Abstract
Background
Skull stripping remains a challenge for neonatal brain MR image analysis. However, little is known about the accuracy of how skull stripping affects the neonatal brain tissue segmentation and subsequent network construction. This paper therefore aimed to clarify this issue by comparing two automatic (FMRIB Software Library’s Brain Extraction Tool, BET; Infant Brain Extraction and Analysis Toolbox, iBEAT) and a semiautomatic (iBEAT with manual correction) processes in constructing 3D T1-weighted imaging (T1WI)-based brain structural network.
Methods
Twenty-two full-term neonates (gestational age, 37–42 weeks; boys/girls, 13/9) without abnormalities on MRI who underwent brain 3D T1WI were retrospectively recruited. Two automatic (BET and iBEAT) and a semiautomatic preprocessing (iBEAT with manual correction) workflows were separately used to perform the skull stripping. Brain tissue segmentation and volume calculation were performed by a Johns Hopkins atlas-based method. Sixty-four gray matter regions were selected as nodes; volume covariance network and its properties (clustering coefficient, Cp; characteristic path length, Lp; local efficiency, Elocal; global efficiency, Eglobal) were calculated by GRETNA. Analysis of variance (ANOVA) was used to compare the differences in the calculated volume between three workflows.
Results
There were significant differences in volumes of 50 brain regions between the three workflows (P < 0.05). Three neonatal brain structural networks presented small-world topology. The semiautomatic workflow showed remarkably decreased Cp, increased Lp, decreased Elocal, and decreased Eglobal, in contrast to the two automatic ones.
Conclusions
Imperfect skull stripping indeed affected the accuracy of brain structural network in full-term neonates.
Similar content being viewed by others
Background
It is known that magnetic resonance imaging (MRI) has become a very important tool for investigating the early brain development and injury in neonates [1,2,3]. In particular, MRI-based brain structural network analysis provides critical ways to understand the topological structure of brain information integration and transmission during the early development [4,5,6,7]. As a basic preprocessing, skull stripping, designed to eliminate skull, scalp, dura, and other non-brain tissues and retain brain parenchyma from brain MRI, is an essential process in brain tissue segmentation and subsequent brain network construction [8]. For neonates, numerous efforts has been made to perform the automated processing [9,10,11,12,13,14,15,16,17,18], such as FMRIB Software Library’s (FSL) Brain Extraction Tool (BET) [19], infant brain extraction and analysis toolbox (iBEAT developed by the IDEA group at the University of North Carolina at Chapel Hill) [20]. In detail, BET performed the brain extraction by using a deformable surface model to detect the brain boundaries, and the results highly depend on the used parameters [19]. Differently, a learning-based method which combines a meta-algorithm and level-set fusion has been employed in iBEAT [20]. iBEAT shows good performance in infant skull stripping with extensive evaluations on more than 200 infants. Despite this, skull stripping remains a challenge for neonatal brain MRI analysis due to its low tissue contrast, large within-tissue intensity variations, and regionally heterogeneous [21]. Besides, inaccurate skull stripping, e.g., unremoved non-brain tissues would result in the overestimation of local brain volume and cortical thickness [22]. And it would further affect the construction of brain structural network. To our knowledge, little is known about the impact of preprocessing accuracy on the accuracy of brain tissue segmentation and structural network construction.
The present study aimed to investigate the effects of skull stripping on brain tissue segmentation and structural network construction based on three-dimensional (3D) T1-weighted imaging (T1WI). Firstly, we constructed the processing flow based on 3D T1WI on brain gray matter in 22 term neonates; secondly, repeatability and consistency of brain region volume’s segmentation were performed to verify the accuracy; in final part, the volumes of 64 brain region and properties of brain structural network were calculated and compared between three workflows, i.e., two automatic (BET, iBEAT) and a semiautomatic iBEAT with manual correction. Such a workflow could be applied to characterize brain structural connectivity and may provide valuable anticipatory information about the potential for encountering abnormalities at a later stage in development.
