Abstract
Background
The SWI/SNF ATP dependent chromatin remodeling complex is a multi-subunit complex, conserved in eukaryotic evolution that facilitates nucleosomal re-positioning relative to the DNA sequence. In recent years the SWI/SNF complex has emerged to play a role in cancer development as various sub-units of the complex are found to be mutated in a variety of tumors. One core-subunit of the complex, which has been well established as a tumor suppressor gene is SMARCB1 (SNF5/INI1/BAF47). Mutation and inactivation of SMARCB1 have been identified as the underlying mechanism leading to Malignant Rhabdoid Tumors (MRT) and Atypical Teratoid/Rhabdoid Tumors (AT/RT), two highly aggressive forms of pediatric neoplasms.
Methods
We present a phosphoproteomic study of Smarcb1 dependent changes in signaling networks. The SILAC (Stable Isotopic Labeling of Amino Acids in Cell Culture) protocol was used to quantify in an unbiased manner any changes in the phosphoproteomic profile of Smarcb1 deficient murine rhabdoid tumor cell lines following Smarcb1 stable re-expression and under different serum conditions.
Results
This study illustrates broad changes in the regulation of multiple biological networks including cell cycle progression, chromatin remodeling, cytoskeletal regulation and focal adhesion. Specifically, we identify Smarcb1 dependent changes in phosphorylation and expression of the EGF receptor, demonstrate downstream signaling and show that inhibition of EGFR signaling specifically hinders the proliferation of Smarcb1 deficient cells.
Conclusions
These results support recent findings regarding the effectivity of EGFR inhibitors in hindering the proliferation of human MRT cells and demonstrate that activation of EGFR signaling in Rhabdoid tumors is SMARCB1 dependent.
Similar content being viewed by others
Introduction
The SWI/SNF ATP dependent chromatin remodeling complex is a multi-subunit complex, conserved in eukaryotic evolution, that facilitates nucleosome re-positioning relative to the DNA sequence [1]. The SWI/SNF complex has been found to play a role in fundamental cellular functions such as transcriptional regulation, DNA replication and DNA repair, but is mainly regarded to as a broad transcriptional co-activator / co-repressor [2].
In recent years various deep sequencing studies have demonstrated repeating mutations in sub-units of the SWI/SNF complex across various types of tumors [3, 4]. One core-subunit of the complex, which has been well established as a tumor suppressor gene is SMARCB1 (SNF5/INI1/BAF47). As more and more tumors are deep sequenced, mutations in SMARCB1 are found across a growing spectrum of cancers. More specifically, inactivating mutations of SMARCB1 are found in all Malignant Rhabdoid Tumors (MRT) and Atypical Teratoid/Rhabdoid Tumors (AT/RT), two highly aggressive forms of pediatric neoplasms [5]. In spite of significant progress in treatment over recent years, long-term prospects for MRT and AT/RT patients remain poor as the tumors demonstrate relative resistance to conventional chemotherapy and radiotherapy and tumor resection is in many cases not possible [6, 7].
MRT which manifests in the kidney and AT/RT of the central nervous system are unique in that apart from the SMARCB1 locus they show unusually low mutation rate. Several recent deep sequencing studies have revealed the poor mutational landscape of these tumors [8–11]. This finding suggests that SMARCB1 inactivation alters multiple pathways that promote cellular transformation, and results in the simultaneous acquisition of the various hallmarks of a transformed cancer cell [12] through a singular mutation.
We have been studying SMARCB1 associated transformation using cell lines derived from rhabdoid tumors which developed in Smarcb1 heterozygous p53 null mice [13]. These tumor cell lines show loss of heterozygosity and lack Smarcb1. Restoration of Smarcb1 expression had a minor effect on cell proliferation in culture but completely ablated the tumorigenic capacity of xenografted tumor cells [14]. This result indicates that by comparing the Smarcb1 deficient and proficient tumor cells one can define Smarcb1 dependent changes which are functionally relevant to transformation. Using this system we previously showed that Smarcb1 deficiency results in persistent AKT activation. Accordingly we found that Smarcb1 deficient tumor cells are specifically vulnerable to AKT or PI3-kinase inhibition [14].
In this study we use a high throughput phosphoproteomic analysis comparing Smarcb1 deficient and proficient tumor cells to further identify aberrant signaling associated with Smarcb1 deficiency. We describe Smarcb1 dependent constitutive phosphorylation of the EGFR, which is also transcriptional elevated in Smarcb1 deficient cells and demonstrate that inhibition of the EGFR/ERBB signaling pathway inhibits proliferation of Smarcb1 deficient tumor cells. We further identify multiple biological networks and kinases whose regulation is altered in Smarcb1 deficient tumor cells in a Smarcb1 dependent manner.
Results
Profound changes in the phosphoproteomic landscape between Smarcb1 proficient and deficient cells
We previously reported persistent activation of AKT in Smarcb1 deficient cells [14], yet we could not identify the cause of this Smarcb1 dependant activation. To better characterize altered signaling pathways in Smarcb1 deficient tumor cells, which may contribute to the transformation process and to AKT activation, we conducted an unbiased quantitative phospho-proteomic analysis designed to identify differentially phosphorylated peptides between Smarcb1 proficient and deficient tumor cells.
The triple - SILAC (Stable Isotopic Labeling of Amino Acids in Cell Culture) protocol [15–18] was used to compare Smarcb1 deficient and proficient tumor cells (Cell line 365 [14] containing an empty retroviral vector as control (pMIG) or pMIG-Smarcb1 respectively) (Fig. 1a, b). Because serum is a rich source for signals, we expected Smarcb1 dependent differential activation of signaling pathways to reflect better under serum starvation conditions, as demonstrated by the differential phosphorylation of AKT (Fig. 1b, c and [14]).
