Abstract
Background
Integrons play important role in the spread and maintenance of antimicrobial resistance among strains of Escherichia coli (E. coli) and other species of Enterobacteriaceae. This study investigated the prevalence of class 1 and 2 integrons among E. coli strains isolated from aquaculture water of fish fields in Iran.
Methods
One hundred and fifty water samples from different geographical regions in Chaharmahal Va Bakhtiari province were examined over a 2 months period. Isolation was through culture and biochemical tests. Integrons were identified through polymerase chain reaction (PCR) using oligonucleotide primers specific for class 1 and 2 integrons. Antimicrobial susceptibility testing was carried out using disc diffusion methods.
Results
Eighteen percent of the water samples were positive for E. coli. All the strains were multi-drug resistant; 100% to ciprofloxacin, chloramphenicol, gentamycin, ampicillin and tetracycline and least resistant to imipenem (7.2%). Ten (50%) of the most resistant strains were positive for class 1 (40%) and class 2 (10%).
Conclusions
Escherichia coli in aquaculture in Iran carried integrons class 1 and 2 which could be of public health concern since they could play a role in the spread and maintenance of antimicrobial resistance among bacterial population in the region and should be constantly monitored.
Similar content being viewed by others
Background
Development of antimicrobial resistance in microorganisms has been attributed to indiscriminate use of antibiotics which poses serious public health concern worldwide [1]. Multi drug resistant strains of Escherichia coli (E. coli) can be found in both human and animal isolates worldwide [2] with multiple drug resistant non-pathogenic E. coli found in the intestine implicated as important reservoir of resistance genes [3]. Acquired multi-drug resistance to antimicrobial agents creates an extensive trouble in case of the management of intra and extra intestinal infections caused by E. coli, which are a major source of diseases, mortality and increased production costs [4]. Pathogenic E. coli strains could acquire resistance genes through horizontal gene transfer of mobile genetic elements like integrons which have been reported to house resistance genes. Mobile genetic elements like plasmids and transposons are known to carry integrons which can contain genes for sit-specific recombination and are capable of capturing and mobilizing gene cassettes [5, 6]. Integrons are categorized into types, the super-integrons and the antibiotic resistance integrons (ARIs) [7, 8]. Dissemination of antibiotic resistance genes among bacteria can occur by mobile genetic elements containing the ARIs (5, 7, 9). There are four classes of integrons namely classes 1, 2, 3 and 4 which are known to carry multi-drug resistance genes [7, 9, 10]. Class 1 integrons are the most widespread and have been frequently found in ESBL producing clinical isolates of Enterobacteriaceae [11, 12]. Class 2 integrons occur less frequently in ESBL producing E. coli and Klebsiella pneumoniae while class 3 integrons are rarely found [13]. Class 4 integrons have only been described in Vibrio cholerae strain [10]. The Prevalence of class 1 and 2 integrons in multi-drug resistant E. coli isolated from aquaculture water was investigated in this study.
Methods
Sample collection
A total of 150 water samples from different fish fields from different geographical regions in Chaharmahal Va Bakhtiari province, Iran were examined over a period of 2 months, from October 2013 to November 2013. Water from each fish field was collected in 1,000 mL glass bottles and was taken to the laboratory immediately for bacterial isolation. All samples were labeled to show serial number, place of water, type of water as well as time and date of collection.
Bacterial isolation and biochemical tests for the identification of E. coli
Examination of the water samples was completed within 24 h after collection using Standard Total Coliform Multiple-Tube (MPN) Fermentation Techniques. After determination of MPN the tubes showing growth were inoculated onto MacConkey agar and positive ones subcultured on Eosin-methylene blue (EMB) plates (Merck). After 24 h incubation at 35 ± 0.5°C for 24 ± 2 h g negative microorganisms were isolated from MacConkey and EMB agar and determined at the species level using cytochrome oxidase, triple sugar iron agar, urea and indole tests as putatively E. coli.
DNA extraction and polymerase chain reaction (PCR) conditions for detection of integrons
Purification of DNA directly from water samples filtered was achieved using a Genomic DNA purification kit (Fermentas, Germany) according to the manufacturer’s instructions. Oligonucleotide primers specific for the E. coli are presented in Additional file 1: Table S1 [13, 14].
