Abstract
Background
Rapid diagnostic tests (RDTs) have become the most common diagnostic tool for detection of Plasmodium falciparum malaria, in particular in remote areas. RDT blood spots provide a source of parasite DNA for molecular analysis. In this study, the utility of RDTs for molecular analysis and the performance of different methods for whole genome amplification were investigated.
Methods
Positive P. falciparum RDTs were collected from Kayin, Myanmar from August 2014 to January 2016. The RDT samples were stored for 6 months, 9 months, 20 months, 21 months, and 32 months before DNA extraction and subsequent molecular analysis of P. falciparum kelch 13 (pfkelch13) mutations, P. falciparum multidrug resistance 1 (pfmdr1), and P. falciparum plasmepsin 2 (pfplasmepsin2) gene amplification. In addition, performance of four whole genome amplification (WGA) kits were compared, including REPLI-g®, MALBACTM, PicoPLEX®, and GenomePlex®, for which DNA quantity and quality were compared between original DNA and post-WGA products.
Results
The proportion of successful amplification of the different molecular markers was similar between blood spots analysed from RDTs stored for 6, 9, 20, 21, or 32 months. Successful amplification was dependent on the molecular markers fragment length (p value < 0.05): 18% for a 1245 bp fragment of pfkelch13, 71% for 364 bp of pfkelch13, 81% for 87 bp of pfmdr1, 81% for 108 bp of pfplasmepsin2. Comparison of the four WGA assay kits showed that REPLI-g®, MALBACTM, and PicoPLEX® increased the quantity of DNA 60 to 750-fold, whereas the ratio of parasite DNA amplification over human DNA was most favourable for MALBAC®. Sequencing results of pfkelch13, P. falciparum chloroquine resistance transporter (pfcrt), P. falciparum dihydrofolate reductase (pfdhfr) and six microsatellite markers assessed from the post-WGA product was the same as from the original DNA.
Conclusions
Blood spots from RDTs are a good source for molecular analysis of P. falciparum, even after storage up to 32 months. WGA of RDT-derived parasite DNA reliably increase DNA quantity with sufficient quality for molecular analysis of resistance markers.
Similar content being viewed by others
Background
Rapid diagnostic test kits (RDTs) have become the most common diagnostic tool for identifying patients with Plasmodium falciparum malaria, in particular in more remote settings where microscopy services are not readily available [1]. RDTs testing positive for P. falciparum are a potential source of P. falciparum DNA. Previous studies have described several techniques to recover Plasmodium DNA from RDT [2,3,4] and different DNA extraction methods vary regarding quality and quantity of obtained parasite DNA [2]. Out of three different extraction techniques evaluated, phenol/chloroform extraction was most efficient in obtaining parasite DNA from RDT samples [2]. RDT derived Plasmodium DNA has been evaluated for assessment of 18S rRNA [2, 4, 5], the P. falciparum histidine rich protein-2 and 3 genes [6], P. falciparum polymorphic microsatellite markers [7], drug resistance markers pfcrt and pfmdr1 [8] and tRNA methionine-based quantification of Plasmodium parasites [9]. Successful assessment varied between 70% and 100% depending on the target gene and method. Previous study shows that the RDT sample stored at room temperature for 36 months (n = 1) was successfully amplified for 18S rRNA gene based on nested PCR assay [2]. However, the limited quality and quantity of extracted DNA from RDT blood spots remains an important issue for its use in molecular genetic investigations.