Phosphoproteomic profiling reveals robust Smarcb1 dependent changes in protein phosphorylation. Matching Smarcb1 proficient and deficient tumor cells were generated by re-introducing SMARCB1 (pMIG- Smarcb1) or an empty vector as control (pMIG) [14]. a The outline of the phosphoproteomic study. Two triple-SILAC experiments were conducted. The first set allowed evaluation of Smarcb1 dependent changes when cultured in normal serum whilst the second set allowed for the evaluation under serum starvation. Smarcb1 proficient cells grown in normal serum and light isotopic labeling were included in both sets to allow comparison between the two sets. b Western blot demonstrating differential AKT phosphorylation in Smarcb1 deficient versus proficient cells. Bar graph shows quantification of western blot presented as fold change in AKT phosphorylation in pMIG/Smarcb1 cells normalized to beta-actin. c Volcano plot depicting Smarcb1 dependent changes in site phosphorylation across the two sets. X-axis is the log2 ratio of the abundances of specific residues between Smarcb1 proficient and deficient cells. Negative values for highly phosphorylated in Smarcb1 deficient cells. Y-axis is the logarithmic scale for the P-value of the fold change. For P.V < 0.05; Violet dataset represents residues altered in high serum; Green dataset represents residues altered under serum starvation. d Venn-diagram portraying the residues found to be differentially phosphorylated in a Smarcb1-dependent manner. Under 10 % serum, 458 residues are highly phosphorylated in Smarcb1 proficient cells (In red) whilst 434 residues are highly phosphorylated in Smarcb1 deficient cells (In blue). Under low serum, 384 residues (roughly two thirds) are highly phosphorylated in Smarcb1 proficient cells, whilst 233 residues are highly phosphorylated in Smarcb1 deficient cells. Overall 205 residues are differentially phosphorylated between Smarcb1 proficient and deficient cells regardless of the serum conditions
All in all 10701 phosphorylation sites from 3655 distinct proteins were identified using high resolution mass spectrometric analysis. 891 sites from 510 distinct proteins were differentially phosphorylated in a statistically significant manner between Smarcb1 deficient and proficient cells under high serum, whilst under serum starvation 616 sites from 407 distinct proteins demonstrated differential phosphorylation (P-value < 0.05). Overall 205 residues from 134 distinct proteins exhibited a differential phosphorylation between Smarcb1 deficient and proficient cells regardless of the growth serum condition (Fig. 1c, Additional file 1: Table S1 and Table S2).
Altered regulation of cell adhesion and cytoskeletal organization in Smarcb1 deficient cells
Across all the mentioned sets a statistically significant enrichment was found for proteins related to several GO annotations including actin cytoskeleton and focal adhesion (Fig. 2a). We previously profiled transcriptional changes brought about following re-introduction of Smarcb1 in the same tumor cell line and found enrichment for cytoskeleton and focal adhesion categories already at transcription level [14]. However, correlating changes in phosphorylation levels with changes in levels of expression (Fig. 2b) demonstrates that only a small fraction of the changes in phosphorylation are correlated to changes in gene expression, suggesting that altered transcription accompanies altered regulation of these cellular functions.
GO annotation enrichment analysis of differentially phosphorylated proteins. a Gene ontology (GO) annotations enrichment among proteins found to be differentially phosphorylated between SMARCB1 proficient and deficient cells. Evaluated using the David Bioinformatics tool, x-axis indicates log base for Benjamini corrected P-value. Violet dataset represents differentially phosphorylated peptides in high serum; green dataset represents differentially phosphorylated peptides in serum starvation. b Scatter plots depicting Smarcb1 dependent differentially phosphorylated peptides and their expression level in high or low serum. Negative values denote highly expressed/phosphorylated in Smarcb1 deficient cells, positive values denote highly expressed/phosphorylated in Smarcb1 proficient cells. Peptides corresponding to enriched GO categories (from panel 2a) are highlighted in red. Group 1 includes GO annotations: Cytoskeleton, Actin cytoskeleton and focal adhesion. Group 2: Cell cycle. Group 3: Small GTPase regulator activity, protein kinase activity and protein tyrosine kinase activity
Proteins that demonstrate differential phosphorylation between Smarcb1 proficient and deficient cells regardless of serum conditions include Paxillin (PAX) and its binding protein Vinculin (VCL), two proteins localized to focal adhesion sites. These genes were found to be transcriptionally up-regulated in Smarcb1 proficient cells [14]. PAX is found to be highly phosphorylated in Smarcb1 proficient cells at residue Y118, whose phosphorylation is associated with altered cell adhesion, motility and cytoskeletal organization [19]. Moreover, Focal adhesion kinase 1 (FAK1) demonstrates elevated levels of phosphorylation in Smarcb1 deficient cells at serine 948, whilst FAK2 has elevated levels of phosphorylation in Smarcb1 proficient cells at serine 375. Despite the fact that the precise nature of the phosphorylation at the observed residues is unclear, these findings suggest that loss of Smarcb1 leads to alteration in the composition and arrangement of focal adhesion sites and in the organization of the cytoskeleton. These alterations can be accompanied by deregulation of focal adhesion related signaling [20].
Indeed, actin staining reveals profound changes in cytoskeletal organization between Smarcb1 proficient and deficient cells. Whilst Smarcb1 proficient cells exhibit actin stress fibers, Smarcb1 deficient cells lack stress fibers and the actin seems diffused throughout the cytoplasm (Fig. 3a). This Smarcb1 dependent remodeling of the actin cytoskeleton was evident in an additional Smarcb1 deficient murine MRT derived cell line; 167 (Fig. 3a). Adhesion assay, which assesses adhesion following cell re-plating [21], showed that Smarcb1 deficient cells adhere less efficiently than their Smarcb1 proficient counterparts (Fig. 3b), indicating a defect in focal adhesion. In accordance with this last finding, Paxillin immunostaining reveals gross changes in adhesion site size, number and appearance between Smarcb1 deficient and proficient cells (Fig. 3c). Paxillin itself is moderately accumulated in Smarcb1 proficient cells (Fig. 3d), as expected in light of the transcriptional up-regulation. All together, these results show that loss of Smarcb1 results in transcriptional and post-transcriptional deregulation of processes related to the actin cytoskeleton and to focal adhesion.