The PCR reactions were performed in a total volume of 25 μL, including 1.5 mM MgCl2, 50 mM KCl, 10 mM Tris–HCl (pH 9.0), 0.1% Triton X-100, 200 μM dNTPs each (Fermentas), 50 pmoL of each of the E. coli specific primers, 1.5 U of Taq DNA polymerase (Fermentas), and 3 μL (40–260 ng/μL) of DNA.
Amplification reactions were carried out using a DNA thermo-cycler (Eppendorf Mastercycler 5330, Eppendorf-Nethel-Hinz GmbH, Hamburg, Germany) and are listed in Additional file 1: Table S1 [13, 14]. Amplified samples were analyzed by electrophoresis in a 1.5% agarose gel in 1× TBE buffer at 80 V for 30 min, stained with solution of Ethidium Bromide and examined under Ultra Violet illumination (Uvitec, UK). Twenty most resistant strains out of the 27 isolates were tested for presence of integrons in their genome using PCR technique described above. For each set of PCR reactions, was positive control for class 1 integrons.
PCR conditions for detection of resistance genes
The presence of genes associated with choramphenicol [cmlA], gentamicin [aac(3)-IIa, tetracycline-[tet(A), ciprofloxacin, norfloxacin and nalidixic acid resistance-(qnrA), and sulphonamides (Sul 1 and Sul 2) was determined by PCR using specific primers are presented in Additional file 2: Table S2 [15–18].
Antimicrobial Susceptibility Testing
Antimicrobial susceptibility tests were performed by the Kirby-Bauer disc diffusion method using Mueller–Hinton agar (HiMedia Laboratories, Mumbai, India, MV1084), according to the Clinical and Laboratory Standards Institute guidelines [19]. After incubating the inoculated plate aerobically at 37°C for 18–24 h in an aerobic atmosphere, the susceptibility of the E. coli isolates to 12 antimicrobial agents was determined, and the results were interpreted in accordance with interpretive criteria provided by CLSI [19]. E. coli ATCC 25922 was used as quality control organisms in antimicrobial susceptibility determination.
The following antibiotic disks (Padtan-Teb, Iran) were used: ceftazidime (CAZ) (30 μg), tetracycline (TE) (30 μg), chloramphenicol (C) (30 μg), imipenem (IMP) (10 μg), ciprofloxacin (CRO) (5 μg), norfloxacin (NOR) (10 μg), cefalotin (CF) (30 μg), nalidixic acid (NA) (30 μg), nitrofurantoin (FM) (300 μg), trimethoprim/sulfamethoxazole (SXT) (30 μg) and gentamicin (GM) (10 μg).
Results
Out of the 150 water samples, 27 harboured E. coli after culture and biochemical tests. Hence the prevalence of E. coli in aquaculture was 18%.
Antimicrobial resistance
All isolates exhibited multi-drug resistance phenotypically. They were most resistant to ciprofloxacin, chloramphenicol, gentamycin, ampicillin and tetracycline (100%) and were least resistant to imipenem (7.4%) Additional file 3: Table S3.
Detection of integrons
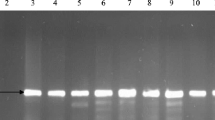
Out of these E. coli isolates, 10 (37%) carried integrons presumably in their plasmids. Eight isolates carried Class 1 integrons while 2 isolates carried class 2 integrons. Class 1 and 2 integrons were not found in any single isolate Additional file 4: Table S4. The results show in Fig. 1.
Detection of resistance genes and integrons
Of the 27 strains that were resistant to chloramphenicol and gentamicin, 23 (85.2%) contained the cmlA and aac(3)IIa genes respectively while 22 (81.5%) contained tetA gene. qnrA gene that is responsible for ciprofloxacin, norfloxacin and nalidixic acid resistance was detected in one (3.7%) of the strains tested. Sul1gene was found in 9 (33.3%) strains while sul 2 gene was present in 7 (25.9%) of the strains (Additional file 5: Table S5, Additional file 6: Table S6).
Discussion
It was observed that aquaculture in Chaharmahal Va Bakhtiari province, Iran provided enabling environment for the growth and survival of microorganisms since 18% of aquaculture samples haboured E. coli. This could be as a result of direct contamination of the water by feacal content due to the abundance of E. coli in mammalian colon [20]. Fecal content could be from the intestine of the fish [21] or from manure feeding which have been reported to increase incidence of E. coli in aquaculture [22]. These organisms could be dangerous to the fish grown in such aquaculture thereby reducing the quantity and quality of the produce. Since E. coli causes disease for both fish and other mammals [23, 24], fish farmers and aquaculture attendants are at the danger of exposure to these organisms since working in aquaculture involves high degree physical contact with the water. This could lead to aquatic infections spreading to the community since E. coli in seafood have been implicated as a source of diarrheagenic infections [25]. To safeguard cross-infections in this instance, microbial water quality maintenance, post-harvest care, handling hygiene and adequate sanitation during processing must be adhered to strictly [25].
