Abstract
Background
Insulin resistance (IR) and fat accumulation in visceral adipose tissue are key players in developing type 2 diabetes (T2D). Several adipose tissue derived-gene polymorphisms are related to higher body mass index (BMI), insulin resistance and T2D. The association of omentin rs2274907 (Val109Asp) and fat-mass and obesity-associated (FTO) rs9939609 gene polymorphisms with overweight/obesity and T2D is controversial. The aim of this study was to determine the association between omentin Val109Asp and FTO rs9939609 polymorphisms and insulin resistance in newly-diagnosed T2D patients.
Methods
The case-control study included 83 newly-diagnosed T2D patients and 85 healthy matched controls, aged 20–80 years. Fasting blood glucose and insulin levels were measured by the enzymatic method and enzyme-linked-immunosorbent assay, respectively. Insulin resistance was calculated using the homeostasis model assessment (HOMA) index. Genotyping was examined using the polymerase chain reaction-restriction fragment length polymorphism (PCR-RFLP).
Results
There are significant differences between both omentin Val109Asp and FTO rs9939609 polymorphisms and studied individuals (P = 0.011 and P = 0.0001, respectively). Both genetic polymorphisms of omentin Val109Asp and FTO rs9939609 (T/A) are significantly related to higher HOMA index (P = 0.030 and P = 0.046, respectively). However, omentin Val109Asp polymorphism was only related to individuals who were overweight/obese. Additionally, both omentin Val109Asp and FTO rs9939609 polymorphisms were significantly positively correlated to familial history of diabetes (P = 0.046 and P = 0.024, respectively).
Conclusions
Omentin V109D and FTO rs9939609 genetic variations may change insulin metabolism and have key roles in developing T2D through insulin resistance. Thus, the evaluation of these polymorphic regions may be helpful for predicting type 2 diabetes.
Similar content being viewed by others
Background
Type 2 diabetes mellitus (T2D) as the most common metabolic disorder arises from the impaired function of pancreatic beta cells and/or decreased sensitivity of peripheral tissues to insulin leading to insulin resistance (IR) and hyperglycemia. These defects might be associated with genetic and environmental factors that cause impairments in glucose, lipids, and amino acid metabolism [1, 2]. The IR is an important feature in T2D and is also implicated in other conditions, such as obesity and hypertension [3].
Obesity and overweight are defined as the body mass index (BMI) of ≥30 and ≥ 25 kg/m2, respectively. These conditions result from adipose tissue accumulation, especially in visceral (omental) and subcutaneous fats, and can change the level of adipokines. Obesity and overweight are closely related to diverse metabolic disorders, including IR, T2D, hypertension, dyslipidemia, and atherosclerosis [4, 5]. Based on the evidence, both lifestyle issues and genetic factors are involved in predisposing individuals to obesity [6, 7].
Omentin (also known as intelectin: ITLN) is one of the most major visceral fat adipokines expressed by genes 1 and 2. [8]. This adipokine is predominantly expressed in the vascular cells of the adipose tissue stroma, in addition to the epicardial fat, lungs, ovary, and placenta [4]. Expression of both Omentin 1 and Omentin 2 genes in visceral adipose tissue decreased with obesity [5]. The Omentin 1 is the main form of Omentin in circulation, and its exact biological role is still not well known [4, 9]. A number of studies have shown that Omentin enhances glucose uptake in human adipocytes by increasing insulin sensitivity [8, 10]. In addition, some studies indicated that plasma Omentin 1 level is inversely correlated with BMI, waist circumference, and IR as measured by homeostasis model assessment (HOMA). Moreover, there are data showing a positive correlation between Omentin and the levels of adiponectin and high-density lipoprotein (HDL) [5, 8].
It has been demonstrated in the literature that A326T (rs2274907) single nucleotide miss-sense polymorphism in the exon-4 of the Omentin 1 gene, which substitutes valine instead of aspartate at position 109 (Val109Asp or V109D), could be accompanied by T2D and obesity [4, 11, 12]. Structurally, position 109 is located outside the fibrinogen domain of Omentin protein. Therefore, Val109Asp and other sequence variations can lead to a real disease-causing mutation [11].
On the other hand, based on a body of evidence, the common rs9939609 polymorphism in the fat mass- and obesity-associated (FTO) gene is associated with higher BMI, risk of obesity, and subsequent T2D in different populations [12,13,14,15,16,17,18].
Sequence analysis of FTO protein has predicted a 2-oxoglutarate oxygenase activity with a central role for transcriptional regulation during the hypoxic response [19, 20]. Moreover, histone demethylation has been found [21], particularly in hypothalamic nuclei, which control energy balance and arcuate nucleus that is regulated by the feeding-fasting process [22].
The polymorphisms of Omentin Val109Asp (rs2274907) and FTO rs9939609 genes are of high importance in overweight, obesity, and T2D. Moreover, T2D is positively associated with visceral adipose tissue hypertrophy. With this background in mind, the present study was conducted to investigate the simultaneous presence of Omentin Val109Asp and FTO rs9939609 polymorphisms in the Iranian population with newly diagnosed T2D and their relationships with IR.