Differential morphology of Smarcb1 proficient and deficient cell lines. a Whilst in Smarcb1 proficient cell lines actin forms stress fibers across the cell, in Smarcb1 deficient cell lines the actin is found diffused throughout the cytoplasm. In red – Phalloidin, In blue – Dapi b Reduced adhesiveness of Smarcb1 deficient cell lines as evident in an adhesion assay. Cells were allowed to adhere for 20/40 min before quantification of adherent fraction was carried out as described in the materials and methods section. T-test; 167 20', P.V = 0.0015; 167 40', P.V = 0.0027; 365 20', P.V = 0.0008; 365 40', P.V = 0.0011. c Differential morphology, size and number of focal adhesion sites as visualized via Paxillin staining. In red – Paxillin, In blue – DAPI. d Western blot showing a slight increase in PAX following Smarcb1 expression, consistent with available expression data
Altered activation of several kinases is reflected in the phosphoproteomic data
Differential phosphorylation profiles may reflect changes in the activity of kinases and/or phosphatases between Smarcb1 deficient and proficient cells. We therefore attempted to determine whether targets of specific kinases are over-represented in one condition over another. Using the kinase target database from Phosphositeplus [22] we constructed sets of known and characterized kinase targets at residue level for each kinase and applied the GSEA algorithm to test for enrichment of any such set in our phosphoproteomic data. Although the information linking kinases to their target sites, particularly in mouse, is very limited, this analysis identified the expected enrichment for Cyclin dependent kinase (CDK1) targets and for AKT1 targets among residues that were phosphorylated in high serum compared to serum starvation conditions in Smarcb1 proficient cells (False Discovery Rate (FDR) = 0.05 for CDK1; FDR = 0.06 for AKT1). Application of the same approach to the Smarcb1 dependent phosphorylation under serum starvation revealed that peptides phosphorylated in Smarcb1 deficient tumor cells are enriched for targets of AKT, a result consistent with our previous findings demonstrating activation of AKT in the same Smarcb1 deficient cell line used for the phosphoproteomic analysis. This activation was found to persist in serum starvation and result in phosphorylation of AKT targets such as ribosomal protein S6 [14]. Differentially phosphorylated Erk1/2 targets did not pass the statistical significance threshold as individual peptides, but as a group, were significantly overrepresented among peptides that showed elevated phosphorylation in Smarcb1 deficient cells (Table 1). These results are consistent with elevated phosphorylation of ERK1/2 proteins at key residues in the Smarcb1 deficient cells. (Y205 in ERK1 and T183 in ERK2 [23, 24]) in Smarcb1 deficient cells under serum starvation found by manual examination of the data.
Further analysis of the phosphoproteomic data revealed additional kinases whose statistically significant differential phosphorylation levels would suggest altered activation state. These include activation of PKACA through phosphorylation of T198 [25] in Smarcb1 proficient cell lines and phosphorylation in JNK1 Y185 [23, 24] and its targets (Table 1) in Smarcb1 deficient cell lines (Additional file 1: Table S1). Moreover, we find persistent phosphorylation of EGFR Y1197 in Smarcb1 deficient cells. Tyrosine 1197 is an autophosphorylation site of the EGFR associated with enzymatic activation.
Differential response to serum reveals altered regulation of ErbB signaling in Smarcb1 deficient tumor cells
We next examined the differential response to serum withdrawal between Smarcb1 proficient and deficient cells. Phosphorylation sites that were regulated in a coordinated manner in Smarcb1 deficient and proficient cells upon serum withdrawal were excluded (Fig. 4a – red dots). Of the remaining peptides, we focused on sites whose phosphorylation level in response to serum withdrawal was the most distinct between the Smarcb1 deficient and proficient cells (Fig. 4a – in blue and gray). The fact that few of the phosphorylation sites revealed in the analysis had any known biological effect limited our ability to deduce any functional significance from the data. Therefore, we focused on the affected proteins and used STRING v9.1 [26] to explore protein neighborhoods (high confidence physical and functional interactions) and define functional protein association networks which are differentially regulated between Smarcb1 proficient and deficient cells upon serum withdrawal (Additional file 2: Figure S1). Several functional networks were identified for proteins, which remain phosphorylated in Smarcb1 deficient cells upon serum withdrawal but lose their phosphorylation in Smarcb1 proficient cells (blue group). These are enriched for proteins localized to the nuclear lumen and chromosome in addition to the cytoskeleton. More striking however is the enrichment in proteins regulating cell cycle, transcriptional initiation and the ErbB signaling pathway. In contrast, for proteins that lose phosphorylation specifically in Smarcb1 deficient cells upon serum withdrawal (gray group), we find functional networks that are enriched for proteins localized to the nuclear lumen and function in RNA splicing and processing (Fig. 4b and Additional file 2: Figure S1). Cell cycle regulators include CDK1 which demonstrates differential phosphorylation of T14 related to cell cycle regulation. Chromatin modifying proteins include ARID1A and SMARCC1 which together with SMARCB1 assemble to form the SWI/SNF complex and regulate transcription. Proteins annotated as ErbB signaling include ERK2, JUN MYC and EGFR. Consistent with this, residue T183 in ERK2 and residue S63 in JUN which are associated with induced enzymatic activity [23, 24], and residue T58 in MYC which is required for protein degradation [27], remain phosphorylated in Smarcb1 deficient cells upon serum withdrawal. Upstream to these affects, EGFR is found to be highly phosphorylated in Smarcb1 deficient cells deprived of serum, in residue Y1197, an autophosphorylation site associated in humans with enzymatic activation of the receptor [28].
Different response to serum withdrawal between Smarcb1 proficient and deficient cells. a Scatter plot depicting each peptides response to serum withdrawal (N = 5518 peptides). X-axis: log 2 (high serum/serum starvation) ratio of phosphopeptide abundance in Smarcb1 proficient cells. Y-axis: log 2 (high serum / serum starvation) ratio of phosphopeptide abundance in Smarcb1 deficient cells. Red – correlated change for Smarcb1 proficient and deficient cells. Blue and gray – anti-correlated peptides analyzed with STRING v9.1. b GO annotation enrichment for networks identified using STRING v9.1. Colors correlate to panel a. X-axis represents log base for Benjamini corrected P-value
Differential EGFR expression and phosphorylation promotes downstream AKT activation and cell proliferation
We previously identified persistent activation of AKT in Rhabdoid tumor cells which was Smarcb1 dependent and central for proliferation and survival of Smarcb1 deficient tumor cells [14]. AKT can be activated through multiple pathways and part of the motivation for performing the phospho-proteomic study was to identify the origin of AKT activation. The phosphoproteomic analysis directly indicated phosphorylation of EGFR in Smarcb1 deficient cells, and the network analysis indicated EGFR pathway to be activated in these cells. As activation of the EGFR and ErbB signaling pathway lay upstream to all the above mentioned signaling effects observed in Smarcb1 deficient cell lines [29], we examined EGFR activation in Smarcb1 deficient cells.
Western blot analysis for an additional auto-phosphorylation site of the EGFR, residue Y1092, showed higher levels of phosphorylation in Smarcb1 deficient tumor cells compared with Smarcb1 proficient cells under low serum. These results reinforce the observations made in the phosphoproteomic study and indicate a Smarcb1 dependent activation of EGFR in tumor cells (Fig. 5a).