Whole genome amplification (WGA) is a technique to increase the quantity of DNA including Plasmodium DNA from RDTs [10]. Several commercial kits are available for WGA, including REPLI-g®, MALBACTM, PicoPLEX®, and GenomePlex®. The REPLI-g® (Qiagen, Hilden, North Rhine-Westphalia, Germany) is based on a multiple displacement amplification (MDA) method utilizing phi29 DNA polymerase in isothermal amplification [11], and has been used successfully to amplify P. falciparum DNA from patient samples [12, 13]. MALBACTM (Yikion Genomic, Nantong, China) is based on multiple annealing and looping-based amplification cycles. Studies on human DNA showed that MALBACTM had very high amplification homogeneity, a low allele dropout rate, good reproducibility and adequate detection of copy number variations (CNVs) and single nucleotide polymorphisms (SNPs) [14]. PicoPLEX® (Takara Bio, Mountain View, California, U.S.A.) and GenomePlex® (Sigma–Aldrich, Foster City, California, U.S.A.) are based on degenerate-oligonucleotide-primed PCR (DOP-PCR). Comparisons between PicoPLEX® and GenomePlex® showed that PicoPLEX® provided higher reproducibility and lower sequencing error frequency than GenomePlex®, whereas Genomplex had lower GC-bias than PicoPLEX® [15]. The size of amplicons varied with different WGA kits: 4.0–10.0 kb in REPLI-g®, 0.5–2.0 kb in MALBACTM, 0.25–1.0 kb in PicoPLEX®, and 0.1–1.0 kb in GenomePlex® [16,17,18]. However, a comparative study of WGA kits focusing on the quality and quantity of P. falciparum DNA obtained from stored RDT samples has not been reported.
The current study assessed the utility for molecular marker analysis of DNA extracted from stored malaria RDTs and the effect of the duration of storage. In addition, four different WGA kits were evaluated for the quantitative and qualitative yield of parasite DNA form stored RDTs, including an assessing concordance of the molecular markers results between original DNA and post-WGA samples. Finally, estimated costs and time consumption were briefly discussed.
Methods
Ethical approvals for the study were obtained from the ethical review committees of the Faculty of Tropical Medicine, Mahidol University (MUTM 2012-045-05) and the Department of Medical Research, Myanmar (Ethics/DMR/2015/109E and Ethics/DMR/2015/113E). Informed consent was obtained from all participants.
RDT samples and DNA extraction
Used RDT kits SD BIOLINE Malaria Ag P.f/P.v kits (Abbott Diagnostics, USA) from confirmed P. falciparum positive cases (n = 108) were collected from Kayin State, Myanmar from August 2014 to January 2016, which obtained from the previous study reviewed and approved by the Ethical Review Committee of the Department of Medical Research, Myanmar (Ethics/DMR/2015/113E, Ethics/DMR/2015/109E). They were used for the evaluation of the impact of RDT samples stored for 6 months (n = 27), 9 months (n = 5), 20 months (n = 36), 21 months (n = 36), and 32 months (n = 4) at room temperature until DNA extraction and molecular analysis. Positive control samples from microscopically confirmed P. falciparum malaria patients from Northeastern Thailand (n = 5) were prepared by loading 5 µl of whole blood samples on the RDT, and stored for 24 h before DNA extraction and molecular analysis.In addition, confirmed P. falciparum positive RDT kits (n = 33) were used for validation of four WGA kits (n = 8) and analysis of the analytical concordance between the molecular marker results of original DNA and post-WGA samples (n = 25).
The RDT samples were extracted using the elution methods adapted from previous study [2, 3]. Crude DNA extraction was performed by cutting the filter paper in the RDTs, elute the DNA using PCR-grade water (100 µl), mixing by vortex, incubating at 95 °C for 10 min, and transferring the supernatant into a DNA collection tube.
Utility of DNA from RDTs for molecular analysis
DNA samples extracted from RDTs were used to analyse the polymorphism of P. falciparum kelch-13 (Pfkelch13), pfmdr1, and pfplasmepsin2 gene amplification. A total of 108 samples were analysed for Pfkelch13 mutations based on PCR assays of 1245 bp and 364 bp, respectively. The samples were also analysed for pfmdr1 (n = 108) and pfplasmepsin2 (n = 108) CNVs based on relative quantitative real-time PCR assay as described previously for pfmdr1 [19] and pfplasmepsin2 [20]. The success rates for each molecular marker was calculated as:
Comparison of success rates between storage duration and between each molecular marker were determined using Chi Square Tests (SPSS version 25).
