Abstract
Background
Malaria is a life-threatening public health problem globally with particularly heavy burden in the sub-Saharan Africa including Sudan. The understanding of feeding preference of malaria vectors on different hosts is a major challenge for hindering the transmission cycle of malaria. In this study, blood meals taken by blood-fed Anopheles mosquitoes collected from the field in malaria endemic areas of Sudan were analysed for source of blood meal and malaria parasite presence.
Methods
Anopheles mosquitoes were collected from different regions in Sudan: Khartoum state, Sennar state, Northern state, and El Gedarif state between September 2020 and February 2021. Anopheles mosquitoes were collected using the standard pyrethrum spray catch and back-pack aspirator. Mosquito samples were sorted and morphologically identified to species level using international identification keys. Morphologically identified mosquito species were also confirmed using PCR. Genomic DNA was extracted from mosquitoes for molecular identification of blood meal source and parasite detection. The presence of Plasmodium species DNA in each mosquito sample was investigated using semi-nested PCR. Frequency of each blood meal source, Anopheles mosquito vector, and malaria parasite detected was calculated. Positivity rate of each fed female Anopheles mosquito was calculated for each species.
Results
A total of 2132 Anopheles mosquitoes were collected. 571 (26.8%) were males and 1561 (73.2%) were females classified based on their abdominal status into 1048 (67.1%) gravid, 274 (17.6%) fed, and 239 (15.3%) unfed females. Among the blood fed Anopheles mosquitoes, 263 (96.0%) were morphologically identified and confirmed using PCR to Anopheles arabiensis, 9 (3.3%) to Anopheles stephensi, and 2 (0.7%) to Anopheles rufipes. Of 274 blood-fed An. arabiensis, 68 (25.9%) fed on mixed blood meals from human and cattle, 8 (3.0%) fed on cattle and goat, and 13 (4.8%) fed on human, cattle and goat. For single blood meal sources, 70 (26.6%) fed on human, 95 (36.1%) fed on cattle, 8 (3.0%) fed on goat, and 1 (0.4%) fed on dog. While An. rufipes and An. stephensi fed on dog (2; 0.75%) and cattle (9; 3.3%), respectively. Plasmodium parasite detection in the blood meals showed that 25/274 (9.1%) An. arabiensis meals were positive for Plasmodium vivax and 19/274 (6.9%) An. arabiensis meals were positive for Plasmodium falciparum. The rate of positivity of An. arabiensis with any Plasmodium species was 16.7%. However, the positivity rate with P. falciparum only was 7.2%, while P. vivax was 9.5%. Both An. rufipes and An. stephensi were having positivity rates of 0.0% each.
Conclusions
This study which was mainly on blood-fed Anopheles mosquitoes showed a diversity in the type of diet from human, cattle, and goat. Anopheles mosquitoes especially An. arabiensis in Sudan, are opportunistic blood feeders and can feed broadly on both human and cattle. The application of blood meal identification is not only important in malaria vector epidemiological surveillance but also is very useful in areas where arthropods exhibit zoophilic feeding behaviour for mammals.
Similar content being viewed by others
Background
Vector-borne diseases (VBDs) are transmitted by arthropod vectors when they feed on vertebrate host blood [1]. The role of a disease vector in the transmission of VBDs largely depends on its host preference [2]. Feeding on a different host and the host preference of the diseases’ vectors constitute a significant challenge for transmitting zoonotic diseases that infect both human and animal populations. These include leishmaniasis, onchocerciasis, and arboviral diseases [3,4,5,6]. Thus, a broader range of hosts’ availability as sources of blood meals contributes substantially to the diseases’ transmission [2]. Malaria is a life-threatening public health problem globally with particularly heavy burden in the sub-Saharan African region including Sudan with frequent outbreaks [7].
Mosquitoes feed on a wide range of vertebrate hosts, including amphibians, reptiles, birds, and mammals [8,9,10,11]. The behaviour of targeting a single species is not the choice of blood-feeding mosquitoes. Despite the trait of host preference being innate and controlled by genes, it is affected by confounding factors, such as host availability and accessibility [12]. The vector’s choice of host is important as it affects host/pathogen relationship, which may differ accordingly, ranging from a pathogen with a wide range of susceptible hosts to a less vector-host specificity [13]. Host feeding preference affects the vectorial capacity of mosquitoes and vectors control programme.
Globally, there were an estimated 241 million malaria cases and 627 thousand malaria deaths in 2021, with more than 95% of the cases being reported in Africa [14]. The main malaria vectors in Africa belong to three major groups of vectors, the Anopheles gambiae complex, the Anopheles funestus group, and the Anopheles nili complex [15, 16]. However, recently the invasive Asian malaria vector, Anopheles stephensi has emerged in the Horn of Africa region and is rapidly spreading in the area [17,18,19,20].