Smarcb1 dependent EGFR phosphorylation and transcriptional de-regulation. a Western blot demonstrating constitutive phosphorylation of Tyr-1092 of the EGFR in Smarcb1 deficient cells, and down-regulation of the total-EGFR protein. b Smarcb1 dependent transcriptional regulation of the ErbB family receptors and the EGF ligand. Graph shows RNA levels relative to beta actin as estimated by qRT-PCR. Note that the Y axis of the right panel is two orders of magnitude lower, indicating that expression levels of ErbB3, 4 and EGF are significantly low. In both cell lines Egfr and ErbB3 showed reduced expression in Smarcb1 proficient cells. * Fold change pMIG/Smarcb1 167 = 2.4, 365 = 4.5; T-test P.V; 167 = 0.00058, 365 = 0.0098. ** Fold change pMIG/Smarcb1 167 = 8.2, 365 = 6.8; T-test P.V; 167 = 0.0078, 365 = 0.013
To address the origin of EGFR activation in Smarcb1 deficient tumor cells we considered various mechanisms that can cause aberrant activation of EGFR and downstream signaling. We find total EGFR levels to be down-regulated in Smarcb1 proficient cells, as evident in western blot (Fig. 5a). Transcriptionally, we find Egfr to be significantly repressed in Smarcb1 proficient relative to deficient cells (Fig. 5b). Examining the expression profile of other ErbB family members we identify ErbB3 (HER3) as an additional repressed target of SMARCB1 but the significance of this result remains to be established since ErbB3 levels are significantly lower than ErbB2 or Egfr (Fig. 5b). Though expression data from MRT and AT/RT tumors and cell lines suggests overexpression of ErbB2/Her2 relative to other central nervous system tumors [30, 31], in our system we detect no Smarcb1 dependent change in the expression of ErbB2. ErbB4, as in most cases [32], is not expressed and unresponsive to Smarcb1. Egf itself is also transcriptionally unresponsive to Smarcb1 and with very low expression level. Two additional proteins that negatively regulate EGFR (Caveolin1 [33–35] and ERRFI1 [36]) are low in Smarcb1 deficient cells and are upregulated upon its re-introduction, but expression of either one of them in Smarcb1 deficient cells was insufficient in diminishing EGFR or AKT activation (Additional file 2: Figure S2 and [14]).
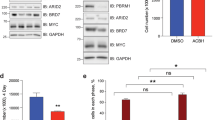
We next inhibited EGFR using Gefitinib, a selective inhibitor of the EGFR kinase activity, or Lapatinib a dual EGFR/ErbB2 kinase inhibitor [37]. Both treatments resulted in inhibition of AKT phosphorylation, implicating EGFR in the activation of AKT in these cells (Fig. 6a). Surprisingly, though both treatments resulted in inhibition of downstream AKT signaling, only Lapatinib treatment led to reduction in EGFR phosphorylation while Gefitinib treatment resulted in apparent elevation of EGFR phosphorylation. These results repeated in multiple experiments and may relate to differences in the preference of the molecules to altered conformations of the EGFR kinase domain [38–41], and their possible effects on the protein stability which results in the accumulation of the receptor [42]. Although the basis for this paradoxical response of EGFR to Gefitinib is unclear, both inhibitors caused reduction of AKT phosphorylation, indicating that the ErbB pathway is responsible for the persistent activation of AKT in Smarcb1 deficient tumor cells.
EGFR activation mediates AKT activation in Smarcb1 deficient cells. a Inhibition of AKT activation upon treatment with the EGFR/HER2 inhibitor Lapatinib (Lap.) and the EGFR inhibitor Gefitinib (Gef.) versus the DMSO (D.) control. b WST1 proliferation assay demonstrating relative proliferation of Smarcb1 deficient and proficient cells following a 6 day treatment with the EGFR inhibitors Lapatinib/Gefitinib, with the AKT inhibitor 1/2, dual inhibition and serum withdrawal
Inhibition of EGFR signaling with Gefitinib or Lapatinib reduced proliferation of Smarcb1 deficient tumor cells as demonstrated by a WST1 proliferation assay. Importantly, Smarcb1 deficient cells demonstrated greater sensitivity to EGFR inhibitors, as their proliferation was hindered to a greater extent than their Smarcb1 proficient counterparts (Fig. 6b). These results implicate EGFR signaling in Smarcb1 mediated tumorigenesis and suggest that Smarcb1 deficient cells are specifically sensitive to EGFR/ErbB2 inhibition.
Discussion
We previously showed that re-introduction of Smarcb1 diminishes the oncogenic capacity of Smarcb1 deficient mouse rhabdoid tumors. By comparing Smarcb1 deficient tumor cells with their Smarcb1 proficient counterparts we identified persistent activation of AKT in Smarcb1 deficient cells, which plays a key role in the survival and proliferation of these tumor cells [14]. To elucidate the source of AKT phosphorylation in Smarcb1 deficient cells and to characterize Smarcb1 dependent effects on post-transcriptional regulation, we conducted a comprehensive proteomic analysis of Smarcb1 dependent changes in protein phosphorylation.
Smarcb1 deficiency affected the phosphorylation of many proteins (Fig. 1). A systematic analysis of the phospho-proteomic data indicated differential activation of multiple kinases and pathways involved in regulation of cell survival and proliferation (Table 1), which are generally in agreement with our initial observation on persistent activation of AKT in Smarcb1 deficient cells. Yet, the analysis of such data at the phosphorylation site level is confined by the limited biological information available on many of the identified the phosphorylation sites, as a result the significance of many intriguing observations remains to be explained (for example: differential phosphorylation of various nuclear pore complex proteins, centromeric proteins or lamins (see supplementary tables and figures)).
Focusing on the differential response to serum starvation between Smarcb1 deficient and proficient cells, we identified several protein networks whose post-transcriptional regulation is altered in Smarcb1 deficient cells (Figs. 2, 4, Additional file 2: Figure S1). The strong enrichment for cell cycle proteins among proteins that remain phosphorylated following serum withdrawal exclusively in Smarcb1 deficient cells (Fig. 4), is in accordance with our findings on sustained proliferation of Smarcb1 deficient cells cultured under serum starvation [14].
Regardless of serum conditions, the analysis reveals Smarcb1 dependent phosphorylation of actin cytoskeleton and of focal adhesion proteins. Correspondingly, we find the actin skeleton of Smarcb1 deficient cells to be diffuse and unstructured and lack stress fibers when compared to Smarcb1 proficient cells, along with a gross difference in the number, size and distribution of focal adhesion sites (Fig. 3). Concurrently, Smarcb1 deficient cells demonstrate an altered morphology and a reduced adhesiveness which are consistent with the changes described. Smarcb1 expression has been previously linked to alterations in the regulation of cytoskeletal components, migration and adhesion [43, 44].
