghapour S, Xiang S, Zhao Q, Li M, Wu X, Du F, Zhao Y, Shen J, Cho CH, Xiao Z. Akt-targeted therapy as a promising strategy to overcome drug resistance in breast cancer - A comprehensive review from chemotherapy to immunotherapy. Pharmacol Res. 2020;156: 104806.
Shi WJ, Liu H, Ge YF, Wu D, Tan YJ, Shen YC, Wang H, Xu H. LINC00673 exerts oncogenic function in cervical cancer by negatively regulating miR-126-5p expression and activates PTEN/PI3K/AKT signaling pathway. Cytokine. 2020;136: 155286.
Wang Q, Yan SP, Chu DX, Xie Y, Wang CF, Zhang JY, Li WC, Guo RX. Silencing of long non-coding RNA RP1–93H18.6 acts as a tumor suppressor in cervical cancer through the blockade of the PI3K/Akt Axis. Mol Ther Nucleic Acids. 2020;19:304–17.
Jayson GC, Kerbel R, Ellis LM, Harris AL. Antiangiogenic therapy in oncology: current status and future directions. Lancet. 2016;388:518–29.
Braicu EI, Gasimli K, Richter R, Nassir M, Kümmel S, Blohmer JU, Yalcinkaya I, Chekerov R, Ignat I, Ionescu A, Mentze M, Fotopoulou C, Pop C, Lichtenegger W, Sehouli J. Role of serum VEGFA, TIMP2, MMP2 and MMP9 in monitoring response to adjuvant radiochemotherapy in patients with primary cervical cancer–results of a companion protocol of the randomized NOGGO-AGO phase III clinical trial. Anticancer Res. 2014;34:385–91.
Tewari KS, Sill MW, Long HJ 3rd, Penson RT, Huang H, Ramondetta LM, Landrum LM, Oaknin A, Reid TJ, Leitao MM, Michael HE, Monk BJ. Improved survival with bevacizumab in advanced cervical cancer. N Engl J Med. 2014;370:734–43.
Minion LE, Tewari KS. Cervical cancer - State of the science: From angiogenesis blockade to checkpoint inhibition. Gynecol Oncol. 2018;148:609–21.
Gao Y, Zou T, Liang W, Zhang Z, Qie M. Long non-coding RNA HAND2-AS1 delays cervical cancer progression via its regulation on the microRNA-21-5p/TIMP3/VEGFA axis. Cancer Gene Ther. 2021;28:619–33.
Zhang W, Xiong Z, Wei T, Li Q, Tan Y, Ling L, Feng X. Nuclear factor 90 promotes angiogenesis by regulating HIF-1α/VEGF-A expression through the PI3K/Akt signaling pathway in human cervical cancer. Cell Death Dis. 2018;9:276.
Acknowledgements
None
Funding
The CSCO-HengRui Tumor Research Fund (grant no. Y-HR2018-200). The CSCO-ShiYao Tumor Research Fund (grant no. Y-SY201901-0127). The Qingdao Medical and Health Research Program Project (grant no. 2021-WJZD095)
Author information
Authors and Affiliations
Contributions
Jing Tian and Lei Cheng Design and research the experiment and receive funding, Enqi Kong and Wenjin Gu and Li Sun write the revision and article, Yuanyuan Jiang, Quan Hao and Lei Cheng Conduct experiments and collect data, Beihua Kong and Jing Tian prepare for the survey, corresponding to Li Sun. All authors read and approved the final version of the manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
This study has been submitted to and approved by Qingdao University Laboratory Animal Welfare Ethics Committee(Approval No. 202208Balb/C12202306025). No human sample was used in this study. All the cell and animal experiments were performed by strictly following the regulations of Qingdao Central Hospital, The Second Affiliated Hospital of Medical College of Qingdao University. In accordance with the Declaration of Helsinki.
Consent for publication
All the authors consented for publication.
Competing interests
All authors declared there are no conflict of interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Tian, J., Cheng, L., Kong, E. et al. linc00958/miR-185-5p/RSF-1 modulates cisplatin resistance and angiogenesis through AKT1/GSK3β/VEGFA pathway in cervical cancer. Reprod Biol Endocrinol 20, 132 (2022). https://doi.org/10.1186/s12958-022-00995-2
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12958-022-00995-2