Materials and methods
Study population
A case-control study was performed in Bojnurd, Northeast of Iran, during December 2016–June 2018. Informed consents were obtained from the individuals included in the study. The study protocol conforms to the ethical guidelines of the Declaration of Helsinki as reflected in a prior approval granted by the Human Research Ethics Committee of North Khorasan University of Medical Sciences in Bojnurd (ethics code of IR.nkums.REC.1396.38).
The eligible participants corresponded to a total of 168 individuals referring to the clinical laboratory of Imam Reza Hospital of Bojnurd city, including 83 cases with T2D and 85 healthy people without any complications considered as the control group. The two groups were matched in terms of age and gender. The study participants had an age range of 20–80 years with no history of hyperglycemia. Based on the clinical and laboratory investigations performed by an endocrinologist, 83 patients with T2D were selected as cases [23]. The case group had a fasting blood sugar level (FBS) of > 7 mmol/L as examined in two different times. In addition, 85 individuals with a normal level of FBS were considered as the control group.
The structured questionnaire covered such information as age, gender, smoking habits, BMI, familial history of diabetes, and history of medication usage, especially antidiabetic, lipid-lowering, and antihypertensive agents. The exclusion criteria included a history of hyperglycemia, or hepatic, cardiovascular, rheumatologic, renal, and thyroid diseases, as well as taking antihypertensive, anti-diabetic, lipid-lowering, and anti-inflammatory medications 1 month prior to sampling.
Laboratory and molecular diagnosis
The FBS, triglyceride (TG), total cholesterol (TC), HDL cholesterol (HDL-C), and low-density lipoprotein cholesterol (LDL-C) were measured by enzymatic methods (Pars Azmun and Pishtaz Teb, Iran) using a biochemistry autoanalyzer (Dirui, China). Fasting serum insulin levels were also detected by enzyme-linked immunosorbent assay (ELISA) method (Demeditec Diagnostics GmbH, Germany) using a plate reader (BioTek, USA).
The HOMA-IR developed by Matthews et al. [24] is the most frequently employed technique both in clinical practice and epidemiological studies. Therefore, in the current study, IR assessment was carried out based on this model [25]. In this regard, HOMA-IR was calculated using the following equation: HOMA-IR = (Fasting insulin × Fasting glucose)/22.5 [26].
To analyze gene polymorphisms, blood samples were collected in ethylenediaminetetraacetic acid (EDTA)-coated vacuum tubes and stored at − 20 °C. The genomic deoxyribonucleic acid (DNA) was extracted using Proteinase K and column method according to the protocol of the manufacturer (Genet Bio, Korea). Afterwards, the DNA samples were randomly assessed by ultraviolet (UV) bio-spectrophotometer (Lambda, Japan), and their purity was checked by A260/ A280 ratio.
Omentin Val109Asp (V109D) and FTO rs9939609 gene polymorphisms were determined by polymerase chain reaction (PCR) using PreMix tubes (Genet Bio, Korea), followed by restriction fragment length polymorphism analysis (RFLP).
The PCR was used to amplify the amplicons of 471 and 182 bp for single-nucleotide polymorphisms (SNPs) in the exon-4 of Omentin and intron-1 of FTO, respectively. The oligonucleotide primers, namely F: 5′-GAGCCTTTAGGCCATGTCTCT-3′ and R: 5′-CTCTCCTTCTTCTCCAGCCCAT-3′, were utilized for Omentin Val 109 Asp polymorphism. In addition, we applied the primers F: 5′-AACTGGCTCTTGAATGAAATAGGATTCAGA-3′ and R: 5′- AGAGTAACAGAGACTATCCAAGTGCAGTAC-3′ for FTO rs9939609 polymorphism.
The PCRs were carried out by a Veriti Thermal Cycler (Applied Biosystems, USA) in which the DNA templates were denatured at 95 °C for 5 min. The amplification step consisted of 40 cycles of 45 s at 95 °C, 60 s at 59 °C (for OMENTIN V109D SNP) and 45 s at 56 °C (for FTO rs9939609 SNP), in addition to 72 °C for 45 s with a final extension of 5 min at 72 °C.
Afterward, the PCR products of Omentin V109D and FTO rs9939609 polymorphic regions were subjected to restriction enzyme analysis by digestion at 37 °C for 8 h with AccI (XmiI) and ScaI restriction endonucleases, respectively (ThermoFisher Scientific, USA). Within the exon-4 of Omentin gene, the Val encoded by polymorphic codon GTC can recognize AccI, whereas the GAC codon encoding Asp eliminates the AccI recognition site. In addition, ScaI enzyme recognized the single nucleotide polymorphism T to A in the first intron of FTO gene.
In the RFLP tests, each restriction endonuclease mixture with a total volume of 20 μl contained 5 μl amplified fragments, 12 μl DNase free distilled water, 2 μl appropriate buffer, and 1 μl of each restriction endonuclease. One unit of restriction enzyme is the amount of enzyme required to digest 1 μg of lambda DNA in 1 h at 37 °C for AccI and ScaI. The restriction products were separated by electrophoresis on a 3% agarose gel (Sinaclon, Iran) in TBE buffer, and then stained with DNA safe stain.