The E. coli isolated were found to be multi-drug resistant, being resistant to more than five antimicrobials commonly used for feed additives and or therapeutics; suggesting a linkage between antibiotic usage and development of resistance [26, 27]. Although these isolates were not proven to be pathogenic, their antimicrobial resistance attributes also make them dangerous as potential source of resistance dissemination among bacterial population in the region. They could constitute environmental hazard since aquaculture water when discarded could become available to other animals and could contaminate sources of water for domestic, industrial and animal husbandry use. Since these resistance attributes have been reported elsewhere to be mediated by specific genes [28] which may be transferable horizontally [29], the possibility of resistance transfer to other E. coli species and other bacterial organisms in a multi culture environment is high. Molecular studies on the underlying resistance mechanisms in these multi-resistant E. coli revealed that resistance phenotypes were mediated by several different genes. This finding suggests that the presence of these multi-resistant E. coli in these aquaculture water were probably due to the acquisition of resistance genes by different isolates from different sources.
This antimicrobial resistance is encoded oftentimes by integrons that occur on plasmids or that are integrated into the bacterial chromosome. We found 50% of the most resistant isolates positive for integrons, class 1 (40%) and class 2 (10%) which was similar to the report of Goldstein et al. [10] that class 1 integrons were present in approximately 46% of the isolates from the family Enterobacteriaceae. In that report, class 2 integrases were present only among E. coli and Salmonella isolates. Kang et al. [30] also reported 44% integrons among E. coli isolates in Korea. Our result was however, lower than 76% occurrence of class 1 integrons reported by van Essen-Zandbergen et al. [31] among isolates of E. coli in Netherlands. Again, our finding goes contrary to the report of Bass et al. [32] which previously determined that multiple-drug resistance exhibited by avian E. coli isolates correlated with the incidence of class 1 integrons. The presence of integrons in these isolates may not have contributed to the multi-drug resistance attributes of the isolates since the isolates without class 1 or 2 integrons exhibited nearly the same phenotypic resistance pattern with the isolates that carried class 1 or 2 integrons. However, most of the genes found in the isolates seem to be carried by class 1 integrons which is expected since class 1 type occurred more with isolates. Multi-drug resistance may also have been conferred by other genetic elements like plasmids, transposons or individual gene cassettes. Among those integron negative strains, with exception of one that was resistant to all the antimicrobials tested, all were resistant to the same class of antimicrobials and none was resistant to naldixic acid, sulfamethoxazole-trimethoprim, nitrofurantoin, imipenem and cephalotin. It seems class 2 integron do not carry the genetic basis for resistance to naldixic acid, sulfamethoxazole-trimethoprin, nitrofurantoin, and imipenem since the class 2 positive strains were susceptible to them.
Although integrons were detected in the strains of E. coli, their carriage did not seem to play a major role in the ability of the strains to become resistant. Similar finding was reported by Gallego and Towner [33] among clinical isolates of Acinetobacter baumannii in Northern Spain. However, the role of integrons in the spread and maintenance of resistance factors among the E. coli population and other species of the Enterobaceriacae cannot be overemphasized [6, 34].
Conclusion
In conclusion, the E. coli strains from aquaculture in Iran carried class 1 and 2 integrons which may be contributing to the resistance of these organisms to common antimicrobials. The potential for horizontal gene transfer among bacterial population is hereby highlighted. There is therefore need for adequate monitoring to forestall perpetuation of antimicrobial resistance in the region.