Validation of four WGA kits
DNA samples (n = 8) were extracted from RDTs kits and positivity for P. falciparum was confirmed using 18S rRNA gene [21, 22]. Before and after whole genome amplification, the DNA samples were purified using DNA clean kit (Zymoresearch, USA). Five microliters of cleaned DNA samples (n = 8) were used following standard manufacturer protocols of the different WGA kits, including REPLI-g® Midi Kits (Qiagen, Germany), MALBACTM WGA kit (Yikon Genomics, China), PicoPLEX® (Takara Bio, U.S.A.) and GenomePlex® (Sigma–Aldrich, U.S.A.) (Fig. 1). Human DNA samples without P. falciparum DNA was used as positive control for WGA and as negative control for parasites DNA amplification.
To assess the efficiency of whole genome amplification, original DNA samples extracted from RDT and post-WGA samples were checked for DNA quality using Nanodrop and DNA quantity using Qubit kit. To measure the quantity of parasites DNA, relative quantitative real-time PCR assay was performed based on amplification of the 18S rRNA gene [21, 22]. The amplification performance of the kits was evaluated based on the quantitative increase in overall DNA, parasite DNA, and the ratio between parasite and human DNA in the amplified sample. Reproducibility of WGA was determined based on the coefficient of variation of the fold-change of DNA concentration for each WGA kit.
Molecular markers analysis of original DNA and post-WGA samples
To evaluate the accuracy of molecular marker assessments from post-WGA samples compared to the original DNA sample. Crude DNA extracts from P. falciparum positive RDTs kits (n = 25) were purified using DNA clean kit, and then 5 µL of each purified DNA was subjected to amplification using both the REPLI-g® and MALBACTM kits. DNA samples of original DNA and post-WGA were accessed using real-time PCR assay for molecular markers, such as pfmdr1 [19] and pfplasmepsin2 CNVs [20]. In addition, the amplicons of pfkelch13 [23], P. falciparum chloroquine resistance transporter gene (pfcrt), P. falciparum dihydrofolate reductase gene (pfdhfr) and six micro-satellite markers (ARA2, PfPK2, TAA81, Poly A, TA1, and TAA87) [24] were sequence-analysed. The analytical concordance between the results obtained pre- and post-WGA were assessed using Kappa analysis (SPSS version 25). The success rate of molecular markers analysis of each WGA samples were calculated using the formula:
The association of molecular marker success rate and the initial parasites DNA concentration were determined using Spearman’s rank-order test using SPSS version 25.
Results
Success of P. falciparum gene amplification from DNA preps isolated from RDT samples upon varied period of storage
The molecular analysis revealed that: (1) DNA isolated from RDT samples can be successfully used for amplification of P. falciparum gene, and no significant difference of success rates exists between storage duration of RDT samples at 6, 9, 20, 21, and 32 months (p-value > 0.05); (2) Success rate did depend on the molecular marker being assessed: with lower success rate (18%) for long fragment of pfkelch13 (1245 bp) compared to higher success rate (71.3%) for short fragment of pfkelch13 (364 bp) (p-value < 0.05, Chi Square); moreover, the success rates for either pfmdr1 fragment (87 bp) or pfplasmepsin2 fragment (108 bp) was 81% (Fig. 2).