With 2 million cases and 5,000 deaths, malaria is a disease of serious public health importance in Sudan [21]. The major mosquito vector species is Anopheles arabiensis [22], but other species, such as An. funestus, Anopheles pharoensis and An. nili, have been identified [23, 24]. Anopheles stephensi is increasingly spreading throughout the country [18, 19].
Different assays have been used to identify the blood meal sources in mosquitoes, such as multiplex PCR [25, 26], microsatellites [27], enzyme-linked immunosorbent assay (ELISA) or precipitin test [28, 29]. In this study, blood meals taken by blood-fed mosquitoes collected in the field from different areas of Sudan were analysed to identify the source of blood meal and the presence of malaria parasite using multiplex PCR.
Methods
Samples collection and study areas
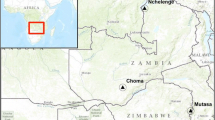
Wild blood-fed samples of Anopheles were collected from different regions in Sudan, namely; Khartoum state (15°55′N 32°53′E), Northern state (19° 37′ N 29° 38′ E), Al Gedarif state (14° 02′ N 35° 23′ E), and Sennar state (12° 58′ N 34° 3′ E) (Fig. 1). These regions were considered as mesoendemic areas with different malaria seasonality [30]. In the studied areas, Plasmodium falciparum is the most common malaria parasite, responsible for 90% of malaria infections, while 10% are caused by Plasmodium vivax [31]. Anopheles collection was carried out simultaneously inside the rooms of 20 houses in each study site for five consecutive days. Anopheles samples were collected using the standard pyrethrum spray catch (PSC) and back-pack aspirator. The collected samples were morphologically identified to species level using international identification keys and sorted out microscopically according to their sex and status of blood feeding [32, 33]. Anopheles males, and unfed and gravid females were excluded from the blood meal analysis due to the lack of feeding on blood for the formers and the low yield of blood source DNA for the later [25]. Anopheles samples were then preserved with silica gel until DNA extraction.
DNA extraction
Genomic DNA was extracted from mosquitoes individually using sodium chloride-Tris-EDTA (STE) solution following the method of Kent and Norris [25] with a minor modification. Briefly, each mosquito was placed into 100 µl STE, crushed in 1.5 ml Eppendorf tube using glass pestle until complete homogenization. Homogenized samples were then incubated at 65° C for a single hour. Following the incubation, 30 µl of potassium acetate 8.0 M was added and then placed at − 20 °C for 1 h. After freezing, samples were allowed to thaw at room temperature and then centrifuged for 15 min at 15,000 rpm. The supernatant was then transferred into a new tube and 1 ml of absolute ethanol was added to precipitate the DNA. Samples were then placed at − 20 °C for 30 min to increase DNA precipitation. Then, the samples were subjected to high-speed centrifugation at 15,000 rpm for 10 min. The supernatant was discarded and the pellet was allowed to dry completely. Dried pellets were then dissolved using 30 µl of deionized distilled water. Extracted DNA was checked for purity and concentration using nanodrop (Implen, Germany) and preserved at − 20 °C until molecular identification of blood meal source and parasite detection.
Molecular confirmation of the species identity
Individual mosquitoes that morphologically identified as An. gambiae sensu lato (s.l.) were further investigated using PCR. Maxime™ i-Taq PCR premix kit was used to prepare the PCR reaction mixture according to the manufacturer instructions (iNtRON Biotechnology, South Korea). Primers and cycling condition used for the confirmation of the An. gambiae s.l. were as previously shown [34]. The PCR reactions were performed using 2027 Thermocycler (Applied Biosystems, Germany). The specific primers of each species produce a band size of 315 in case of An. arabiensis, and a product size of 395 bp in case of An. gambiae.