Abbreviations
- E. coli :
-
Escherichia coli
- PCR:
-
polymerase chain reaction
- ARIs:
-
antibiotic resistance integrons
- MPN:
-
Standard Total Coliform Multiple-Tube
- EMB:
-
Eosin-methylene blue
- CLSI:
-
Clinical and Laboratory Standards Institute
- BP:
-
base pair
- CAZ:
-
ceftazidime
- TE:
-
tetracycline
- CP:
-
chloramphenicol
- IMP:
-
imipenem
- CF:
-
ciprofloxacin
- NOR:
-
norfloxacin
- C:
-
cefalotin
- NA:
-
nalidixic acid
- NF:
-
nitrofurantoin
- SXT:
-
trimethoprim/sulfamethoxazole
- GM:
-
gentamicin
References
World Health Organisation (2010) WHO global strategy for containment of antimicrobial resistance. WHO, Geneva, Switzerland, p 105
Amara A, Ziani Z, Bouzoubaa K (1995) Antibioresistance of Escherichia coli strains isolated in Morocco from chickens with colibacillosis. Vet Microbiol 43:325–330
Osterblad M, Hakanen A, Manninen R, Leistevuo T, Peltonen R, Meurman O et al (2000) A between-species comparison of antimicrobial resistance in enterobacteria in faecal flora. Antimicrob Agents Chemother 44:1479–1484
Gupta K, Hooton TM, Stamm WE (2001) Increasing antimicrobial resistance and the management of uncomplicated community-acquired urinary tract infections. Ann Int Med 135:41–50
Hall RM, Collis CM (1995) Mobile gene cassettes and integrons: capture and spread of genes by site-specific recombination. Mol Microbiol 15:593–600
Stokes HW, Hall RM (1989) A novel family of potentially mobile DNA elements encoding site-specific gene-integration functions: integrons. Mol Microbiol 3:1669–1683
Cambray G, Guerout AM, Mazel D (2010) Integrons. Annu Rev Genet 44:141–166
Fluit AC, Schmitz FJ (2004) Resistance integrons and super-integrons. Clin Microbiol Infect 10:272–288
Hall RM (1997) Mobile gene cassettes and integrons: moving antibiotic resistance genes in gram-negative bacteria. Ciba Found Symp 207:192–205
Goldstein C, Lee MD, Sanchez S, Hudson C, Phillips B, Register B et al (2001) Incidence of class 1 and 2 integrases in clinical and commensal bacteria from livestock, companion animals, and exotics. Antimicrob Agents Chemother 45:723–726
Jones LA, Mclver CJ, Rawlinson WD, White PA (2003) Polymerase chain reaction screening for integrons can be used to complement resistance surveillance programs. Commun Dis Intel 27:103–110
Yao F, Quian Y, Chen S, Wang P, Huang Y (2007) Incidence of extended spectrum β-lactamases and characterization of integrons in extended spectrum β-lactamase producing Klebsiella pneumoniae isolated in Shantou, China. Acta Biochim Biophys Sin 39:527–532
Machado E, Cantón R, Baquero F, Galán JC, Rollán A, Peixe L et al (2005) Integron content of extended spectrum β-lactamase producing Escherichia coli strains over 12 years in a single hospital in Madrid, Spain. Antimicrob Agents Chemother 49:1823–1829
Li D, Liu B, Chen M, Guo D, Guo X, Liu F et al (2010) A multiplex PCR method to detect 14 Escherichia coli serogroups associated with urinary tract infections. J Microbiol Methods 82(1):71–77
Maynard C, Bekal S, Sanschagrin F, Levesque RC, Brousseau R, Masson L et al (2004) Heterogeneity among virulence and antimicrobial resistance gene profiles of extraintestinal Escherichia coli isolates of animal and human origin. J Clin Microbiol 42(12):5444–5452
Ahmed MO, Clegg PD, Williams NJ, Baptiste KE, Bennett M (2010) Antimicrobial resistance in equine faecal Escherichia coli isolates from North West England. Ann Clin Microbiol Antimicrob 9:12
Soleimani-Asl Y, Zibaei M, Firoozeh F (2013) Detection of qnrA gene among quinolone-resistant Escherichia coli isolated from urinary tract infections in Khorram Abad during 2011-2012. Feyz 17(5):488–495
Kerrn MB, Klemmensen N, Frimodt-Møller N, Espersen F (2002) Susceptibility of Danish Escherichia coli strains isolated from urinary tract infections and bacteraemia, and distribution of sul genes conferring sulphonamide resistance. J Antimicrob Chemotherapy 50:513–516
Clinical and laboratory standards institute (CLSI) (ed) (2012) Performance standards for antimicrobial susceptibility testing: twenty-second informational supplement M100-S21. CLSI, Wayne