Statistical analyses
Statistical analysis was performed in SPSS software (version 18). The demographic and metabolic data were compared between the study groups using the t-test. All the data were expressed as mean and standard deviation. Comparison of the allele frequencies and genotype distribution between the case and control groups was performed using the Pearson’s Chi-squared test. In addition, the correlation between genotype alleles with chemicals and insulin levels was examined by Pearson correlation test. The Hardy-Weinberg equilibrium was assessed using http://scienceprimer.com/hardy-weinberg-equilibrium-calculator.
The Chi-square goodness of fit test was carried out to evaluate allele and genotype frequencies in the case and control groups. Moreover, the allele frequency of SNPs in the study groups was evaluated using odds ratio (OR) without adjusting for confounding factors, such as BMI. P-value less than 0.05 was considered statistically significant. The power calculation of this study was estimated at about 85%.
Results
Demographic and laboratory analysis
The demographic characteristics and biochemical profile for both healthy controls (n = 85) and T2D patients (n = 83) are shown in Table 1. The individuals in the case group had a familial history of diabetes and significantly higher weight, FBS, HOMA-IR, and TG, compared to the controls. In addition, the case group was hypertensive (Table 1). Considering the optimal HOMA-IR threshold of 1.85–1.95 reported by Esteghamati et al. [27] for the Iranian population, in the present study, 50.6 and 45.8% of the diabetic patients had moderate and severe IR with the HOMA-IRs of 2–4.5 and > 4.5, respectively. In addition, 36.5% of the healthy controls had a HOMA index of 2–4.5, and 6% of them had HOMA-IR of > 4.5 known as severe IR (OR: 34.78, 95% CI: 22.28–46.75, P = 0.0001; Table 2).
Distribution of Omentin Val109Asp and FTO rs9939609 polymorphisms
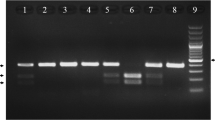
Regarding the presence of two alleles in Omentin gene, in the determination of Omentin V109D polymorphism, the undigested fragment (471 bp) was detected as a healthy homozygote for D allele (genotype DD). In addition, the digested fragments (274 and 197 bp) were identified as homozygotes for V allele (genotype VV). Moreover, both digested and undigested fragments (471, 274, and 191 bp) were detected as heterozygotes (genotype DV; Fig. 1a).
The electrophoretic patterns for determination of omentin Val 109 Asp and FTO rs9939609 polymorphisms. a Omentin V109D genotypes include DD (normal), DV (polymorphism in one allele), VV (polymorphism in two alleles). b FTO rs9939609 genotypes include TT (normal), TA (polymorphism in one allele), AA (polymorphism in two alleles)
Genotype distribution of Omentin V109D polymorphism was in the Hardy-Weinberg equilibrium. Out of 85 controls, 70.6, 27.1, and 2.3% of the individuals were found to have DD, DV, and VV genotypes, respectively. On the other hand, among the 83 cases, DD, DV, and VV genotypes were observed in 48.2, 49.4, and 2.4% of the participants, respectively.
Based on the results, V allele had the frequencies of 0.158 and 0.271 in the healthy controls and diabetic cases, respectively. There was a significant difference between the case and control groups in terms of relative genotype (P = 0.011) and allele frequency (P = 0.011; Table 3, Fig. 2). In addition, the presence of the DV and VV genotypes of Omentin V109D SNP was associated with a higher risk of T2D (OR = 1.98, 95% CI = 1.35–2.64, P = 0.001; Table 3).
In terms of FTO rs9939609 polymorphism, the undigested fragment (182 bp) was found to be a healthy homozygote (genotype TT) and the digested fragments (154 and 28 bp) were detected as polymorphic homozygotes (genotype AA). Moreover, both digested and undigested fragments (182, 154, and 28 bp) were identified as heterozygotes (genotypes TA) (Fig. 1b).
The FTO rs9939609 genotype frequency, as well as allele frequency is shown in Table 3. The genotype frequency of heterozygosity for TA alleles in the case and control groups was 51.8 and 37.6%, respectively. Moreover, the frequencies of two-allele polymorphism (AA) in the case and control groups were obtained as 13.3 and 1.2%, respectively. Regarding the frequency of A allele, it was estimated at 0.392 and 0.2 in the diabetic cases and healthy controls, respectively (OR = 2.57, 95% CI = 1.87–3.04, P = 0.0001; Table 3, Fig. 3).
Concerning the higher prevalence of Omentin and FTO gene polymorphisms in the subjects with higher fat mass, the investigation of the genotype distribution of these polymorphic regions in the study population would be helpful. Table 4 presents the combinational Omentin-FTO genotypes in the study population. In this regard, the included genotypes were DD/TT (DT), DV/TT (DVT), VV/TT (VT), DD/TA (DTA), DV/TA (DVTA), VV/TA (VTA), DD/AA (DA), DV/AA (DVA), and VV/AA (VA). The results showed a significant difference between the two study groups in this respect so that the polymorphic genotypes were more frequent in the diabetic individuals than in the healthy controls (P < 0.0001).
Relationship of Omentin V109D and FTO rs9939609 polymorphisms with IR and other biochemical parameters
Table 5 demonstrates the distribution of genotype/allele frequencies for Omentin and FTO polymorphisms in the participants according to HOMA-IR index for determining their genetic polymorphism relationship with IR. In terms of Omentin V109D SNP, 17.3 and 4.2% of the subjects with increased and normal HOMA index had V polymorphic allele, respectively (OR = 1.78, 95% CI = 1.57–1.96, P = 0.03).