This paper is organized as follows. In “Results” section, we report the results on repeatability and reliability, and comparisons of brain structural network between automatic and semiautomatic workflows. In “Discussion” section, we provide additional discussions and followed by conclusion in “Conclusion” section. The methods for repeatability and reliability test, and construction of brain structural network were provided in “Methods” section.
Results
Repeatability and reliability
The Bland–Altman graph of two repeated measurements indicated that 95% of the differences between two measurements located in the mean ± 1.96 SD (standard deviation) range that suggested good repeatability (Fig. 1a). Meanwhile, the ICC analysis indicated the average correlation coefficient was 0.894. Most of values were greater than 0.8, while those of a few brain regions, e.g., caudate nucleus, precuneus, superior occipital gyrus, and inferior occipital gyrus were relatively lower, but were greater than 0.7 (Fig. 1b).
Results of repeatability and reliability of brain region volume calculated by iBEAT with correction. a Bland–Altman graph of whole brain volume between two measurements with 95% of the differences located in the mean ± 1.96 SD (standard deviation) range showing the good repeatability; b intraclass correlation coefficient (ICC) of volume of 64 brain regions between two repeated measurements
Comparisons of brain structural network between three workflows
Brain region volume
The differences of volume of 64 brain regions performed through ANOVA analysis among the three workflows are shown below (Fig. 2). There were significant differences of volume between three workflows in 50 brain regions, while, no significant difference was found in the remaining 14 brain regions, such as bilateral superior frontal gyrus, bilateral pre- and post-central gyrus, bilateral superior parietal gyrus, bilateral precuneus, bilateral superior occipital gyrus and so on.
Small-world properties
In the defined threshold range, the neonatal brain network exhibited high-efficiency small-world topology (Fig. 3).
Properties of brain structural network
The corrected workflow, compared with the others, showed a significantly decreased Cp and increased Lp. With regard to network efficiency, the corrected workflow showed a significantly decreased Elocal and Eglobal (Fig. 4).
Discussion
To investigate the impacts of skull stripping on the accuracy of brain tissue segmentation and structural network construction, three workflows (BET, iBEAT and iBEAT with manual correction) were compared to perform the 3D T1WI-based brain structural network. Our results indicated that the iBEAT with manual correction showed a more accurate consistency and repeatability in brain segmentation. Besides, significant differences in calculations of brain volume and structural network properties between the three workflows further implied the importance of accurate skull stripping in brain segmentation and subsequent brain network construction.
The ICC results indicated that the majority of brain regions showed good repeatability and reliability, while the ICC of some regions, such as caudate nucleus, gyrus rectus, left superior parietal gyrus, left precuneus, left superior occipital gyrus and right inferior occipital gyrus, were relatively lower. One reason may be rooted in the inherently low spatial resolution, insufficient tissue contrast, and ambiguous tissue intensity distributions in neonatal MRI [21]. On the other hand, the adult head coil used in this study may affect the MR image quality in neonates [23, 24]. In this regard, MRI acquisition settings including the specific head coil and scanning parameters should be adjusted for neonates. In detail, a neonatal head coil with appropriate size and high signal-to-noise ratio, as well as the use of appropriate scanning parameters (e.g., smaller FOV, longer repetition time and echo time than adult scanning) would facilitate the signal acquisition [23, 25].
By comparing the three workflows, we found that slight adjustment for skull stripping would remarkably affect the calculations of regional brain volume. Of 64 regions, significant differences existed in brain volume calculations of 50 regions between the three workflows. This further confirmed the difficulty of brain segmentation in neonatal MRI. Specifically, most brain volumes calculated by BET workflow were smaller than those by iBEAT and iBEAT with manual correction. This may be linked to the difference of skull stripping in neonatal MRI between BET and iBEAT [21]. It is worth noting that more unremoved skull components were found in BET than iBEAT; while unremoved components by iBEAT were mainly located in the base of skull (Fig. 5). It may be such facts that led to the larger brain volumes by iBEAT than iBEAT with manual correction. From the above, the accuracy of skull stripping would have considerable effects on the neonatal brain segmentation.