Molecular identification of the blood meal
Sources of blood meals were identified from the extracted DNA using the previously published primers [25, 35, 36]. To identify blood meal source of human, cattle, goat, dog, bird, and reptile, the following primers were used; Human-F: 5′ CCT ACT CCT GCT CGC ATC TG ‘3 and Human-R: 5′ AGA ATG GGG TCT CCT CCT CC ′3, Cattle-F: 5’ CCC ATC CTA TTG GCC GTA GC ′3, Cattle-R: 5’ GAT GTA GCG GGT CGT AGT GG ‘3, Goat-F: 5’ ACG TAG AAT ATG CCG CAG GG ‘3, Goat-R: 5’ CGT AAC GGA ATC GGG GGT AG ‘3, Dog-F: 5’ GCC TTC CTG ACC CTT GTT GA ‘3, Dog-R: 5’ TTA CTG CGT CTG CGA TTG GT ‘3, Bird-F: 5’ CGC CTG TTT ATC AAA AAC AT ′3, Bird-R: 5′ CCG GTC TGA ACT AGA TCA CGT ‘3, Reptile-F: 5’ TNT TMT CAA CNA ACC ACA AAG A ‘3, and Reptile-R: 5’ ACT TCT GGR TGK CCA AAR AAT CA ‘3 [36]. The PCR reaction mixture to identify the blood meal source was in total of 25 µl containing 10 mM Tris-HCl (pH 8.3), 2 mM MgCl2, 200 µM of each dNTP, 0.1 U Taq polymerase (i-Taq Plus™, DNA Polymerase, iNtRON Biotechnology, South Korea). Adding to 21 µl of the PCR reaction mixture, 1 µl 10 pmol of each of the forward and reverse primers of each source separately and 2 µl of the extracted DNA and incubated in the PCR amplification condition as followed, an initial denaturation step at 95° C for 5 min followed by 14 cycles of denaturation at 95 °C for 30 s, annealing at 60 °C for 30 s with decrement of 0.5 °C each cycle, and extension at 72 °C for 30 s followed by another 19 cycles of denaturation at 95 °C for 30 s, annealing at 53 °C for 30 s, and extension at 72 °C for 30 s. Finally, a final extension step at 72 °C for 10 min and then the PCR products were cooled down to 4 °C. Positive DNA samples of human, cattle (Bos taurus), goat (Capra hircus), dog (Canis familiaris), bird (Columba livia domestica), and reptile (Hemidactylus species) were used as positive controls for each specific primer, while double distilled water was used as a negative control in each run. Molecular identification of blood meal was interpreted according to the specific band sizes produced for each blood meal primers. Blood meals of human, cattle, goat, and dog sources were presented with PCR product sizes of 363 bp, 164 bp, 213 bp, and 109 bp, respectively.
Molecular detection of Plasmodium species
To investigate the positivity rate of Anopheles with malaria parasites, the presence of the DNA of Plasmodium species in each mosquito sample was investigated. Using the semi-nested PCR, the primers used for the detection of the 4 human malaria parasites in Sudan including P. falciparum, P. vivax, Plasmodium ovale, and Plasmodium malariae were adopted from Rubio et al. [37] including; UN-R: 5′ GAC GGT ATC TGA TCG TCT T 3′, UN-F: 5′ AGT GTG TAT CAA TCG AGT TT 3′, for the first PCR reaction and UN-F primer and Fal-R: 5′ AGT TCC CCT AGA ATA GTT ACA 3′, Viv-R: 5′ AGG ACT TCC AAG CCG AAG 3′, Ova-R: 5′ GCA TAA GGA ATG CAA AGA ACA G 3′, and Mal-R: 5′ GCC CTC CAA TTG CCT TCT 3′, for the second PCR reaction to confirm the presence of Plasmodium species. PCR cycling condition was adjusted according to Mohamed et al. [38]. Positive DNA of P. falciparum, P. vivax, P. ovale, and P. malariae were used as positive controls in each PCR run. Double distilled water was used as a negative control.
Gel electrophoreses and amplicons interpretation
Following PCR amplification, PCR products were visualized using 2.5% agarose gel. Electrophoresis was made in comparison to a 100 bp DNA ladder in 5X Tris Borate EDTA running buffer in 100 V and 25 A for 1 h. Lengths of amplicons were read by placing the agarose gel on UV-transilluminator (Major Sciences, USA).
Statistical analysis
Data analysis of the blood meal and parasite positivity rate was made using the Statistical Package for Social Sciences (SPSS version 20.0). Frequency of each blood meal source, Anopheles vector species, and malaria-positive mosquitoes was calculated. Positivity rate of each fed female Anopheles was calculated for each species by dividing the number of blood meals detected positive for parasite presence on the total number of mosquitoes belonging to that species. Chi-Square test was calculated to test the significance association of Anopheles blood meal sources with host preference, and the presence of Plasmodium species. A P value less than 0.05 was considered statistically significant.