Success of whole genome amplification of P. falciparum DNA preps isolated from RDT samples
The total DNA quantity of original DNA extracted from RDT samples was low (1.05 ng/µl on average). After WGA using REPLI-g®, MALBACTM, PicoPLEX®, and GenomePlex® kits, DNA concentrations increased to 415 ng/µl, 15.30 ng/µl, 32.35 ng/µl, and 2.85 ng/µl, respectively (Table 1). The fold-change in DNA concentration was calculated using the formula:
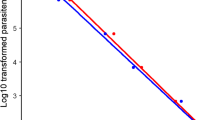
The average fold-change in DNA concentrations using REPLI-g®, MALBACTM, PicoPLEX®, and GenomePlex® were 745.00, 30.51, 60.31 and 3.64, respectively (p-value < 0.05 for all kits, except for GenomePlex®: p-value = 0.167; Table 1). REPLI-g® provided the highest fold increase in DNA concentrations (Additional file 1).Quantitative real-time PCR estimates of P. falciparum DNA (genome/µl) showed a significant increase in parasite DNA quantity between original DNA and post-WGA samples using MALBACTM kit (Table 1). In contrast, no significant differences were observed using REPLI-g®, PicoPLEX®, and GenomePlex® (0.167 ≤ p-value ≤ 0.401) (Table 1). 18S rRNA gene amplification results showed that all four WGA kits could amplify parasite DNA but MALBACTM yielded the highest fold-change in parasite DNA. The reproducibility expressed as coefficient of variation in WGA yield using the REPLI-g®, MALBACTM, PicoPLEX® and GenomePlex® kits ranged from 1.04 to1.14 (Table 1). The ratio of P. falciparum DNA quantity (genome/µl) to total DNA (ng/µl) before and after WGA decreased significantly when using REPLI-g®, PicoPLEX®, and GenomePlex® (Related-Samples Wilcoxon Signed Rank Test, p-value ≤ 0.05), but increased significantly using MALBACTM (Related-Samples Wilcoxon Signed Rank Test, p-value ≤ 0.05) (Table 1). There was a positive correlation between DNA quantity of original DNA and post-WGA using REPLI-g® (rs = 0.631, p-value < 0.05) and MALBACTM (rs = 0.662, p-value < 0.05) (Fig. 3). There was also a correlation between the estimated parasites DNA quantity for original DNA samples and post-WGA using REPLI-g® (rs = 0.834, p-value < 0.05®) and MALBACTM (rs = 0.919, p-value < 0.05) (Fig. 3).
Relationship between DNA concentration of original DNA samples and post-WGA samples using a REPLI-g® (n = 25) and b MALBACTM (n = 25). The relationship between parasites DNA quantity estimated by real-time PCR of original DNA samples and post-WGA samples using c REPLI-g® (n = 23) and d MALBACTM (n = 24)
Success of assessment on the quality of P. falciparum DNA preps isolated from RDT samples using molecular marker typing
The quality of WGA samples using REPLI-g® and MALBACTM were assessed regarding accuracy for molecular marker typing. These kits were selected since REPLI-g® yielded the highest fold-change in DNA concentration and MALBACTM yielded the highest fold-change in estimated parasites concentration.
WGA samples amplified using REPLI-g® (n = 25) and MALBACTM (n = 25) were used for molecular markers typing, including three real-time PCR assays, four nested PCR assays, and six microsatellite markers. The success rates of molecular marker analysis using REPLI-g® and MALBACTM kits ranged from 0–100% and 7.69–100%, respectively. There was an association between the success of molecular marker typing and the initial parasites DNA concentrations, both with REPLI-g® (rs = 0.898, p-value < 0.05) and MALBACTM (rs = 0.929, p-value < 0.05).
With both kits, pfmdr1 and pfplasmepsin2 CNVs was successfully assessed from post-WGA samples in the majority of samples. Concordance with the assessment from original DNA using REPLI-g® kit was high (κ = 1) for pfmdr1 CNV (n = 15) assessment, but lower (κ = 0.582) for pfplasmepsin2 CNVs (n = 14). With the MALBACTM kit (n = 17), concordance for pfmdr1 CNVs (n = 16) and pfplasmepsin2 CNVs (n = 17) assessments was low (κ = 0.776 and κ = 0.605, respectively) (Additional file 2). Amplification of pfkelch13, pfdhfr, pfcrt, and microsatellite markers was successful in 32-72% of WGA samples with either REPLI-g® or MALBACTM WGA kit. The genotyping was accurate in all samples where the target gene was successfully amplified (κ = 1.000).
Cost and time consumption of four WGA kits, namely REPLI-g®, MALBACTM, PicoPLEX® and GenomePlex®, were estimated and summarized. Performance of MALBACTM WGA kit was relatively low time-consuming and less labour intensive (Table 2).