By studying the brain structural network, we found the highly small-world topology in full-term neonates. This may suggest that a highly efficient brain network, serving for brain information integration and transfer, has been constructed in the early development. This is also in agreement with prior findings in neonates [7]. Besides, significant differences were found in local (Cp and Elocal) and global (Eglobal) topological properties of brain structural network between the three workflows. These may indicate that these parameters were sensitive to the accuracy of preprocessing (skull stripping).
The study had several limitations. First, the sample size was relatively small in this study, and more data should be acquired to further improve the accuracy. Second, the template and atlas we used are made by foreign neonates. Given the differences of brain morphology between Western and Eastern neonates [26,27,28,29], it may lead to certain errors in the initial estimation of tissue intensity distributions and thus resulted in the large deformations in the registration procedure. In this regard, a dedicated atlas and template appropriate for Chinese neonates need to be developed in further study. Besides, based on the above, further studies regarding the differences of brain structural network constructed by iBEAT and iBEAT with manual correction between Western and Eastern neonates are also required to facilitate the understandings of preprocessing’s impacts on the brain network construction. Third, skull stripping by the time-consuming semi-automated workflow, i.e., iBEAT with manual correction was implemented; an automated method is further required to perform a more accurate skull stripping.
Conclusions
In this study, we quantitatively analyzed the influences of skull stripping on brain volume calculation and topological properties of brain structural network in neonates. Our results indicated that the brain networks had robust small-world configuration; in addition, there were significant differences in both local and global topological parameters between the three workflows. These findings enhanced the importance of accurate skull stripping in calculation of brain tissue volume and brain structural network construction.
Methods
This study was approved by the local institutional review board of the first author’s affiliation. The parents of the neonates were informed regarding the goals and risks of the MRI scan, and requested for the written consent.
Subjects
This study recruited 22 full-term neonates (13 males and 9 females; gestational age range: 37–42 weeks) without any MRI abnormalities or evidences of any clinical episodes that might cause cerebral damages.
MRI data acquisition
All data were acquired on a 3.0-Tesla scanner (Signa HDxt, General Electric Medical System, Milwaukee, WI, USA) with an 8-channel phase array radio-frequency head coil. To reduce the head movement and complete the MRI procedure, the subjects were sedated with a relatively low dose of oral chloral hydrate (25–50 mg/kg). The potential risks of the chloral hydrate were fully considered. The selection, monitoring, and management of subjects were strictly performed following the guidelines [30]. Neonates were laid in a supine position and snugly swaddled in blankets. A pediatrician was present during the MRI scan. Micro-earplugs were inserted into the external auditory canal for hearing protection. Heads of the subjects were immobilized by molded foam, which was placed around the head. The temperature, heart rate, and oxygen saturation were monitored throughout the procedure.
Three-dimensional fast spoiled gradient-recalled echo (3D-FSPGR) T1-weighted magnetic resonance images were acquired with the parameters: repetition time/echo time = 10.28 ms/4.62 ms, inversion time = 400 ms, field of view = 240 mm, voxel size = 0.94 × 0.94 × 1 mm3.
Construction of brain structural network
Figure 6 provides the workflow for constructing the neonatal brain structural network. The three steps for the workflow were detailed as follows.
Data preprocessing
To investigate the impacts of data preprocessing on network construction, three preprocessing workflows were performed by BET, iBEAT and iBEAT with manual correction.
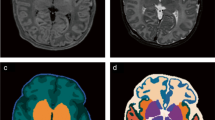
BET workflow: non-brain tissues in each T1WI image were automatically removed by BET. iBEAT workflow: three automatic preprocessing steps, including brain contrast enhancement and skull stripping were sequentially performed. iBEAT with manual correction: (1) manually align the AC–PC line; (2) strip the skull by using iBEAT; (3) identify the remaining non-brain tissues by the boundaries of brain gray matter and manually remove these tissues. The manual correction was performed by two pediatric radiologists with 5 years of experience and differences in identifying the remaining non-brain tissues were resolved by consensus.