Results
Distribution, morphological and molecular identification of Anopheles mosquitoes
A total of 2132 Anopheles were collected. Of these, 571 (26.8%) were males and 1561 (73.2%) were females. Of the 1561 female Anopheles, 1048 (67.1%) were gravid, 274 (17.6%) were blood fed and 239 (15.3%) were blood unfed. Based on site of collection, Sennar state constituted the majority of the collected samples; 1126 (52.8%), while El Gedarif contributed the least collected samples; 86 (4.0%) (Table 1). Of the 274 fed Anopheles females, 263 (96.0%) were An. arabiensis, 9 (3.3%) were An. stephensi, and 2 (0.7%) were Anopheles rufipes. The molecular confirmation of An. arabiensis was confirmed by the presence of a PCR product band size of 315 bp. According to site of collection, An. arabiensis was found in all the study sites except in El Gedarif in which only 9 fed females of An. stephensi were collected. The 2 (0.7%) collected An. rufipes fed females were found in Sennar state (Table 2).
Molecular identification of blood meals sources
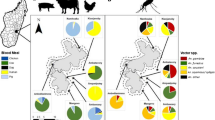
A total of 193 (70.4%) blood-fed Anopheles fed on cattle, 151 (55.1%) fed on human, 29 (10.6%) fed on goat, and 3 (1.1%) fed on dog. There were no mosquitoes with blood meals from birds or reptiles. When illustrating the source of blood meal based on multiple or single source, multiple blood meals of human and cattle, cattle and goat, and human, cattle and goat were detected; 68 (24.8%), 8 (2.9%), and 13 (4.7%), respectively. Also, single blood meal source was detected among 70 (25.5%) fed on human, 104 (38.0%) fed on cattle, 8 (2.9%) fed on goat, and 3 (1.1%) fed on dog. According to species stratification, the two An. rufipes fed on dogs and the nine An. stephensi fed on cattle. The association of blood meal source with the collected Anopheles mosquitoes was found to be statistically significant for meals detected from human, cattle, and goat, P values; 0.001, 0.014, and 0.001, respectively (Table 3).
Plasmodium species detection and the rate of mosquitoes’ positivity
The results of molecular detection of Plasmodium species showed the presence of Anopheles mosquitoes infected with P. falciparum and P. vivax. However, none of the samples harboured P. ovale or P. malariae. Also, no Plasmodium species co-infection was detected. Plasmodium parasite detection in the blood meals showed that 25 (9.1%) were harbouring P. vivax and 19 (6.9%) were harbouring P. falciparum. The remaining 230 (83.9%) were found negative. All the P. falciparum and P. vivax detected were only present in blood meals of An. arabiensis. Plasmodium falciparum positive blood meals reported from cattle and goat mixed meals were 8 (42.1%) and 3 (15.8%), respectively. Also, among the single blood meal of human, P. vivax infection was detected; 12 (48.0%). However, the presence of P. vivax among multiple blood meals from human, cattle and goat was 13 (52.0%). In contrast, P. falciparum was noted among multiple blood meals of cattle and goat; 8 (42.1%) (Table 4).
The positivity rate of An. arabiensis with either P. falciparum and P. vivax was 16.7% (44/263). However, the positivity rate with P. falciparum was 7.2% (19/263), while positivity rate with P. vivax was 9.5% (25/263). Both An. rufipes and An. stephensi were having positivity rates of 0.0% each (Table 4). The presence of Plasmodium parasites among the different meal sources was statistically significant among meals of human, cattle, and goat, P values 0.001, 0.052, 0.001, respectively. Although 3 (1.1%) dog meals were detected, none of them showed presence of Plasmodium species, P value 0.748 (Table 5).
Discussion
Identifying the source of mosquito blood meals is of paramount importance in malaria epidemiological studies [39]. The correct identification of the preferred host for malaria vectors determines the major hosts in the area that support the sustainability of vector population. It is also useful in the estimation of vectorial capacity and identifying the role of each mosquito species in malaria transmission [40]. Studies on feeding behaviour of Anopheles mosquitoes have been implemented in Kenya [41], Ethiopia [42], Mali [25], Sri Lanka [26], and Sudan [43]. In this study, blood meal sources of human, cattle, goat, dog have been identified. The low prevalence of fed females (12.9%) reported in this study compared to gravid females (49.1%), supports the assumption that mosquito collection for blood meal analysis is mainly affected by the resting position especially after taking the blood meal [42]. Recently, a study conducted by Finney et al. indicated that most of the human blood meals were from Anopheles mosquitoes trapped outdoors, while many livestock blood meals were from Anopheles mosquitoes trapped indoors [44].
The abundance of An. arabiensis in Sennar compared to the other sites was similar to a previous study in which the majority of the collected mosquitoes were An. arabiensis; 88.5% [45]. This difference in mosquito abundance can be explained by interference of many factors such as seasonality and the availability of suitable resources for survival in the environment [46].