Regarding FTO rs9939609 SNP, 23% of the participants with a high HOMA index had A polymorphic allele, while 6.6% of the individuals with a normal HOMA index had A polymorphic allele (OR = 2.05, 95% CI = 1.88–2.21, P = 0.046). The correlation tests indicated that Omentin D109V polymorphism was correlated with higher HOMA-IR (P = 0.011), while FTO polymorphism was not significantly correlated with IR according to HOMA-IR (P = 0.062). Furthermore, there was no correlation between Omentin V109D and FTO polymorphisms (P = 0.084).
The results showed that 21.8% of the participants with overweight/obesity had FTO A allele, while 7.8% of the individuals with normal BMI had FTO A allele. There was no significant genotype-related difference between the study groups with high BMI concerning the FTO polymorphism (P = 0.383). In addition, 17 and 4.5% of the subjects with overweight/obesity and normal BMI had Omentin V allele, Omentin respectively (P = 0.048). The results of the Pearson correlation test showed that FTO polymorphism was not correlated with high BMI (P = 0.054), while Omentin polymorphism was correlated with higher BMI (P = 0.002).
The frequency distributions of FTO rs9939609 polymorphic A allele were estimated at 19.4 and 10.1% in the individuals with a familial history of diabetes and those without the history, respectively (P = 0.024). Moreover, the Omentin V allele was significantly correlated with the familial history of diabetes (14.3% for cases vs. 7.2% for controls; P = 0.046). On the other hand, the frequency distributions of FTO A allele were 12.5 and 16.9% in the hypertensive and normotensive participants, respectively (P = 0.099).
Furthermore, the Omentin V allele did not have a significant correlation with hypertension (9.2% for cases vs. 12.2% for controls; P = 0.179). Concerning the relationship between the Omentin and FTO genotypes, as well as between allele frequencies and lipid profile (e.g., TG, TC, HDL-C, and LDL-C), no significant difference was found between the polymorphic alleles and the lipid profile.
Discussion
Central obesity and accumulation of visceral adipose tissue are known as risk factors for IR development and subsequent T2D [4, 5]. Diabetes mellitus and IR are affected by genetic factors, as well as acquired overweight or obesity. As a result, it is important to find the gene polymorphisms related to adipose tissues, especially in people with a familial history of diabetes.
The relationship of Omentin Val 109 Asp and FTO rs9939609 genetic variations with newly diagnosed diabetes is not well determined yet. Consequently, we investigated these polymorphisms in some patients with T2D from the Northeast of Iran for the first time through clinical examinations and laboratory tests. The results of the present study revealed significant relationship between both FTO rs9939609 and Omentin Val 109 Asp polymorphisms with T2D in the studied population (P = 0.0001 and P = 0.011, respectively).
Moreover, our findings through HOMA-IR indicated that both gene polymorphisms of FTO rs9939609 (T/A) and Omentin rs2274907 (Val 109 Asp) were significantly related to IR (P = 0.046, P = 0.03, respectively). In terms of BMI, the Omentin V109D polymorphism was observed to have a significant relationship with overweight/obesity. In addition, both FTO rs9939609 and Omentin Val 109 Asp polymorphisms had a significant positive correlation with a familial history of diabetes (P = 0.024 and P = 0.046, respectively).
Several studies have investigated the relationship of Val 109 Asp polymorphism in Omentin gene and/or Omentin-1 levels with different diseases, such as diabetes [11], rheumatoid arthritis [28], psoriasis [29], osteoporosis [30], and coronary artery disease [31]. Some studies have shown that circulating Omentin-1 may enhance glucose uptake in human adipocytes by insulin via Akt signaling [8,9,10].
de Souza Batista et al. suggested that the plasma level of Omentin-1 was inversely correlated with BMI, waist circumference, leptin levels, and HOMA-IR. However, it was positively correlated with adiponectin and HDL levels. Moreover, they reported the downregulation of both Omentin 1 and Omentin 2 genes in obese people [5]. Although we did not measure the plasma Omentin-1 in the present study, it was observed that the A326T (rs2274907) genetic variation in the Omentin 1 gene could have a relationship with higher BMI, as well as IR.
Some studies have revealed an inverse relationship between plasma Omentin-1 levels and the obesity and IR [5]. Moreover, some researches showed that Omentin Val 109 Asp gene polymorphism is correlated with obesity and diabetes mellitus [11, 32]. It seems that Asp substitution by Val in position 109 of Omentin-1 might diminish the activity and/or lifetime of circulating Omentin-1. Therefore, concerning the positive effect of Omentin on glucose uptake by insulin receptors [8], the Val 109 Asp gene polymorphism may be inversely related to plasma Omentin and insulin levels/functions.
Various studies indicated that Omentin Val109Asp gene polymorphism is associated with T2D [11, 33]. Nonetheless, this genetic variation and its correlation with IR have not been investigated in patients newly diagnosed with T2D. According to the results of the present study, the polymorphic V allele of Omentin gene was correlated to two critical factors for T2D development, namely IR and BMI. Consequently, this polymorphism was found to be associated with T2D occurrence.