Calculations of brain region volume
To calculate the volume of each brain region, the preprocessed data were firstly registered to a standard 3D-T1WI-based template (Johns Hopkins University) by linear (rigid transformation) and nonlinear (affine transformation) registrations. And then the volume of each brain region can be estimated by Eq. (1):
where Vi is the brain volume of the ith region of individual subject, Vt represents the corresponding area in the template. \(\left| {{\text{A}}_{3 \times 3}^{\text{rigid}} } \right|\) is the determinant of 3 × 3 sub-matrix in upper left corner of rigid transformation matrix, and \(\left| {{\text{A}}_{3 \times 3}^{\text{affine}} } \right|\) is the determinant of 3 × 3 sub-matrix in upper left corner of affine transformation matrix. JF is Jacobian, for linear transformation, JF is a constant which is equal to the inverse of the determinant of the 3 × 3 sub-matrix in the upper left corner of the transformation matrix; for nonlinear transformation, JF is a three-dimensional function.
Construction of brain structural network
By using the graph theoretic approaches, the cortical and subcortical regions were used as nodes to construct the brain networks, with connections between nodes defined as correlations between regional brain volumes. Here, 64 brain regions that mainly involved the gray matters and several important subcortical regions (thalamus, hippocampus and cerebellum) were selected as network nodes (Table 1). GRETNA (http://www.nitrc.org/projects/gretna/) was used to construct the network. Partial correlations between all nodes’ volumes were firstly estimated as edges of the network, and then network was constructed by binary connective matrices. The network properties, such as clustering coefficient (Cp), characteristic path length (Lp), global (Eglobal) and local efficiency (Elocal) were calculated by the following Eqs. (2–5).
The clustering coefficient of a node i (C(i)) is defined as the likelihood that the neighborhoods of a given node i are connected to each other:
where ki represents the number of edges connected to the node i, and \(\varpi_{ij}\) is equal to the weight between node i and j. The clustering coefficient, Cp, of a network is the average of the clustering coefficient over all nodes:
where N is the number of nodes in the network, and Lij is the shortest path length between nodes i and j in a network G. Lp is the average of the shortest path length between all pairs of nodes in the network:
where Lij is the shortest path length between node i and node j in G, and N represents all nodes in the network:
where Eglob (Gi) is the global efficiency of Gi, the subgraph of the neighbors of node i.
The Cp of a network is defined by the average of the clustering coefficients across nodes, where the Cp of a node is the ratio of the number of actual connections nearest neighbors of this node to the maximum number of possible connections [31]. Cp quantifies the local interconnectivity of a graph. The Lp of a graph is the average of the shortest path length between all pairs of nodes in the network, and it is an indicator of overall routing efficiency of a graph [32]. The Elocal of a network is the average of the local efficiency over all nodes. It measures the mean local efficiency of the network. The Eglobal of a network is defined by the mean shortest path length [33]. It measures the extent of information propagation through the whole network. Typically, a small-world network should fulfill the following conditions: Cp/Crand > 1 and Lp/Lrand≈ 1.
Statistical analysis
Regarding the manual corrections for anterior commissure–posterior commissure (AC–PC) alignment and skull stripping, the repeatability and consistency of brain volume calculations between two repeated measurements were evaluated by the Bland–Altman graph and intraclass correlation coefficient (ICC). Analysis of variance (ANOVA) was used to compare the differences in the volumes of 64 brain regions between the three workflows.
All the segmentation, calculation of brain region volume and network parameters, and statistical analysis were performed by using the MATLAB R2016b (Mathworks Inc, Natick, MA, USA).
Availability of data and materials
The codes and datasets of this study are available from the corresponding authors upon reasonable request.
References
Dubois J, Dehaene-Lambertz G, Kulikova S, Poupon C, Hüppi PS, Hertz-Pannier L. The early development of brain white matter: a review of imaging studies in fetuses, newborns and infants. Neuroscience. 2014;276:48–71.
Holland D, Chang L, Ernst TM, Curran M, Buchthal SD, Alicata D, Skranes J, Johansen H, Hernandez A, Yamakawa R, Kuperman JM, Dale AM. Structural growth trajectories and rates of change in the first 3 months of infant brain development. JAMA Neurol. 2014;71(10):1266–74.