The results of molecular analysis of blood meal source revealed that Anopheles feeding from cattle; 70.4%, was relatively higher than feeding from human; 55.1%, and higher compared to other hosts investigated, including goat and dog; 10.6% and 1.1%, respectively. These results disagree with a previous report from Burkina Faso, where human blood meal source was more than 80% [47]. However, this variation can be as a result of host availability in that specific region [44].
The malaria parasite detected in this study shows the importance of mosquito blood meal analysis in terms of determination of parasite reservoir host. Although, this assumption is not fully supported by this study, either through animal screening or sporozoite detection in the salivary glands of the infected vector. However, it can hint to the need for a wide animal screening especially during the non-transmission seasons of malaria [44]. These results should be interpreted with caution. It has been reported previously [25] that identification of the source of blood meals in mosquitoes was only possible up to 48 h post-feeding; excluding gravid females from the analysis could, therefore, have been one limitation of this study that could have affected frequencies of blood meals source detected.
The chances of Anopheles to encounter the parasite in the blood meal of infected human can be very high during the peak transmission season [30, 38, 47]. This can affect significantly the role of vector population in transmitting malaria, such as described by Guelbéogo et al. [47], who reported a high frequency of Anopheles coluzzii engorged females with human-animal mixed blood meals. The ability of Anopheles to feed on multiple hosts has been also reported previously [48,49,50]. In this study, the results of multiple blood meals detected are similar to reports from Ethiopia, where multiple feeding on human and cattle was present when both hosts share the same house or closely present in the area [51]. Furthermore, the results of An. rufipes and An. stephensi feeding on dog and cattle, indicate that a wide range of host preference can occur in Sudan. This also emphasizes a need for wider-scale studies in which An. stephensi can be studied extensively, since no previous studies of An. stephensi feeding behaviour has been reported [17, 52].
The positivity rate of An. arabiensis with P. falciparum and P. vivax reported in this study has similarities to mosquito positivity rates reported from Burkina Faso, where mosquito positivity rates during the period of malaria transmission season were 5.1%, 13.9%, 6.5%, during the start, the peak, and the end of transmission season, respectively [47]. Although, the study period was performed during the transmission season, the calculated positivity rate might be confounded by the time of collection. Higher positivity rates can be seen during the transmission season where chances of Anopheles mosquitoes to encounter the parasite is much higher. Although, no Plasmodium species co-infection was detected and all the detected P. falciparum and P. vivax were only present in blood meals of An. arabiensis. This is worrying since the presence of one of the Plasmodium species in a vector leads to increase vector ability to multiple feeding on hosts [53]. Further, the introduction of An. stephensi since 2016 is quite alarming since its known to be a notorious vector in transmitting both P. falciparum and P. vivax, as well as being capable of transmitting zoonotic malaria parasites, such as Plasmodium knowlesi [54]. This urges for improving the vector control strategies in any area with change in the vector composition or change in the vector behaviour [17].
The most prevalent fed Anopheles in this study was An. arabiensis compared to An. rufipes and An. stephensi; although, this can be due to relative vector abundance, it might also be explained by the variation in mosquito survival and age and their relation with malaria transmission. In a similar scenario, the abundance of An. gambiae sensu stricto (s.s.) was linked to its importance for malaria transmission. In contrast, longevity was lower for other species with less contribution to malaria transmission leading to infrequent capture during vector surveillance [47]. Additionally, the contribution of other Anopheles species in transmitting malaria depends on the successful of the Anopheles species to inoculate the parasite in more than one host through multiple feeding [55, 56]. All this together, shows the importance of using blood meal analysis escorted with assessment of mosquito positivity rate in malaria epidemiological studies can significantly improve the understanding of identifying specific reservoirs, which harbour the parasite until the next malaria transmission season.
Conclusions
This study which was mainly on blood-fed Anopheles mosquitoes showed a diversity in the type of diet from human, cattle and goat. With the exclusion of the three blood meal samples from dogs, most analysed blood meals were identified as cattle, human, and goat. This study might give the assumption that Anopheles mosquitoes in Sudan, especially An. arabiensis, are intrinsic unprincipled feeders and can feed broadly on both humans and cattle. However, these results need to be considered carefully given the fact that limitation of the study on the active biting ratio analysis was not applicable. These findings might also indicate that humans, cattle, and goat were only the most abundant host species exists, and this by any means does not inevitably mean that humans and cattle are the preferred hosts in Sudan. Also, the application of blood meal identification is not only important in malaria vector epidemiological surveillance but also is very useful in areas where arthropods exhibit zoophilic feeding behaviour for mammals, such as Rift Valley fever virus where mosquito vectors affect cattle and humans.
Availability of data and materials
All datasets generated for this study are included in the manuscript.