Schäffler et al. conducted a study on Caucasian patients with T2D or chronic inflammatory bowel disease (IBD). They stated that V allele was present in 26% of the healthy controls, while 30 and 31% of the patients with T2D and IBD had V allele, respectively. On the other hand, they did not show any significant difference in genotype distribution between the study groups. Furthermore, in the mentioned study, no significant relationship was found between the genotype subgroups and the anthropometric and laboratory parameters in diabetic patients [11].
Mrozikiewicz-Rakowska et al. reported that the rs2274907 (V 109 D) variant of Omentin gene is associated with the elevated prevalence of diabetic foot in patients with diabetes mellitus. However, they did not observe a significant difference in the distribution of alleles between the diabetic individuals with and without diabetic foot [33]. Our results also showed a significant difference between the study groups in terms of Omentin V allele.
In this study, the genotype distributions of Asp/Val were 50 and 34.3% in the diabetic patients and healthy controls, respectively. In addition, the genotype distributions of Val/Val were obtained as 3.8 and 1.4% in the diabetic patients and healthy controls, respectively. Moreover, statistical analysis confirmed that the probability risk for T2D in individuals who had V allele was 1.98 times higher than that for people with normal allele.
Bahadori et al. concluded that Iranian women with Val/Val genotype had a greater risk of obesity complications in comparison to those with Asp/Asp [32]. Isakova et al. investigating Kyrgyz population reported that the Val/Val genotype was significantly more prevalent in the patients with abdominal obesity than in the healthy controls (OR = 3.12). However, the allelic variants of Omentin Val 109 Asp polymorphism did not correlate with abdominal obesity [34].
In line with the latter results, our data indicated that Val/Val homozygous genotype was more frequent in overweight or obese people. In addition, there was a significant relationship between the allelic genotypes of Omentin Val 109 Asp polymorphism and BMI. Kohan et al. investigated the influence of Omentin V109D polymorphism on genetic susceptibility to nonalcoholic fatty liver disease in Iranian population. They demonstrated that the frequency of Omentin V 109 D gene polymorphism in these patients was significantly different from that of the healthy controls (OR = 2.3) [35].
It is hypothesized that Omentin Val 109 Asp gene polymorphism may have a role in lipid accumulation in the liver. Furthermore, Omentin levels and the V 109 D polymorphism are in association Omentin with BMI and fatty liver disease. Therefore, further studies concerning the unknown mechanism of Omentin effect on lipogenesis and lipid accumulation may render helpful results.
Several studies indicated that the fat mass and obesity-associated (FTO) gene, especially the FTO rs9939609 variant, has a relationship with body weight and fat mass [12, 15, 36,37,38,39]. However, some studies in Asia did not find any relationship between FTO rs9939609 A polymorphic allele and overweight [16, 40]. The FTO rs9939609 polymorphism is less frequent in East Asians (12.6%) [38], compared to that in Europeans (45%) and West Africans (52%) [12, 14].
Although some studies showed a significant relationship between FTO rs9939609 variant and T2D [41], others did not find any relationship [38]. In addition, the FTO variant is correlated with a higher occurrence of T2D. However, this effect can be entirely explained by the BMI differences between the diabetic and healthy controls [12, 42, 43].
Chey et al. performed a study on Malaysian multi-ethnic population, including Malays, Chinese, and Indians. They concluded that the genotype distribution and allele frequencies of FTO rs9939609 gene variant were significantly different among the ethnicities. On the other hand, they did not observe any significant relationship between FTO polymorphism and obesity [16].
In the present study, the relationship between FTO rs9939609 gene variant and higher BMI was not significant, which is consistent with the findings of the mentioned study. However, our results showed that this polymorphism was correlated with IR. Moreover, the results of the current study demonstrated that the participants with A allele had a ~ 2.6-fold higher susceptibility to T2D than the control subjects.
Freathy et al. indicated that FTO rs9939609 A allele is correlated with higher fasting insulin, FBS, TG, and lower HDL-C. They concluded that FTO polymorphism has a relationship with metabolic syndrome (OR = 1.17). It is noteworthy that they found no evidence of the mentioned relationships when adjusted for BMI. Moreover, no significant relationship was observed between FTO polymorphism and serum alanine aminotransferase, γ-glutamyl-transpeptidase, LDL-C, HbA1c, and systolic and diastolic blood pressure [7]. In accordance with the mentioned study, our findings demonstrated that FTO A allele is correlated with higher FBS in newly diagnosed T2D patients (P = 0.0001). However, there was no significant relationship between this polymorphic allele and lipid profile.
Chang et al. confirmed the role of FTO genetic variants in obesity and T2D in the Chinese population by a genome-wide study on 19 SNPs. They found that the rs9939609 A allele was strongly correlated with obesity and higher BMI. However, they did not observe significant relationships between 19 SNPs with T2D or other obesity-related traits [14]. In contrast to the results obtained by Chang et al., our findings demonstrated no significant correlation between rs9939609 A allele and higher BMI. In addition, our data showed that FTO A allele is associated with IR and T2D. These controversial data confirm the effect of ethnicity in distinct populations on FTO rs9939609 polymorphism.