Kline-Fath BM, Horn PS, Yuan W, Merhar S, Venkatesan C, Thomas CM, Schapiro MB. Conventional MRI scan and DTI imaging show more severe brain injury in neonates with hypoxic-ischemic encephalopathy and seizures. Early Human Dev. 2018;122:8–14.
Cao M, Huang H, He Y. Developmental connectomics from infancy through early childhood. Trends Neurosci. 2017;40(8):494–506.
Gao W, Lin W, Grewen K, Gilmore JH. Functional connectivity of the infant human brain: plastic and modifiable. Neuroscientist. 2017;23(2):169–84.
Zhao T, Mishra V, Jeon T, Ouyang M, Peng Q, Chalak L, Wisnowski JL, Heyne R, Rollins N, Shu N, Huang H. Structural network maturation of the preterm human brain. Neuroimage. 2019;185:699–710.
Brown CJ, Miller SP, Booth BG, Andrews S, Chau V, Poskitt KJ, Hamarneh G. Structural network analysis of brain development in young preterm neonates. Neuroimage. 2014;101:667–80.
Huang H, Shu N, Mishra V, Jeon T, Chalak L, Wang ZJ, Rollins N, Gong G, Cheng H, Peng Y, Dong Q, He Y. Development of human brain structural networks through infancy and childhood. Cereb Cortex. 2015;25(5):1389–404.
Weisenfeld NI, Warfield SK. Automatic segmentation of newborn brain MRI. Neuroimage. 2009;47(2):564–72.
Jaware TH, Khanchandani KB, Zurani A. An accurate automated local similarity factor-based neural tree approach toward tissue segmentation of newborn brain MRI. Am J Perinatol. 2019;36(11):1157–70.
Shi F, Fan Y, Tang S, Gilmore JH, Lin W, Shen D. Neonatal brain image segmentation in longitudinal MRI studies. Neuroimage. 2010;49(1):391–400.
Gui L, Lisowski R, Faundez T, Hüppi PS, Lazeyras F, Kocher M. Morphology-driven automatic segmentation of MR images of the neonatal brain. Med Image Anal. 2012;16(8):1565–79.
Gao Y, Li J, Xu H, Wang M, Liu C, Cheng Y, Li M, Yang J, Li X. A multi-view pyramid network for skull stripping on neonatal T1-weighted MRI. Magn Reson Imaging. 2019;63:70–9.
Mahapatra D. Skull stripping of neonatal brain MRI: using prior shape information with graph cuts. J Digit Imaging. 2012;25(6):802–14.
Anbeek P, Išgum I, van Kooij BJ, Mol CP, Kersbergen KJ, Groenendaal F, Viergever MA, de Vries LS, Benders MJ. Automatic segmentation of eight tissue classes in neonatal brain MRI. PLoS ONE. 2013;8(12):e81895.
Gousias IS, Alexander H, Counsell SJ, Latha S, Rutherford MA, Heckemann RA, Hajnal JV, Daniel R, David EA. Magnetic resonance imaging of the newborn brain: automatic segmentation of brain images into 50 anatomical regions. PLoS ONE. 2013;8(4):e59990.
Jaware Tushar, Khanchandani Kamlesh, Badgujar Ravindra. A novel hybrid atlas-free hierarchical graph-based segmentation of newborn brain MRI using wavelet filter banks. Int J Neurosci. 2019;12(1):1–16.
Cardoso MJ, Melbourne A, Kendall GS, Modat M, Robertson NJ, Marlow N, Ourselin S. AdaPT: an adaptive preterm segmentation algorithm for neonatal brain MRI. Neuroimage. 2013;65(1):97–108.
Smith SM. Fast robust automated brain extraction. Hum Brain Mapp. 2002;17(3):143–55.
Dai Y, Shi F, Wang L, Wu G, Shen D. iBEAT: a toolbox for infant brain magnetic resonance image processing. Neuroinformatics. 2013;11:211–25.
Li G, Wang L, Yap PT, Wang F, Wu Z, Meng Y, Dong P, Kim J, Shi F, Rekik I, Lin W, Shen D. Computational neuroanatomy of baby brains: a review. Neuroimage. 2019;185:906–25.