The phosphoproteomic results suggested that in Smarcb1 deficient tumor cells phosphorylation of ErbB signaling cascade and EGFR itself persists even upon serum withdrawal (Fig. 4 and Table 1). These results were confirmed by western blot that demonstrated higher EGFR phosphorylation specifically in Smarcb1 deficient cells (Fig. 5a). Accordingly, higher levels of total EGFR correlating to transcriptional de-repression of Egfr are observed in Smarcb1 deficient cells (Fig. 5b). These findings suggest that EGFR activation is mediated by transcriptional upregulation of the receptor. Moreover we find Smarcb1 mediate transcriptional inhibition of the ErbB3/HER3 receptor. This receptor is a kinase dead receptor, incompetent in promoting downstream signaling, yet heterodimers of ErbB2/HER2-ErbB3/HER3 have a potent signaling competence observed in many neoplasms [32]. As such, this de-repression of ErbB3/HER3 in Smarcb1 deficient cells may be an additional mechanism for ErbB downstream signaling in MRT and AT/RT. Inhibition of EGFR kinase activity reduced AKT phosphorylation, indicating that it drives the activation of AKT in Smarcb1 deficient cells. We further demonstrate the effectiveness of selective EGFR signaling inhibitors on the proliferation of Smarcb1 deficient cells, which show increased sensitivity to Lapatinib and Gefitinib compared to Smarcb1 proficient cells (Fig. 6). Several studies in human Rhabdoid tumor cells have demonstrated Lapatinib and Gefitinib as highly effective in inhibition of proliferation, consistent with high levels of EGFR / ErbB expression and signaling [30, 45, 46]. Taken together, our results reproduce these findings and reinforce the possibility of targeted EGFR/ErbB therapy in MRT and AT/RT patients. Moreover, we demonstrate that EGFR activation is a consequence of Smarcb1 deficiency, suggesting that additional tumors with a mutation in Smarcb1 or in other SWI/SNF subunits may be susceptible to EGFR/ErbB inhibitors.
Oncogenic transformation is considered to occur through a stepwise multiple-hit process, however several recent studies that examined the genome of MRT and AT/RT demonstrated exceptionally low mutation rates in both neoplasms. Indeed, when analyzing point mutations, copy number alterations or chromosomal rearrangements, all recurrent genetic aberrations were found to be limited to the SMARCB1 locus [8–11]. Because SMARCB1 is a core component of the SWI/SNF chromatin remodeling complexes which function as transcriptional co-regulators, the low mutation rate, together with the very early onset of these tumors, raise the possibility that SMARCB1 inactivation alone may be sufficient to drive multiple changes that promote cell transformation.
The networks we identify here and the experimental findings from our system are in line with this intriguing idea. This as they demonstrate how deficiency for Smarcb1 results in profound transcriptional and post transcriptional deregulation, which alter the cell's response to external stimuli, its proliferative capacity and the way it interacts with the environment, in so promoting the acquisition of cancer hallmarks.
Conclusions
The results demonstrate activation of EGFR in Smarcb1 deficient murine rhabdoid cells lines which stems from Smarcb1 dependent transcriptional de-repression of Egfr and possibly ErbB3/HER3. Concurrently, downstream activation of the AKT and ERK signaling cascades is evident in the tumor cells, in line with our previous findings. In accordance we find that small molecule EGFR inhibitors (specifically Gefitinib and Lapatinib) hinder the proliferation of Smarcb1 deficient rhabdoid cells and may prove beneficial in clinical settings.
Materials and methods
Cell line establishment and culture
The establishment and characterization of Rhabdoid tumor cell lines 167 and 365, as well as the re-introduction of Smarcb1 was previously described [14]. Cells were grown in DMEM supplemented with 10 % Hyclone fetal bovine serum (FBS), penicillin (50 mg/ml), streptomycin (50 mg/ml), 2 mM L-Glutamine, 0.1 nM non-essential amino acids, 0.1 mM β-Mercaptoethanol and 1 mM sodium pyruvate. For serum starvation conditions, cells were washed twice in PBS before being transferred to medium containing 0.1 % FBS. Gefitinib (Cell signaling, Cat. No. #4765), Lapatinib (Santa Cruz, Cat. No. sc-202205) and AKT inhibitor 1/2 (Calbiochem, Darmstadt, Germany, AKT inhibitor VIII No. 124018) were added in the indicated concentrations.
Growth curves
WST-1 (Roche, Cat. No. 11–644–807–001) reagent was used with the standard protocol. Briefly, 1000 cells were plated in triplicates in a 96-well plate and cultivated for the indicated time. At each time point, 10 μl of WST-1 were added to 100 μl of growth medium and incubated for an hour. Plate was read at 480 nm with the background absorbance at 690 nm.
Phosphoproteomic analysis
365 Smarcb1 proficient and 365 pMIG deficient cells were SILAC labeled by culturing them for 10 population doublings in SILAC-DMEM (deprived of lysine and arginine), supplemented with 10 % dialyzed FCS and heavy, medium or light labeled lysine and arginine (lys0/arg0; lys4/arg6; lys8/arg10). Following verification of amino acid incorporation, during the experiments, cells were transferred to the same SILAC culture medium, supplemented with 10 % FCS or 0.1 % FCS over-night as illustrated in Fig. 1a. Proteins were extracted using SDS lysis buffer containing; 4 % SDS, 0.1 M DTT, 0.1 M Tris–HCl pH 7.5. Trypsin digestion was performed following the FASP protocol [47] and was followed by strong cation exchange and titanium-dioxide phosphopeptide enrichment as previously described [48].
Mass spectrometric analysis was performed on the EASY-nLC high performance liquid chromatography coupled to the LTQ-Orbitrap Velos mass spectrometer (Thermo Scientific), using data-dependent HCD fragmentation of the top 10 peptides from each MS scan. Raw MS files were analyzed with the MaxQuant software and included phospho(STY) as a variable modification. Data were filtered to have 1 % FDR on the peptide and protein levels. Data analysis was performed on the phospho(STY) sites table. Significance B calculation (based on overall distribution of the SILAC ratios and peptide intensity) was used to extract significantly changing phosphosites, with a p-value threshold of 0.05.
Network analysis
Proteins found to differentially respond to serum withdrawal between v proficient and deficient cells were inputted to identify functional networks. Networks were predicted using the String database [49] with a cut-off for high confidence interactions (>0.9) based on co-occurrence, co-expression, experiments and databases. Resulting networks were visualized using the Cytoscape platform [50].