The results of the present study demonstrated that FTO rs9939609 polymorphism has a relationship with the familial history of diabetes (P = 0.024), IR (P = 0.046), and T2D (P = 0.0001) in Northeast Iranian population. However, no relationship was detected between FTO A allele and higher BMI.
Our data showed that each additional FTO A allele could alter the risk of diabetes with an OR of 2.6 and 95% CI of 1.87–3.04 when the case and control subjects are not matched in terms of BMI. The FTO A allele had a frequency of 39.2 and 20% FTO in diabetic patients and healthy people, respectively. In addition, 23% of the individuals with a higher HOMA index considered as moderate or severe IR had A allele. This might be indicative of the role of FTO rs9939609 A allele in IR. Therefore, further studies are required to examine whether FTO polymorphism alters insulin secretion or targets tissue response to insulin.
One of the limitations of the present study is its relatively small sample size. As a result, the frequency of genetic polymorphisms should be confirmed by further investigations in a larger population. It should be mentioned that in the current study, sample size was selected according to the frequency of T2D among the population residing in Bojnurd. In addition, it was difficult to find new diabetic patients to exclude the probable effects of anti-diabetic drugs on study gene polymorphisms and other clinical or biochemical variables, compared to other studies focused on gene polymorphisms in patients with overt diabetes.
Conclusion
The findings of this study demonstrated that 96.4% of the newly diagnosed Iranian patients with T2D had IR as estimated by HOMA-IR index. Moreover, the statistical analysis showed that the probability risk of disease in individuals with IR was 34.8 times higher than that in the subjects with normal HOMA index. In the current study, both Omentin V109D and FTO rs9939609 polymorphisms had a relationship with IR and familial history of diabetes in the newly diagnosed T2D patients.
In addition, the Omentin V109D polymorphism (not FTO rs9939609) was related to higher BMI. Moreover, combinational genotyping confirmed that DV/TA (one polymorphic allele in each studied gene), DD/AA (two polymorphic alleles of FTO versus normal Omentin genotype), and DV/AA (one polymorphic allele of Omentin and two polymorphic alleles of FTO) genotypes were more frequent in the diabetic patients than in the healthy controls.
These results suggested that Omentin V 109 D and FTO rs9939609 genetic variations may change insulin metabolism, especially via the target tissue receptors leading to T2D development through IR. Therefore, the evaluation of these polymorphic regions may be helpful for predicting T2D and the related morbidities in normal subjects with a familial history of diabetes and high HOMA index. As a result, the implementation of genome-wide association studies on more gene polymorphisms related to IR and familial histories of diabetes is an efficient strategy by determining the newly diagnosed diabetic patients and prevent diabetic morbidities.
Availability of data and materials
The authors confirm that all of the data and material of the manuscript are available for more consideration by the journal.
Abbreviations
- BMI:
-
Body mass index
- FBS:
-
Fasting blood sugar
- FTO:
-
Fat mass and obesity associated
- HDL-C:
-
High density lipoprotein-cholesterol
- HOMA-IR:
-
Homeostasis model assessment - estimated insulin resistance
- IR:
-
Insulin resistance
- LDL-C:
-
Low density lipoprotein-cholesterol
- PCR:
-
Polymerase chain reaction
- RFLP:
-
Fragment length polymorphism analysis
- T2D:
-
Type 2 diabetes
- TC:
-
Total cholesterol
- TG:
-
Triglycerides
References
Conget I. Diagnosis, classification and pathogenesis of diabetes mellitus. Rev Esp Cardiol. 2002;55:528–35 (In Spanish).
Marchetti P, Lupi R, Federici M, Marselli L, Masini M, et al. Insulin secretory function is impaired in isolated human islets carrying the Gly972→ Arg IRS-1 polymorphism. Diabetes. 2002;51:1419–24.
Gayoso-Diz P, Otero-González A, Rodriguez-Alvarez MX, Gude F, García F, et al. Insulin resistance (HOMA-IR) cut-off values and the metabolic syndrome in a general adult population: effect of gender and age: EPIRCE cross-sectional study. BMC Endocr Disord. 2013;13(1):47.
Halabis M, Dziedzic M, Warchulinska J, Kaznowska-Bystryk I, Solski J. Omentin-a new adipokine with many roles to play. Curr Issues Pharm Med Sci. 2015;28:176–80.
de Souza Batista CM, Yang RZ, Lee MJ, Glynn NM, Yu DZ, et al. Omentin plasma levels and gene expression are decreased in obesity. Diabetes. 2007;56:1655–61.
Must A, Spadano J, Coakley EH, Field AE, Colditz G, Dietz WH. The disease burden associated with overweight and obesity. JAMA. 1999;282(16):1523–9.
Freathy RM, Timpson NJ, Lawlor DA, Pouta A, Ben-Shlomo Y, Ruokonen A, Ebrahim S, Shields B, Zeggini E, Weedon MN, Lindgren CM. Common variation in the FTO gene alters diabetes-related metabolic traits to the extent expected given its effect on BMI. Diabetes. 2008;57(5):1419-26.
Yang RZ, Lee MJ, Hu H, Pray J, Wu HB, et al. Identification of omentin as a novel depot-specific adipokine in human adipose tissue: possible role in modulating insulin action. Am J Physiol Endocrinol Metab. 2006;290:E1253–61.