Abbreviations
- DNA:
-
Deoxyribonucleic acid
- EDTA:
-
Ethylene diamine tetra acetic acid
- PCR:
-
Polymerase chain reaction
- STE:
-
Sodium chloride-Tris-EDTA
References
WHO. Vector borne diseases factsheet. Geneva: World Health Organization; 2020.
Chan EYY, Sham TST, Shahzada TS, Dubois C, Huang Z, Liu S, et al. Narrative review on Health-EDRM primary prevention measures for vector-borne diseases. Int J Environ Res Public Health. 2020;17:5981.
Mohamed NS, Osman HA, Muneer MS, Samy AM, Ahmed A, Mohammed AO, et al. Identifying asymptomatic Leishmania infections in non-endemic villages in Gedaref state, Sudan. BMC Res Notes. 2019;12:566.
Ahmed A, Dietrich I, LaBeaud AD, Lindsay SW, Musa A, Weaver SC. Risks and challenges of arboviral diseases in Sudan: the urgent need for actions. Viruses. 2020;12:81.
Ahmed A, Ali Y, Elduma A, Eldigail MH, Mhmoud RA, Mohamed NS, et al. Unique outbreak of Rift valley fever in Sudan, 2019. Emerg Infect Dis. 2020;26:3030.
Ahmed A, Elbashir A, Mohamed AA, Alim AA, Mubarak A, Abdelrahman D, et al. Socioeconomic impacts of elimination of onchocerciasis in Abu-Hamed focus, northern Sudan: lessons after elimination. BMC Res Notes. 2020;13:256.
Mohamed NS, Ali Y, Muneer MS, Siddig EE, Sibley CH, Ahmed A. Malaria epidemic in humanitarian crisis settings the case of South Kordofan state, Sudan. J Infect Dev Ctries. 2021;15:168–71.
WHO. Anopheline species complexes in south-east Asia. WHO Regional Office for South-East Asia; 1998.
Reeves LE, Holderman CJ, Blosser EM, Gillett-Kaufman JL, Kawahara AY, Kaufman PE, et al. Identification of Uranotaenia sapphirina as a specialist of annelids broadens known mosquito host use patterns. Commun Biol. 2018;1:92.
Miyake T, Aihara N, Maeda K, Shinzato C, Koyanagi R, Kobayashi H, et al. Bloodmeal host identification with inferences to feeding habits of a fish-fed mosquito, Aedes baisasi. Sci Rep. 2019;9:4002.
Perkins SL, Austin CC. Four new species of Plasmodium from New Guinea lizards: integrating morphology and molecules. J Parasitol. 2009;95:424–33.
Molaei G, Andreadis TG, Armstrong PM, Diuk-Wasser M. Host-feeding patterns of potential mosquito vectors in Connecticut, USA: molecular analysis of bloodmeals from 23 species of Aedes, Anopheles, Culex, Coquillettidia, Psorophora, and Uranotaenia. J Med Entomol. 2008;45:1143–51.
Santos CS, Pie MR, da Rocha TC, Navarro-Silva MA. Molecular identification of blood meals in mosquitoes (Diptera, Culicidae) in urban and forested habitats in southern Brazil. PLoS ONE. 2019;14:e0212517.
WHO. World malaria report 2021. Geneva, World Health Organization.; 2021.
Dia I, Diop T, Rakotoarivony I, Kengne P, Fontenille D. Bionomics of Anopheles gambiae Giles, An. arabiensis Patton, An. funestus Giles and An. nili (Theobald)(Diptera: Culicidae) and transmission of Plasmodium falciparum in a Sudano-Guinean zone (Ngari, Senegal). J Med Entomol. 2003;40:279–83.
Adja A, N’goran E, Koudou B, Dia I, Kengne P, Fontenille D, et al. Contribution of Anopheles funestus, An. gambiae and An. nili (Diptera: Culicidae) to the perennial malaria transmission in the southern and western forest areas of Côte d’Ivoire. Ann Trop Med Parasitol. 2011;105:13–24.
Ahmed A, Abubakr M, Ali Y, Siddig EE, Mohamed NS. Vector control strategy for Anopheles stephensi in Africa. Lancet Microbe 2022 (online ahead of print).
Ahmed A, Pignatelli P, Elaagip A, Hamid MMA, Alrahman OF, Weetman D. Invasive malaria vector Anopheles stephensi mosquitoes in Sudan, 2016–2018. Em Infect Dis. 2021;27:2952–4.
Ahmed A, Khogali R, Elnour M-AB, Nakao R, Salim B. Emergence of the invasive malaria vector Anopheles stephensi in Khartoum State, Central Sudan. Parasit Vectors. 2021;14:511.