Kinase target enrichment analysis
We utilized the data available in the kinase target database from Phosphositeplus [22] to define kinase target sets at residue level, this for residues that are defined as phosphorylated by a specific mouse kinase in mouse cells. We then applied the GSEA algorithm to search for leading edge enrichment of kinase target sets in the pre-ranked phosphoproteomic data from Smarcb1 proficient versus deficient cells under low serum or from Smarcb1 proficient cells grown under high serum versus serum starvation.
Protein extraction and Western blot analysis
proteins were extracted using a Triton based buffer (0.5 % Triton, 300 mM Sucrose, 100 mM NaCl, 10 mM PIPES, 3 mM MgCl2*6H2O, 5 mM EDTA) supplemented with 1 μM DTT, 1 μM PMSF, 1 μM Pepstatin, 1 μg/ml Aprotenin, 0.5 μg/ml Leupeptin and phosphatase inhibitor cocktail 2 (Sigma, Cat. No. P5726). Following 10 min on ice the lysate was centrifuged at 14,000 rpm and the pellet discarded. Antibodies used for detection in western blot are as follows: Anti-phospho EGFR Y1092 (Abcam, 1:1000 Cat. No. ab40815), Anti-EGFR (Abcam, 1:1000 Cat. No. ab2430), anti-phospho AKT S473 (Cell signaling, 1:1000, Cat. No. 4058), Anti-AKT (Cell signaling, 1:1000, Cat. No. 11E7), anti-paxillin (Santa Cruz, 1:200, Cat. No. sc-136297), anti-beta-Actin (Abcam, 1:1000,Cat. No. ab6276), Strepavidin coupled HRP (Jackson Immunoresearch Laboratories, 1:1000). Secondary antibodies coupled to horseradish peroxidase (Jackson Immunoresearch Laboratories).
Adhesion assay
As described in [21]. 80,000 cells from each cell line were plated in a 24 well plate in triplicates and allowed to adhere for the indicated time. The cells were then gently washed and stained with 0.5 ml 0.1 % crystal violet dissolved in 10 % acetic acid. The portion of the adhered cells was extrapolated from a standard curve prepared for each cell line concomitantly, where relative fractions from 0 % – 100 % of 80,000 cells were plated and allowed to adhere for several hours before staining.
Immunostaining
Cells were plated on 18 mm sterile coverslips and allowed to adhere overnight. For Phalloidin cell were gently washed in PBS++ before being fixed in 3.7 % paraformaldehyde (PF) in PBS for 10 min. Following fixation cells were permeabilized in 0.5 % triton in PBS. Following several washed coverslips were stained with the Texas-Red-phalloidin (Invitrogen, 0.5U/ml, Cat. No. T7471) and DAPI (Roche, 6 μg/ml, Cat. No. 10–236–276–001) for two hours before being mounted on slides with vecta-shield. For Paxillin immunostaining, permeabilization with 0.5 % triton in 3.7 % PF in PBS with 5 % sucrose for 5 min preceded fixation for 25 min in 3.7 % PF in PBS. Following several washes and blocking with 10 % FCS in PBS for an hour, coverslips were stained with anti-paxillin (Santa Cruz, 1:200, Cat. No. sc-136297) followed by fluorescent secondary (Jackson Immunoresearch Laboratories) and DAPI before being mounted. Images were collected on a Nikon TE-2000 (Nikon, Melville, NY, USA) inverted microscope and processed using NIS-elements software (Nikon). Identical camera and microscope settings were employed to allow valid comparison between images of Smarcb1 deficient and proficient cells.
RNA extraction, reverse transcription and real-time PCR
All performed using standard techniques and kits as described in [51]. Primers used for expression analysis are as follows: Egfr; F': ACACTGCTGGTGTTGCTGAC R': TTGGGTGAGCCTGTTACTTG Erbb2; F': GCAGTGATCATCATGGAGCTG R': AGGTGGGTCTCAGGACTGG Erbb3; F': GTGCTGGGTTTCCTTCTCAG R': TCTGGTACTGGTTGTCAGCATC Erbb4; F': GACTTGCCAAAAATGAAGCTG R': TGCTGTTCCAGGTCAGAGAG Egf; F': CAAACGCCGAAGACTTATCC R': TTTGGCCAGTCCTCTTGTTC Errf1; F': AGCGAGCAGAGAGAAAGAGC R': ACTCTGGGATGCCTTCAAAT Beta-Actin; F': TTTTGTGTCTTGATAGTTCGCCA R': GCCGTTGTCGACGACCAG
Errfi1 cloning
The MS2-HBTH Biotin tag was cloned by PCR from the pQCPX MS2-HBTH vector, generously provided by M. Waterman [52], using the primers: F': ACTGGCTAGCTCTCATTAATGATGGGTGG and R': ACTGGCTAGCATCCGCGGCCGCGCATG. The PCR product was restricted with NheI and ligated into the SpeI site in the pSIN-EF2-Nanog vector, which was formerly restricted with BamHI and self-ligated in-order to excise Nanog. The MS2-HBTH biotin tag was subsequently cloned from the pSin EF2-MS2-HBTH constructed backbone plasmid using the primers: F': ACTGGTCGACCATCATCACCACCATCATGAC and R': ACTGCTCGAGCTCATTAATGATGGTGGTGATG. The PCR product was restricted with SalI and XbaI and inserted into the pHAGE retroviral vector restricted with SalI.
cDNA from SMARCB1 proficent 167 cell line was used to PCR amplify Errfi1 transcript using the following primers; F': ATGCGCGGCCGCATGTCAACAGCAGGAGTTGC R': ATGCGTCGACTGGAGAAACCACGTAGGATAA. The resulting amplicon was inserted into the pHAGE-HBTH vector (described previously) between the Not1 and Sal1 restriction sites. The resulting plasmid was sequenced to ensure correct amplification and insertion. For generation of viral vectors, plasmids were co-transfected with VSVG and PHR into 293 T cells using the jetPEI® transfection reagent (Polyplus, CA, USA). Infections were carried out for 2 sequential days with 8 μg/ml Polybrene followed by selection with Blasticidin.
References
Narlikar GJ, Sundaramoorthy R, Owen-Hughes T. Mechanisms and functions of ATP-dependent chromatin-remodeling enzymes. Cell. 2013;154(3):490–503.
Biggar SR, Crabtree GR. Continuous and widespread roles for the Swi-Snf complex in transcription. EMBO J. 1999;18(8):2254–64.
Lu P, Roberts CW. The SWI/SNF tumor suppressor complex: Regulation of promoter nucleosomes and beyond. Nucleus. 2013;4(5):374–8.