Zhong X, Zhang HY, Tan H, Zhou Y, Liu FL, et al. Association of serum omentin-1 levels with coronary artery disease. Acta Pharmacol Sin. 2011;32(7):873.
Tan BK, Adya R, Randeva HS. Omentin: a novel link between inflammation, diabesity, and cardiovascular disease. Trends Cardiovasc Med. 2010;20:143–8.
Schäffler A, Zeitoun M, Wobser H, Buechler C, Aslanidis C, et al. Frequency and significance of the novel single nucleotide missense polymorphism Val109Asp in the human gene encoding omentin in Caucasian patients with type 2 diabetes mellitus or chronic inflammatory bowel diseases. Cardiovasc Diabetol. 2007;6:3.
Frayling TM, Timpson NJ, Weedon MN, Zeggini E, Freathy RM, Lindgren CM, Perry JR, Elliott KS, Lango H, Rayner NW, Shields B. A common variant in the FTO gene is associated with body mass index and predisposes to childhood and adult obesity. Science. 2007;316(5826):889-94.
Scuteri A, Sanna S, Chen WM, Uda M, Albai G, Strait J, Najjar S, Nagaraja R, Orrú M, Usala G, Dei M. Genome-wide association scan shows genetic variants in the FTO gene are associated with obesity-related traits. PLoS Genet. 2007;3(7):e115.
Chang YC, Liu PH, Lee WJ, Chang TJ, Jiang YD, Li HY, Kuo SS, Lee KC, Chuang LM. Common variation in the fat mass and obesity-associated (FTO) gene confers risk of obesity and modulates BMI in the Chinese population. Diabetes. 2008;57(8):2245-52.
Karasawa S, Daimon M, Sasaki S, Toriyama S, Oizumi T, Susa S, et al. Association of the common fat mass and obesity associated (FTO) gene polymorphism with obesity in a Japanese population. Endocr J. 2010;57(4):293–301.
Chey WW, Fan SH, Say YH. Association of fat Mass and Obesity-Associated (FTO) gene rs9939609 variant with obesity among multi-ethnic Malaysians in Kampar, Perak. Sains Malaysiana. 2013;42(3):365–71.
Quan LL, Wang H, Tian Y, Mu X, Zhang Y, Tao K. Association of fat-mass and obesity-associated gene FTO rs9939609 polymorphism with the risk of obesity among children and adolescents: a meta-analysis. Eur Rev Med Pharmacol Sci. 2015;19(4):614–23.
Raza ST, Abbas S, Siddiqi Z, Mahdi F. Association between ACE (rs4646994), FABP2 (rs1799883), MTHFR (rs1801133), FTO (rs9939609) genes polymorphism and type 2 diabetes with dyslipidemia. Int J Mol Cell Med. 2017;6(2):121.
Ivan M, Kondo K, Yang H, Kim W, Valiando J, Ohh M, et al. HIFα targeted for VHL-mediated destruction by proline hydroxylation: implications for O2 sensing. Science. 2001;292(5516):464–8.
Jaakkola P, Mole DR, Tian YM, Wilson MI, Gielbert J, Gaskell SJ, et al. Targeting of HIF-α to the von Hippel-Lindau ubiquitylation complex by O2-regulated prolyl hydroxylation. Science. 2001;292(5516):468–72.
Klose RJ, Yamane K, Bae Y, Zhang D, Erdjument-Bromage H, Tempst P, et al. The transcriptional repressor JHDM3A demethylates trimethyl histone H3 lysine 9 and lysine 36. Nature. 2006;442(7100):312.
Gerken T, Girard CA, Tung YC, Webby CJ, Saudek V, Hewitson KS, et al. The obesity-associated FTO gene encodes a 2-oxoglutarate-dependent nucleic acid demethylase. Science. 2007;318(5855):1469–72.
American Diabetes Association. Diagnosis and classification of diabetes mellitus. Diabetes Care. 2010;33(Supplement 1):S62–9.
Matthews DR, Hosker JP, Rudenski AS, Naylor BA, Treacher DF, et al. Homeostasis model assessment: insulin resistance and β-cell function from fasting plasma glucose and insulin concentrations in man. Diabetologia. 1985;28:412–9.
Salgado AL, Carvalho LD, Oliveira AC, Santos VN, Vieira JG, et al. Insulin resistance index (HOMA-IR) in the differentiation of patients with non-alcoholic fatty liver disease and healthy individuals. Arq Gastroenterol. 2010;47:165–9.
Wallace TM, Levy JC, Matthews DR. Use and abuse of HOMA modeling. Diabetes Care. 2004;27(6):1487–95.
Esteghamati A, Ashraf H, Esteghamati AR, Meysamie A, Khalilzadeh O, et al. Optimal threshold of homeostasis model assessment for insulin resistance in an Iranian population: the implication of metabolic syndrome to detect insulin resistance. Diabetes Res Clin Pract. 2009;84:279–87.
Yaykasli KO, Yaykasli E, Ataoglu S, Ozsahin M, Memisogullari R, Celebi E, Ucgun T, Ozcan ME, Uslu M, Yuce H. The frequency of omentin Val109Asp polymorphism and the serum level of omentin in patients with rheumatoid arthritis. Acta Medica Mediterranea. 2013;29(3):521–6.