Discussion
Used RDTs provide a valuable source of parasite DNA that can be used for molecular analysis [2,3,4, 7, 8]. The current study investigated the utility of DNA preps from RDT samples for pfkelch13 SNPs, pfmdr1 CNV and pfplasmepsin2 CNV analysis. Even though theDNA preps were of limited quantity and with degradation, molecular typing was successful in 17.59–81.48% of samples. Storage up to 32 months did not negatively affect the accuracy of typing, showing that stored RDTs can be used as a source for Plasmodium DNA typing as also suggested in earlier studies [2, 4, 7, 8]. Comparing short (364 bp) and long (1,245 bp) fragment Pfkelch13 amplicons suggested that PCR product size affects the success rate of molecular analysis using RDT blood spots.
Inadequate amounts of parasite DNA is a limiting factor for conducting molecular assays from RDT blood spots of around 5 µl, in particular when several markers are evaluated [25, 26]. RDT samples were collected from symptomatic cases from the area where the observed geometric mean P. falciparum parasitaemia was reported as 6 Plasmodium genome equivalents/µL (95% CI 2 to 16), so around 30 (95% CI 10 to 80) genomes per blood spot [27]. The purity and amount of DNA will affect outcomes of downstream molecular analysis. Although the WGA using any of the four kits increased parasite DNA, only WGA with REPLI-g® and MALBACTM yielded sufficient P. falciparum DNA purity and quantity needed for further molecular analysis. MALBACTM yielded higher concentrations of parasite DNA compared to REPLI-g®, which makes this kit preferable for amplification using RDT blood spots with low parasites content. A high concordance in microsatellite marker and SNP typing was previously reported using the MDA® kit to amplify parasite DNA from whole blood samples of P. falciparum-infected patients [13]. The current study shows reliable results for molecular marker typing, DNA sequencing and microsatellite marker analysis using REPLI-g® and MALBACTM WGA of RDT blood spots. However, CNV estimates for pfmdr1 and Pfplasmepsin2 were less reliable, suggesting that WGA amplified DNA should not be used for the purpose of CNV assessment. A previous study showed that PicoPLEX® and MALBACTM provided reliable CNVs estimates based on an ion proton platform, whereas CNV estimates were not accurate using MDA or GenomePlex® [16]. As expected from the working principles of WGA and confirmed by the current study, WGA interferes with the assessment of CNV in the parasite genome. This study showed that the success rate of molecular marker analysis was affected by the initial parasite DNA concentration, and WGA with MALBACTM achieved a higher success rate for molecular marker typing than REPLI-g® in concordance. Although an estimated cost of MALBACTM is higher than REPLI-g®, the use of MALBACTM was less time consuming compared to REPLI-g® and provided higher yield of parasite DNA. Therefore, MALBACTM shall be recommended for wide use to generate high-quality parasite DNA in sufficient quantities using stored RDT blood spots containing low parasite DNA quantities. Undoubtedly, good laboratory practices and good laboratory facilities are important to implement these techniques.
Conclusions
This study demonstrates the successful utility of RDT blood spots for molecular analysis and whole genome amplification. WGA from RDT blood spots using the MALBACTM kit yields high-quality parasite DNA in sufficient quantities for preforming molecular assays.
Availability of data and materials
All data generated or analysed during this study are included in this published article and its supplementary information files.
Abbreviations
- DNA:
-
Deoxyribonucleic acid
- RDTs:
-
Malaria rapid diagnostic tests
- WGA:
-
Whole genome amplification
- 18S rRNA :
-
18s ribosomal RNA gene
- Pfmdr1 :
-
P. falciparum multidrug resistance 1 gene
- Pfplasmepsin2 :
-
P. falciparum plasmepsin2 gene
- Pfkelch13 :
-
P. falciparum kelch 13 gene
- Pfcrt :
-
P. falciparum chloroquine resistance transporter gene
- Pfdhfr :
-
P. falciparum dihydrofolate reductase gene
- bp:
-
Base pair
- ng:
-
Nanogram
- µl:
-
Microlitre
References
Mouatcho JC, Goldring JP. Malaria rapid diagnostic tests: challenges and prospects. J Med Microbiol. 2013;62:1491–505.
Veron V, Carme B. Recovery and use of Plasmodium DNA from malaria rapid diagnostic tests. Am J Trop Med Hyg. 2006;74:941–3.