Wang X, Haswell JR, Roberts CW. Molecular pathways: SWI/SNF (BAF) complexes are frequently mutated in cancer--mechanisms and potential therapeutic insights. Clin Cancer Res. 2014;20(1):21–7.
Kim, KH, Roberts, CW. Mechanisms by which SMARCB1 loss drives rhabdoid tumor growth. Cancer Genet. 2014;207(9):365-72.
Ostrom QT, Chen Y, M de Blank P, Ondracek A, Farah P, Gittleman H, et al. The descriptive epidemiology of atypical teratoid/rhabdoid tumors in the United States, 2001–2010. Neuro Oncol. 2014;16(10):1392-9.
Kerl K, Holsten T, Fruhwald MC. Rhabdoid tumors: clinical approaches and molecular targets for innovative therapy. Pediatr Hematol Oncol. 2013;30(7):587–604.
Hasselblatt M, Isken S, Linge A, Eikmeier K, Jeibmann A, Oyen F, et al. High-resolution genomic analysis suggests the absence of recurrent genomic alterations other than SMARCB1 aberrations in atypical teratoid/rhabdoid tumors. Genes Chromosomes Cancer. 2013;52(2):185–90.
Hoell JI, Gombert M, Bartenhagen C, Ginzel S, Husemann P, Felsberg J, et al. Whole-genome paired-end analysis confirms remarkable genomic stability of atypical teratoid/rhabdoid tumors. Genes Chromosomes Cancer. 2013;52(10):983–5.
Lee RS, Stewart C, Carter SL, Ambrogio L, Cibulskis K, Sougnez C, et al. A remarkably simple genome underlies highly malignant pediatric rhabdoid cancers. J Clin Invest. 2012;122(8):2983–8.
Takita J, Chen Y, Kato M, Ohki K, Sato Y, Ohta S, et al. Genome-wide approach to identify second gene targets for malignant rhabdoid tumors using high-density oligonucleotide microarrays. Cancer Sci. 2014;105(3):258–64.
Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144(5):646–74.
Klochendler-Yeivin A, Picarsky E, Yaniv M. Increased DNA damage sensitivity and apoptosis in cells lacking the Snf5/Ini1 subunit of the SWI/SNF chromatin remodeling complex. Mol Cell Biol. 2006;26(7):2661–74.
Darr J, Klochendler A, Isaac S, Eden A. Loss of IGFBP7 expression and persistent AKT activation contribute to SMARCB1/Snf5-mediated tumorigenesis. Oncogene. 2014;33(23):3024–32.
Blagoev B, Ong SE, Kratchmarova I, Mann M. Temporal analysis of phosphotyrosine-dependent signaling networks by quantitative proteomics. Nat Biotechnol. 2004;22(9):1139–45.
Mann M. Functional and quantitative proteomics using SILAC. Nat Rev Mol Cell Biol. 2006;7(12):952–8.
Olsen JV, Blagoev B, Gnad F, Macek B, Kumar C, Mortensen P, et al. Global, in vivo, and site-specific phosphorylation dynamics in signaling networks. Cell. 2006;127(3):635–48.
Ong SE, Blagoev B, Kratchmarova I, Kristensen DB, Steen H, Pandey A, et al. Stable isotope labeling by amino acids in cell culture, SILAC, as a simple and accurate approach to expression proteomics. Mol Cell Proteomics. 2002;1(5):376–86.
Schaller MD. Paxillin: a focal adhesion-associated adaptor protein. Oncogene. 2001;20(44):6459–72.
Mitra SK, Hanson DA, Schlaepfer DD. Focal adhesion kinase: in command and control of cell motility. Nat Rev Mol Cell Biol. 2005;6(1):56–68.
Humphries MJ. Cell adhesion assays. Methods Mol Biol. 2009;522:203–10.
Hornbeck PV, Kornhauser JM, Tkachev S, Zhang B, Skrzypek E, Murray B, et al. PhosphoSitePlus: a comprehensive resource for investigating the structure and function of experimentally determined post-translational modifications in man and mouse. Nucleic Acids Res. 2012;40(Database issue):D261–70.
Boulton TG, Nye SH, Robbins DJ, Ip NY, Radziejewska E, Morgenbesser SD, et al. ERKs: a family of protein-serine/threonine kinases that are activated and tyrosine phosphorylated in response to insulin and NGF. Cell. 1991;65(4):663–75.
Davis RJ. Signal transduction by the JNK group of MAP kinases. Cell. 2000;103(2):239–52.
Cauthron RD, Carter KB, Liauw S, Steinberg RA. Physiological phosphorylation of protein kinase A at Thr-197 is by a protein kinase A kinase. Mol Cell Biol. 1998;18(3):1416–23.
Franceschini A, Szklarczyk D, Frankild S, Kuhn M, Simonovic M, Roth A, et al. STRING v9.1: protein-protein interaction networks, with increased coverage and integration. Nucleic Acids Res. 2013;41(Database issue):D808–15.
Yeh E, Cunningham M, Arnold H, Chasse D, Monteith T, Ivaldi G, et al. A signalling pathway controlling c-Myc degradation that impacts oncogenic transformation of human cells. Nat Cell Biol. 2004;6(4):308–18.
Sorkin A, Helin K, Waters CM, Carpenter G, Beguinot L. Multiple autophosphorylation sites of the epidermal growth factor receptor are essential for receptor kinase activity and internalization. Contrasting significance of tyrosine 992 in the native and truncated receptors. J Biol Chem. 1992;267(12):8672–8.
Yarden Y, Sliwkowski MX. Untangling the ErbB signalling network. Nat Rev Mol Cell Biol. 2001;2(2):127–37.
Patereli A, Alexiou GA, Stefanaki K, Moschovi M, Doussis-Anagnostopoulou I, Prodromou N, et al. Expression of epidermal growth factor receptor and HER-2 in pediatric embryonal brain tumors. Pediatr Neurosurg. 2010;46(3):188–92.
Satomi K, Morishita Y, Murata Y, Shiba-Ishii A, Sugano M, Noguchi M. Epidermal growth factor receptor abnormalities in atypical teratoid/rhabdoid tumors and an unusual case with gene amplification. Pathol Res Pract. 2013;209(8):521–6.
Citri A, Yarden Y. EGF-ERBB signalling: towards the systems level. Nat Rev Mol Cell Biol. 2006;7(7):505–16.