Turan H, Yaykasli KO, Soguktas H, Yaykasli E, Aliagaoglu C, et al. Omentin serum levels and omentin gene Val109Asp polymorphism in patients with psoriasis. Int J Dermatol. 2014;53:601–5.
Boron D, Czerny B, Bartkowiak-Wieczorek J, Sieron D, Wolski H. Omentin polymorphism and its relations to bone mineral density in women. Arch Med Res. 2015;46:173–80.
Yörük Ü, Yaykasli KO, Özhan H, Memisogullari R, Karabacak A, et al. Association of omentin Val109Asp polymorphism with coronary artery disease. Anadulu Kardiyoloji Derg. 2014;14:511.
Bahadori M, Kohan L, Jafari N. Association of assessment between Val109Asp omentin gene and obesity in Iranian women. Iran J Diabetes Metab. 2015;14(2):127–32.
Mrozikiewicz-Rakowska B, Sobczyk-Kopcioł A, Szymański K, Nehring P, Szatkowski P, Bartkowiak-Wieczorek J, Bogacz A, Aniszczuk A, Drygas W, Płoski R, Czupryniak L. Role of the rs2274907 allelic variant of the ITLN1 gene in patients with diabetic foot. Pol Arch Intern Med. 2017;127:319–27.
Isakova J, Talaibekova E, Vinnikov D, Aldasheva N, Mirrakhimov E, et al. The association of Val109Asp polymorphic marker of intelectin 1 gene with abdominal obesity in Kyrgyz population. BMC Endocr Disord. 2018;18:15.
Kohan L, Safarpur M, Abdollahi H. Omentin-1 rs2274907 and resistin rs1862513 polymorphisms influence genetic susceptibility to nonalcoholic fatty liver disease. Mol Biol Res Commun. 2016;5(1):11.
Legry V, Cottel D, Ferrières J, Arveiler D, Andrieux N, Bingham A, et al. Effect of an FTO polymorphism on fat mass, obesity, and type 2 diabetes mellitus in the French MONICA study. Metabolism. 2009;58(7):971–5.
Peeters A, Beckers S, Verrijken A, Roevens P, Peeters P, Van Gaal L, et al. Variants in the FTO gene are associated with common obesity in the Belgian population. Mol Genet Metab. 2008;93(4):481–4.
XI B, MI J. FTO polymorphisms are associated with obesity but not with diabetes in east Asian populations: a meta-analysis. Biomed Environ Sci. 2009;22(6):449–57.
Xi B, Shen Y, Zhang M, Liu X, Zhao X, Wu L, et al. The common rs9939609 variant of the fat mass and obesity-associated gene is associated with obesity risk in children and adolescents of Beijing, China. BMC Med Genet. 2010;11(1):107.
Li H, Wu Y, Loos RJ, Hu FB, Liu Y, Wang J, et al. Variants in the fat mass–and obesity-associated (FTO) gene are not associated with obesity in a Chinese Han population. Diabetes. 2008;57(1):264–8.
Binh TQ, Phuong PT, Nhung BT, Thoang DD, Lien HT, Van Thanh D. Association of the common FTO-rs9939609 polymorphism with type 2 diabetes, independent of obesity-related traits in a Vietnamese population. Gene. 2013;513(1):31–5.
Zeggini E, Weedon MN, Lindgren CM, Frayling TM, Elliott KS, Lango H, et al. Replication of genome-wide association signals in UK samples reveals risk loci for type 2 diabetes. Science. 2007;316(5829):1336–41.
Saxena R, Voight BF, Lyssenko V, Burtt NP, de Bakker PI, Chen H, et al. Genome-wide association analysis identifies loci for type 2 diabetes and triglyceride levels. Science. 2007;316(5829):1331–6.
Acknowledgments
Authors thank Research center of North Khorasan University of Medical Sciences because of financial support. We also thank Clinical Laboratory of Imam Reza Hospital, Bojnurd for specimen preparation and biochemical analysis. Authors have a special appreciation to Dr. Amir Azimian for primer design.
Funding
This study was supported by the Research center of North Khorasan University of Medical Sciences by the foundation under No: 96/1017.
Author information
Authors and Affiliations
Contributions
AK, as the study designer, supervised the laboratory and molecular testing. MKB, as the laboratory technologist, performed the biochemical analyses and molecular testing for omentin gene polymorphism. HS, as the clinical endocrinologist, selected study population including diabetic patients who were newly diagnosed and healthy control subjects and referred them to the clinical laboratory. GG performed the molecular testing for the FTO gene polymorphism. FA, as an intern medical student, assisted in the selection of diabetic patients. All authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Ethics approval and consent to participate
This study approved by the human research ethics committee of North Khorasan University of Medical Sciences (ethics code: IR.nkums.REC.1396.38).
Consent for publication
The authors are consent this manuscript has not been published elsewhere.
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Khoshi, A., Bajestani, M.K., Shakeri, H. et al. Association of Omentin rs2274907 and FTO rs9939609 gene polymorphisms with insulin resistance in Iranian individuals with newly diagnosed type 2 diabetes. Lipids Health Dis 18, 142 (2019). https://doi.org/10.1186/s12944-019-1085-5
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12944-019-1085-5