Cnops L, Boderie M, Gillet P, Van Esbroeck M, Jacobs J. Rapid diagnostic tests as a source of DNA for Plasmodium species-specific real-time PCR. Malar J. 2011;10:67.
Ishengoma DS, Lwitiho S, Madebe RA, Nyagonde N, Persson O, Vestergaard LS, et al. Using rapid diagnostic tests as source of malaria parasite DNA for molecular analyses in the era of declining malaria prevalence. Malar J. 2011;10:6.
Papa Mze N, Ahouidi AD, Diedhiou CK, Silai R, Diallo M, Ndiaye D, et al. Distribution of Plasmodium species on the island of Grande Comore on the basis of DNA extracted from rapid diagnostic tests. Parasite. 2016;23:34.
Beshir KB, Sepulveda N, Bharmal J, Robinson A, Mwanguzi J, Busula AO, et al. Plasmodium falciparum parasites with histidine-rich protein 2 (pfhrp2) and pfhrp3 gene deletions in two endemic regions of Kenya. Sci Rep. 2017;7:14718.
Nabet C, Doumbo S, Jeddi F, Sagara I, Manciulli T, Tapily A, et al. Analyzing deoxyribose nucleic acid from malaria rapid diagnostic tests to study Plasmodium falciparum genetic diversity in Mali. Am J Trop Med Hyg. 2016;94:1259–65.
Morris U, Aydin-Schmidt B, Shakely D, Martensson A, Jornhagen L, Ali AS, et al. Rapid diagnostic tests for molecular surveillance of Plasmodium falciparum malaria -assessment of DNA extraction methods and field applicability. Malar J. 2013;12:106.
Robinson A, Busula AO, Muwanguzi JK, Powers SJ, Masiga DK, Bousema T, et al. Molecular quantification of Plasmodium parasite density from the blood retained in used RDTs. Sci Rep. 2019;9:5107.
Telenius H, Carter NP, Bebb CE, Nordenskjold M, Ponder BA, Tunnacliffe A. Degenerate oligonucleotide-primed PCR: general amplification of target DNA by a single degenerate primer. Genomics. 1992;13:718–25.
Dean FB, Hosono S, Fang L, Wu X, Faruqi AF, Bray-Ward P, et al. Comprehensive human genome amplification using multiple displacement amplification. Proc Natl Acad Sci USA. 2002;99:5261–6.
Carret CK, Horrocks P, Konfortov B, Winzeler E, Qureshi M, Newbold C, et al. Microarray-based comparative genomic analyses of the human malaria parasite Plasmodium falciparum using Affymetrix arrays. Mol Biochem Parasitol. 2005;144:177–86.
Wang Y, Nair S, Nosten F, Anderson TJ. Multiple displacement amplification for malaria parasite DNA. J Parasitol. 2009;95:253–5.
Yao YX, La YF, Di R, Liu QY, Hu WP, Wang XY, et al. Comparison of different single cell whole genome amplification methods and MALBAC applications in assisted reproduction. Yi Chuan. 2018;40:620–31.
Chen D, Zhen H, Qiu Y, Liu P, Zeng P, Xia J, et al. Comparison of single cell sequencing data between two whole genome amplification methods on two sequencing platforms. Sci Rep. 2018;8:4963.
Zhang X, Liang B, Xu X, Zhou F, Kong L, Shen J, et al. The comparison of the performance of four whole genome amplification kits on ion proton platform in copy number variation detection. Biosci Rep. 2017;37:e20170252.
Li N, Wang L, Wang H, Ma M, Wang X, Li Y, et al. The performance of whole genome amplification methods and next-generation sequencing for pre-implantation genetic diagnosis of chromosomal abnormalities. J Genet Genomics. 2015;42:151–9.
Treff NR, Su J, Tao X, Northrop LE, Scott RT Jr. Single-cell whole-genome amplification technique impacts the accuracy of SNP microarray-based genotyping and copy number analyses. Mol Hum Reprod. 2011;17:335–43.