Abubakr MSH, Mahdi I, Altahir O, Abdelbagi H, Mohamed NS, Ahmed A. The phylodynamic and spread of the invasive Asian malaria vectors, Anopheles stephensi, in Sudan. Biology. 2022;11:409.
WHO. World malaria report 2020: 20 years of global progress and challenges. Geneva, Woorld Health Organization.; 2020.
Abdelwhab OF, Elaagip A, Albsheer MM, Ahmed A, Paganotti GM, Hamid MMA. Molecular and morphological identification of suspected Plasmodium vivax vectors in Central and Eastern Sudan. Malar J. 2021;20:132.
Cobani MY, Bashir NH, Abd Elrahman SH. Mapping of Anopheles mosquitoes (Diptera: Culicidae) in Gedarif State, Eastern Sudan. Int J Mosq Res. 2017;4:28–32.
Abdalla AI, Hassan AE, Kehail MA. Population density, developmental period and fecundity of Anopheles mosquito (Diptera: Culicidae) in the Gezira State, Sudan. Gezira J Health Sci. 2017;13:1.
Kent RJ, Norris DE. Identification of mammalian blood meals in mosquitoes by a multiplexed polymerase chain reaction targeting cytochrome B. Am J Trop Med Hyg. 2005;73:336–42.
Gunathilaka N, Denipitiya T, Hapugoda M, Abeyewickreme W, Wickremasinghe R. Determination of the foraging behaviour and blood meal source of malaria vector mosquitoes in Trincomalee District of Sri Lanka using a multiplex real time polymerase chain reaction assay. Malar J. 2016;15:242.
Keven JB, Walker ED, Venta PJ. A microsatellite multiplex assay for profiling pig DNA in mosquito bloodmeals. J Med Entomol. 2019;56:907–14.
Burkot T, Goodman W, DeFoliart G. Identification of mosquito blood meals by enzyme-linked immunosorbent assay. Am J Trop Med Hyg. 1981;30:1336–41.
Gomes LA, Duarte R, Lima DC, Diniz BS, Serrão ML, Labarthe N. Comparison between precipitin and ELISA tests in the bloodmeal detection of Aedes aegypti (Linnaeus) and Aedes fluviatilis (Lutz) mosquitoes experimentally fed on feline, canine and human hosts. Mem Inst Oswaldo Cruz. 2001;96:693–5.
Mohamed NS, Albsheer MMA, Abdelbagi H, Siddig EE, Mohamed MA, Ahmed AE, et al. Genetic polymorphism of the N-terminal region in circumsporozoite surface protein of Plasmodium falciparum field isolates from Sudan. Malar J. 2019;18:333.
Mohamed NS, Abdelbagi H, Osman HA, Ahmed AE, Yousif AM, Edris YB, et al. A snapshot of Plasmodium falciparum malaria drug resistance markers in Sudan: a pilot study. BMC Res Notes. 2020;13:512.
Gillies MT, Coetzee M. A supplement to the anophelinae of Africa South of the Sahara. S Afr Inst Med Res Annu Rep. 1987;55:1–141.
Coetzee M. Key to the females of Afrotropical Anopheles mosquitoes (Diptera: Culicidae). Malar J. 2020;19:70.
Scott JA, Brogdon WG, Collins FH. Identification of single specimens of the Anopheles gambiae complex by the polymerase chain reaction. Am J Trop Med Hyg. 1993;49:520–9.
Kocher TD, Thomas WK, Meyer A, Edwards SV, Pääbo S, Villablanca FX, et al. Dynamics of mitochondrial DNA evolution in animals: amplification and sequencing with conserved primers. Proc Natl Acad Sci USA. 1989;86:6196–200.
Nagy ZT, Sonet G, Glaw F, Vences M. First large-scale DNA barcoding assessment of reptiles in the biodiversity hotspot of Madagascar, based on newly designed COI primers. PLoS ONE. 2012;7:e34506.
Rubio J, Benito A, Berzosa P, Roche J, Puente S, Subirats M, et al. Usefulness of seminested multiplex PCR in surveillance of imported malaria in Spain. J Clin Microbiol. 1999;37:3260–4.
Mohamed NS, AbdElbagi H, Elsadig AR, Ahmed AE, Mohammed YO, Elssir LT, et al. Assessment of genetic diversity of Plasmodium falciparum circumsporozoite protein in Sudan: the RTS, S leading malaria vaccine candidate. Malar J. 2021;20:436.
Service M. The importance of ecological studies on malaria vectors. Bull Soc Vector Ecol. 1989;14:26–38.