Shi F, Wang L, Dai Y, Gilmore JH, Lin W, Shen D. LABEL: pediatric brain extraction using learning-based meta-algorithm. Neuroimage. 2012;62(3):1975–86.
Hillenbrand CM, Reykowski A. MR imaging of the newborn: a technical perspective. Magn Reson Imaging Clin N Am. 2012;20(1):63–79.
Hughes EJ, Winchman T, Padormo F, Teixeira R, Wurie J, Sharma M, Fox M, Hutter J, Cordero-Grande L, Price AN, Allsop J, Bueno-Conde J, Tusor N, Arichi T, Edwards AD, Rutherford MA, Counsell SJ, Hajnal JV. A dedicated neonatal brain imaging system. Magn Reson Med. 2017;78(2):794–804.
Lopez Rios N, Foias A, Lodygensky G, Dehaes M, Cohen-Adad J. Size-adaptable 13-channel receive array for brain MRI in human neonates at 3T. NMR Biomed. 2018;31(8):e3944.
Tang Y, Hojatkashani C, Dinov ID, Sun B, Fan L, Lin X, Qi H, Hua X, Liu S, Toga AW. The construction of a Chinese MRI brain atlas: a morphometric comparison study between Chinese and Caucasian cohorts. NeuroImage. 2010;51(1):33–41.
Sivaswamy J, Thottupattu JA, Mehta R, Sheelakumari R, Kesavadas C. Construction of Indian Human Brain Atlas. Neurol India. 2019;67(1):229–34.
Lee JS, Lee DS, Kim J, Kim YK, Kang E, Kang H, Kang KW, Lee JM, Kim JJ, Park HJ, Kwon JS, Kim SI, Yoo TW, Chang KH, Lee MC. Development of Korean standard brain templates. J Korean Med Sci. 2005;20(3):483–8.
Bai J, Abdul-Rahman MF, Rifkin-Graboi A, Chong YS, Kwek K, Saw SM, Godfrey KM, Gluckman PD, Fortier MV, Meaney MJ, Qiu A. Population differences in brain morphology and microstructure among Chinese, Malay, and Indian neonates. PLoS ONE. 2012;7(10):e47816.
Coté CJ, Wilson S. Guidelines for monitoring and management of pediatric patients before, during, and after sedation for diagnostic and therapeutic procedures. Pediatr Dent. 2019;41(4):26E–52E.
Watts DJ, Strogatz SH. Collective dynamics of ‘small-world’ networks. Nature. 1998;393(6684):440–2.
Rubinov M, Sporns O. Complex network measures of brain connectivity: uses and interpretations. Neuroimage. 2010;52(3):1059–69.
Latora V, Marchiori M. Efficient behavior of small-world networks. Phys Rev Lett. 2001;87(19):198701.
Acknowledgements
The authors appreciate Prof. Li Liu, Prof. Xihui Zhou, Dr. Hongxia Song and Gailian Li from the Neonatology Department for preparing and monitoring the neonates before and during imaging. We also thank all participants and their parents for their loyalty and cooperation.
Funding
This study was funded by National Natural Science Foundation of China (Nos. 51706178, 81171317, 81971581), National Key Research and Development Program of China (2016YFC0100300), the 2011 New Century Excellent Talent Support Plan of the Ministry of Education, China (NCET-11-0438). All authors read and approved the final manuscript.
Author information
Authors and Affiliations
Contributions
GLW and YJH performed the data acquisition, analysis and manuscript writing. XJL performed the data processing method. MMW and CCL performed the data acquisition. JY and CJ designed the study, analyzed the data and wrote the manuscript. All authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Ethics approval and consent to participate
All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional and/or national research committee and with the 1964 Helsinki declaration and its later amendments or comparable ethical standards. Informed consent was obtained from all individual participants included in the study.
Consent for publication
All authors confirmed the consent for publication.
Competing interests
The authors declare that they have no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Wang, G., Hu, Y., Li, X. et al. Impacts of skull stripping on construction of three-dimensional T1-weighted imaging-based brain structural network in full-term neonates. BioMed Eng OnLine 19, 41 (2020). https://doi.org/10.1186/s12938-020-00785-0
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12938-020-00785-0