Price RN, Uhlemann AC, Brockman A, McGready R, Ashley E, Phaipun L, et al. Mefloquine resistance in Plasmodium falciparum and increased pfmdr1 gene copy number. Lancet. 2004;364:438–47.
Amato R, Lim P, Miotto O, Amaratunga C, Dek D, Pearson RD, et al. Genetic markers associated with dihydroartemisinin-piperaquine failure in Plasmodium falciparum malaria in Cambodia: a genotype-phenotype association study. Lancet Infect Dis. 2017;17:164–73.
Imwong M, Hanchana S, Malleret B, Renia L, Day NP, Dondorp A, et al. High-throughput ultrasensitive molecular techniques for quantifying low-density malaria parasitemias. J Clin Microbiol. 2014;52:3303–9.
Kamau E, Tolbert LS, Kortepeter L, Pratt M, Nyakoe N, Muringo L, et al. Development of a highly sensitive genus-specific quantitative reverse transcriptase real-time PCR assay for detection and quantitation of plasmodium by amplifying RNA and DNA of the 18S rRNA genes. J Clin Microbiol. 2011;49:2946–53.
Ashley EA, Dhorda M, Fairhurst RM, Amaratunga C, Lim P, Suon S, et al. Spread of artemisinin resistance in Plasmodium falciparum malaria. N Engl J Med. 2014;371:411–23.
Anderson TJ, Su XZ, Bockarie M, Lagog M, Day KP. Twelve microsatellite markers for characterization of Plasmodium falciparum from finger-prick blood samples. Parasitology. 1999;119:113–25.
Berzosa P, de Lucio A, Romay-Barja M, Herrador Z, Gonzalez V, Garcia L, et al. Comparison of three diagnostic methods (microscopy, RDT, and PCR) for the detection of malaria parasites in representative samples from Equatorial Guinea. Malar J. 2018;17:333.
Das S, Peck RB, Barney R, Jang IK, Kahn M, Zhu M, et al. Performance of an ultra-sensitive Plasmodium falciparum HRP2-based rapid diagnostic test with recombinant HRP2, culture parasites, and archived whole blood samples. Malar J. 2018;17:118.
Chaumeau V, Kajeechiwa L, Fustec B, Landier J, Naw Nyo S, Nay Hsel S, et al. Contribution of asymptomatic Plasmodium infections to the transmission of malaria in Kayin State Myanmar. J Infect Dis. 2019;219:1499–509.
Acknowledgements
We would like to thank Ms. Jureeporn Duanguppama, Mr. Chanon Kunason, Ms. Watcharee Pagornrat, and Ms. Wanassanan Madmanee for their help.
Funding
The research was funded by Thailand Science Research and Innovation (TSRI), Grant No. RTA6280006 and the Wellcome Trust of Great Britain, UK.
Author information
Authors and Affiliations
Contributions
SS and MI contributed to study design. KS, FS, and FN collected samples. SS and KS undertook laboratory work. SS, KS, and MI analysed data. SS, VM, NW, AD and MI drafted the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Ethical approvals for the study were obtained from the ethical review committees of the Faculty of Tropical Medicine, Mahidol University (MUTM 2012-045-05) and the Department of Medical Research, Myanmar (Ethics/DMR/2015/109E and Ethics/DMR/2015/113E). Informed consent was obtained from all participants.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Additional file 1.
Bar Chart of DNA concentrations comparing original DNA and post WGA samples using four WGA kits: REPLI-g®, MALBACTM, PicoPLEX®, and GenomePlex®.
Additional file 2.
Analytical concordance (based on kappa statistic) of Pfmdr1 CNVs and Pfplasmepsin2 CNVs results obtained from original DNA samples extracted from RDT and post-WGA samples using REPLI-g® and MALBACTM kits.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Srisutham, S., Suwannasin, K., Mathema, V.B. et al. Utility of Plasmodium falciparum DNA from rapid diagnostic test kits for molecular analysis and whole genome amplification. Malar J 19, 193 (2020). https://doi.org/10.1186/s12936-020-03259-9
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12936-020-03259-9