Garrett-Jones C, Boreham P, Pant C. Feeding habits of anophelines (Diptera: Culicidae) in 1971–78, with reference to the human blood index: a review. Bull Entomol Res. 1980;70:165–85.
Githeko A, Service M, Mbogo C, Atieli F, Juma F. Origin of blood meals in indoor and outdoor resting malaria vectors in western Kenya. Acta Trop. 1994;58:307–16.
Animut A, Balkew M, Gebre-Michael T, Lindtjørn B. Blood meal sources and entomological inoculation rates of anophelines along a highland altitudinal transect in south-central Ethiopia. Malar J. 2013;12:76.
Abdalla H, Matambo TS, Koekemoer LL, Mnzava AP, Hunt RH, Coetzee M. Insecticide susceptibility and vector status of natural populations of Anopheles arabiensis from Sudan. Trans R Soc Trop Med Hyg. 2008;102:263–71.
Finney M, McKenzie BA, Rabaovola B, Sutcliffe A, Dotson E, Zohdy S. Widespread zoophagy and detection of Plasmodium spp. in Anopheles mosquitoes in southeastern Madagascar. Malar J. 2021;20:25.
Toto H. Up to date Anopheline mosquitoes distribution in Sennar and South Darfur states during dry season (Sudan) 2009. Sudan J Public Health. 2014;9:53–8.
Mayi MPA, Bamou R, Djiappi-Tchamen B, Fontaine A, Jeffries CL, Walker T, et al. Habitat and seasonality affect mosquito community composition in the west region of Cameroon. Insects. 2020;11:312.
Guelbéogo WM, Gonçalves BP, Grignard L, Bradley J, Serme SS, Hellewell J, et al. Variation in natural exposure to anopheles mosquitoes and its effects on malaria transmission. eLife. 2018;7:e32625.
Boreham P, Lenahan J, Boulzaguet R, Storey J, Ashkar T, Nambiar R, et al. Studies on multiple feeding by Anopheles gambiae s.l. in a Sudan savanna area of north Nigeria. Trans R Soc Trop Med Hyg. 1979;73:418–23.
Klowden MJ, Briegel H. Mosquito gonotrophic cycle and multiple feeding potential: contrasts between Anopheles and Aedes (Diptera: Culicidae). J Med Entomol. 1994;31:618–22.
Wekesa J, Yuval B, Washino R. Multiple blood feeding in Anopheles freeborni (Diptera: Culicidae). Am J Trop Med Hyg. 1995;52:508–11.
Massebo F, Balkew M, Gebre-Michael T, Lindtjørn B. Blood meal origins and insecticide susceptibility of Anopheles arabiensis from Chano in South-West Ethiopia. Parasit Vectors. 2013;6:44.
El-Sadig SM, Mohamed NS, Ahmed ES, Alayeib MA, Tahir LH, Edris AMM, et al. Obstacles faced by healthcare providers during COVID-19 pandemic in Sudan. J Infect Dev Ctries. 2021;15:1615–7.
Koella JC, Sörensen FL, Anderson R. The malaria parasite, Plasmodium falciparum, increases the frequency of multiple feeding of its mosquito vector, Anopheles gambiae. Proc Royal Soc B. 1998;265:763–8.
Hawking F, Mellanby H, Terry EJ, Webber WAF. Transmission of Plasmodium knowlesi by Anopheles stephensi. Trans R Soc Trop Med Hyg. 1957;51:397–402.
Shililu JI, Maier WA, Seitz HM, Orago AS. Seasonal density, sporozoite rates and entomological inoculation rates of Anopheles gambiae and Anopheles funestus in a high-altitude sugarcane growing zone in Western Kenya. Trop Med Int Health. 1998;3:706–10.
Bøgh C, Clarke SE, Pinder M, Sanyang F, Lindsay SW. Effect of passive zooprophylaxis on malaria transmission in The Gambia. J Med Entomol. 2001;38:822–8.
Acknowledgements
We are grateful to the Integrated Vector Management staff for their help and support during the study.
Funding
Not applicable.
Author information
Authors and Affiliations
Contributions
OA, NSM, and AA: Study designing. MA: Field work support. OA, HA, and EES: Performed experiments. NSM: Data analysis. OA, EES, AA, and NSM: Manuscript preparation. All authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Ethics approval and consent to participate
Ethical approval for conducting the study was obtained from the Integrated Vector Management, Sudan Federal Ministry of Health.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Altahir, O., AbdElbagi, H., Abubakr, M. et al. Blood meal profile and positivity rate with malaria parasites among different malaria vectors in Sudan. Malar J 21, 124 (2022). https://doi.org/10.1186/s12936-022-04157-y
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12936-022-04157-y