Couet J, Sargiacomo M, Lisanti MP. Interaction of a receptor tyrosine kinase, EGF-R, with caveolins. Caveolin binding negatively regulates tyrosine and serine/threonine kinase activities. J Biol Chem. 1997;272(48):30429–38.
Park WY, Park JS, Cho KA, Kim DI, Ko YG, Seo JS, et al. Up-regulation of caveolin attenuates epidermal growth factor signaling in senescent cells. J Biol Chem. 2000;275(27):20847–52.
Okamoto T, Schlegel A, Scherer PE, Lisanti MP. Caveolins, a family of scaffolding proteins for organizing "preassembled signaling complexes" at the plasma membrane. J Biol Chem. 1998;273(10):5419–22.
Segatto O, Anastasi S, Alema S. Regulation of epidermal growth factor receptor signalling by inducible feedback inhibitors. J Cell Sci. 2011;124(Pt 11):1785–93.
Roskoski Jr R. ErbB/HER protein-tyrosine kinases: Structures and small molecule inhibitors. Pharmacol Res. 2014;87C:42–59.
Park JH, Liu Y, Lemmon MA, Radhakrishnan R. Erlotinib binds both inactive and active conformations of the EGFR tyrosine kinase domain. Biochem J. 2012;448(3):417–23.
Yun CH, Boggon TJ, Li Y, Woo MS, Greulich H, Meyerson M, et al. Structures of lung cancer-derived EGFR mutants and inhibitor complexes: mechanism of activation and insights into differential inhibitor sensitivity. Cancer Cell. 2007;11(3):217–27.
Wood ER, Truesdale AT, McDonald OB, Yuan D, Hassell A, Dickerson SH, et al. A unique structure for epidermal growth factor receptor bound to GW572016 (Lapatinib): relationships among protein conformation, inhibitor off-rate, and receptor activity in tumor cells. Cancer Res. 2004;64(18):6652–9.
Eck MJ, Yun CH. Structural and mechanistic underpinnings of the differential drug sensitivity of EGFR mutations in non-small cell lung cancer. Biochim Biophys Acta. 2010;1804(3):559–66.
Scaltriti M, Verma C, Guzman M, Jimenez J, Parra JL, Pedersen K, et al. Lapatinib, a HER2 tyrosine kinase inhibitor, induces stabilization and accumulation of HER2 and potentiates trastuzumab-dependent cell cytotoxicity. Oncogene. 2009;28(6):803–14.
Medjkane S, Novikov E, Versteege I, Delattre O. The tumor suppressor hSNF5/INI1 modulates cell growth and actin cytoskeleton organization. Cancer Res. 2004;64(10):3406–13.
Caramel J, Quignon F, Delattre O. RhoA-dependent regulation of cell migration by the tumor suppressor hSNF5/INI1. Cancer Res. 2008;68(15):6154–61.
Singh A, Lun X, Jayanthan A, Obaid H, Ruan Y, Strother D, et al. Profiling pathway-specific novel therapeutics in preclinical assessment for central nervous system atypical teratoid rhabdoid tumors (CNS ATRT): favorable activity of targeting EGFR- ErbB2 signaling with lapatinib. Mol Oncol. 2013;7(3):497–512.
Kuwahara Y, Hosoi H, Osone S, Kita M, Iehara T, Kuroda H, et al. Antitumor activity of gefitinib in malignant rhabdoid tumor cells in vitro and in vivo. Clin Cancer Res. 2004;10(17):5940–8.
Wisniewski JR, Zougman A, Nagaraj N, Mann M. Universal sample preparation method for proteome analysis. Nat Methods. 2009;6(5):359–62.
Macek B, Mann M, Olsen JV. Global and site-specific quantitative phosphoproteomics: principles and applications. Annu Rev Pharmacol Toxicol. 2009;49:199–221.
Snel B, Lehmann G, Bork P, Huynen MA. STRING: a web-server to retrieve and display the repeatedly occurring neighbourhood of a gene. Nucleic Acids Res. 2000;28(18):3442–4.
Saito R, Smoot ME, Ono K, Ruscheinski J, Wang PL, Lotia S, et al. A travel guide to Cytoscape plugins. Nat Methods. 2012;9(11):1069–76.
Barzily-Rokni M, Friedman N, Ron-Bigger S, Isaac S, Michlin D, Eden A. Synergism between DNA methylation and macroH2A1 occupancy in epigenetic silencing of the tumor suppressor gene p16(CDKN2A). Nucleic Acids Res. 2011;39(4):1326–35.
Tsai BP, Wang X, Huang L, Waterman ML. Quantitative profiling of in vivo-assembled RNA-protein complexes using a novel integrated proteomic approach. Mol Cell Proteomics. 2011;10(4):M110 007385.
Subramanian A, Tamayoa P, Moothaa VK, Mukherjeed S, Eberta BL, Gillette MA, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102(43):15545–50.
Mootha VK, Lindgren CM, Eriksson KF, Subramanian A, Sihag S, Lehar J, et al. PGC-1alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat Genet. 2003;34(3):267–73.
Acknowledgments
We thank Prof. Matthias Mann for his support with the phosphoproteomic study. We thank Prof. Yosef Yarden and Dr. Sharona Even-Ram for helpful discussions and reagents. This study was supported by a Project Grant from the Israel Cancer Research Fund and by Worldwide Cancer Research grant 13–0209.
Author information
Authors and Affiliations
Corresponding author
Additional information
Competing interests
The authors declare that they have no competing interests.
Authors’ contribution
AE, AK, JD and TG conceptually designed the experiments and methodology. AK established the mouse model and cell lines. JD carried out the SILAC protocol, subsequent analysis of phosphoproteomic data and experimental validation of the findings. TG collected and analyzed phosphoproteomic data described in the manuscript. SI conducted the inhibitor assays. Manuscript writing was done by JD together with AE and reviewed by all the listed authors. All authors read and approved the final manuscript.
Additional files
Additional file 1: Table S1 and S2.
Mass-spectroscopy peptide ratios, attached as Excel files. (ZIP 5595 kb)
Additional file 2: Figure S1 and S2.
Functional protein networks based on differential response to serum withdrawal between Smarcb1 proficient and deficient cells. Figure S2. ERRFI1 over-expression in Smarcb1 deficient cells is insufficient in inhibition of EGFR or AKT activation. (PDF 879 kb)
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Darr, J., Klochendler, A., Isaac, S. et al. Phosphoproteomic analysis reveals Smarcb1 dependent EGFR signaling in Malignant Rhabdoid tumor cells. Mol Cancer 14, 167 (2015). https://doi.org/10.1186/s12943-015-0439-5
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12943-015-0439-5