Kaper JB, Nataro JP, Mobley HLT (2004) Pathogenic Escherichia coli. Nat Rev 2:123–140
Carson CA, Shear BL, Ellersieck MR, Asfaw A (2001) Identification of fecal Escherichia coli from humans and animals by ribotyping. Appl Environ Microbiol 67(4):1503–1507
Dang ST, Dalsgaard A (2012) E. coli contamination of fish raised in integrated pig-fish aquaculture systems in Vietnam. J Food Prot 75(7):1317–1319
Nataro J, Pand Kaper JB (1998) Diarrheagenic Escherichia coli. Clin Microbiol Rev 11(1):142–201
Guerra B, Junker E, Schroeter A, Malorny B, Lehmann S, Helmuth R (2003) Phenotypic and genotypic characterization of antimicrobial resistance in German Escherichia coli isolates from cattle, swine and poultry. J Antimicrob Chemother 52:489–492
Renata AC (2013) Escherichia coli in seafood: a brief review. Adv Biosci Biotechnol 4:450–454
Carneiro DO, Figueiredo HCP, Pereira Júnior DJ, Leal AG, Logato PVR (2007) Perfil de susceptibilidade a antimicrobianos de bactérias isoladas em diferentes sistemas de cultivo de tilápia-do-Nilo (Oreochromis niloticus). Arq Bras Med Vet Zootec 59(4):869–876
Kemper N (2008) Veterinary antibiotics in the aquatic and terrestrial environment. Ecol Indic 8(1):1–13
Recchia GD, Hall RM (1995) Gene cassettes—a new class of mobile element. Microbiology 141:3015–3027
Leverstein-vanHall MA, Paauw A, Box ATA, Blok HEM, Verhoef J, Fluit AC (2002) Presence of integrons-associated with resistance in the community is widespread and contributes to multidrug resistance in the hospital. J Clin Microbiol 40:3038–3040
Kang HY, Jeong YS et al (2005) Characterization of antimicrobial resistance and class 1 integrons found in Escherichia coli isolates from humans and animals in Korea. J Antimicrob Chemother 55(5):639–644
van Essen-Zandbergen A, Smith H, Veldman K, Mevius D (2007) Occurrence and characteristics of class 1, 2 and 3 integrons in Escherichia coli, Salmonella and Campylobacter spp. in the Netherlands. J Antimicrob Chemother 59(4):746–750
Bass L, Liebert CA, Lee MD, Summers AO, White DG, Thayer SG, Maurer JJ (1999) Incidence and characterization of integrons, genetic elements mediating multiple-drug resistance, in avian Escherichia coli. Antimicrob Agents Chemother 43:2925–2929
Gallego L, Towner K (2001) Carriage of class 1 integrons and antibiotic resistance in clinical isolates of Acinetobacter baumannii from northern Spain. J Med Microbiol 50:71–77
Chah KF, Agbo IC, Eze DC, Somalo S, Estapa V, Torres C (2010) Antimicrobial resistance, integrons and plasmid replicon typing in multi-resistant clinical E. coli strains fron Enugu state, Nigeria. J Basic Microbiol 50:1–7
Authors’ contributions
All authors contributed equally to this work. All authors read and approved the final manuscript.
Acknowledgements
The authors would like to thanks Professor Ebrahim Rahimi and Dr. Chidozie Clifford for their support.
Compliance with ethical guidelines
Competing interests The authors declare that they have no competing interests.
Author information
Authors and Affiliations
Corresponding author
Additional files
Additional file 1:
Table S1. Primers used for the PCR to detect integrons.
Additional file 2:
Table S2. Primers used for the PCR to detect Resistance genes.
Additional file 3:
Table S3. Antimicrobial resistance profile of E. coli strains from aquaculture.
Additional file 4:
Table S4. Antimicrobial resistance pattern and integron carriage among E. coli strains from aquaculture water.
Additional file 5:
Table S5. Antimicrobial resistance of E. coli strains from aquaculture by PCR and by antibiotic disks methods.
Additional file 6:
Table 6. Multiple drug resistance (MDR), Antimicrobial resistance gene and integron carriage among E. coli strains from aquaculture water.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Tajbakhsh, E., Khamesipour, F., Ranjbar, R. et al. Prevalence of class 1 and 2 integrons in multi-drug resistant Escherichia coli isolated from aquaculture water in Chaharmahal Va Bakhtiari province, Iran. Ann Clin Microbiol Antimicrob 14, 37 (2015). https://doi.org/10.1186/s12941-015-0096-y
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12941-015-0096-y