Abstract
Background
Accurate diagnosis of Plasmodium infection is crucial for prompt malaria treatment and surveillance. Microscopic examination has been widely applied as the gold standard for malaria diagnosis in most part of malaria endemic areas, but its diagnostic value has been questioned, particularly in submicroscopic malaria. In this study, the diagnostic performance of microscopic examination and nested polymerase chain reaction (PCR) was evaluated to establish optimal malaria diagnosis method in Myanmar.
Methods
A total of 1125 blood samples collected from residents in the villages and towns located in Naung Cho, Pyin Oo Lwin, Tha Beik Kyin townships and Mandalay of Upper Myanmar were screened by microscopic examination and species-specific nested PCR method.
Results
Among the 1125 blood samples, 261 samples were confirmed to be infected with malaria by microscopic examination. Evaluation of the 1125 samples by species-specific nested PCR analysis revealed that the agreement between microscopic examination and nested PCR was 87.3% (261/299). Nested PCR successfully detected 38 Plasmodium falciparum or Plasmodium vivax infections, which were missed in microscopic examination. Microscopic examinations also either misdiagnosed the infected Plasmodium species, or did not detect mixed infections with different Plasmodium species in 31 cases.
Conclusions
The nested PCR method is more reliable than conventional microscopic examination for the diagnosis of malaria infections, and this is particularly true in cases of mixed infections and submicroscopic infections. Given the observed higher sensitivity and specificity of nested PCR, the molecular method holds enormous promise in malaria diagnosis and species differentiation, and can be applied as an effective monitoring tool for malaria surveillance, control and elimination in Myanmar.
Similar content being viewed by others
Background
Accurate diagnosis of malaria is important for providing realistic estimates of malaria burden and preventing misinformed interventions [1, 2]. For over a century, microscopic examination of thick and thin blood smears has been considered the gold standard for the diagnosis of malaria in most malaria-endemic regions due to its ability to identify infected Plasmodium species and quantify parasitaemia levels at a low cost [3]. However, this method has several limitations in that it is laborious and time-consuming, and requires the availability of well-trained and highly qualified microscopists. Misdiagnosis can occur frequently in cases of low parasitaemia and incorrect species identification, and this can lead to incorrect treatment [4]. To overcome these limitations, several alternative methods for malaria diagnosis have been developed. One of these is a molecular detection method based on the amplification of parasitic DNA [5,6,7,8,9]. Polymerase chain reaction (PCR)-based diagnostic protocols have been recognized as powerful tools to detect mixed Plasmodium species infections and differentiate the infected species with high specificity and sensitivity [10,11,12,13]. Nevertheless, PCR methods have some limitations, such as high cost and low applicability in rural areas or field-based settings without adequate laboratory equipment [14]. Rapid diagnostic tests (RDTs) for parasite antigen detection provide reliable results in a short time-period and offer a useful alternative to microscopy in conditions where microscopic examination is not feasible [15,16,17]. Due to these advantages, RDTs have been introduced as a diagnostic tool in many malaria-endemic areas. However, several important issues remain to be addressed, including diagnostic accuracy, high cost, and performance efficacy under unfavorable field conditions. The performance of RDTs can also be affected by the detection of residual parasite antigens from previous infections, leading to a false positive result, and deletions or mutations within the parasite antigen, resulting in a false negative [18, 19]. Moreover, currently available RDTs do not allow quantification and differentiation of Plasmodium species other than Plasmodium falciparum and Plasmodium vivax.
The incidence of malaria in Myanmar has decreased remarkably in recent years, but malaria is still one of the major public health concerns in the country. Myanmar accounts for approximately 77% of the estimated cases and 79% of the estimated malaria deaths in the Greater Mekong Subregion [20]. Moreover, the malaria burden and the emergence of drug resistant malaria parasites along the international border areas of Myanmar are significant challenges for the National Malaria Control Programme [21,22,23,24]. All Plasmodium species associated with human infection, P. falciparum, P. vivax, Plasmodium malariae, and Plasmodium ovale, are considered to occur in Myanmar [25, 26]. Plasmodium knowlesi infections have also occurred in some areas, predominantly as a co-infection with either P. falciparum or P. vivax [27]. The combined use of microscopic examination and RDTs has been widely implemented as a mean to diagnose malaria in endemic areas of Myanmar. However, it is not easy to clearly identify infected Plasmodium species and mixed infections with different malaria parasites using these two diagnostic methods. Moreover, information on the diagnostic efficiency of these diagnostic methods is also highly limited.
In this study, the accuracy of routine microscopic examination was analysed by comparing to species-specific nested PCR detection method. PCR method produced conflicting results, particularly in cases of mixed infections with different Plasmodium species and submicroscopic infections. These results suggest that molecular diagnostic approach is more reliable than microscopic examination for the accurate diagnosis of Plasmodium species as a part of the malaria surveillance programs in Myanmar.
Methods
Study areas and microscopic examination
Between August 2013 and December 2015, field surveys for malaria were conducted in towns and villages located in the regions of Naung Cho, Pyin Oo Lwin, Tha Beik Kyin townships and Mandalay in Upper Myanmar (Fig. 1). These regions were identified as malaria endemics in field surveys over the last few years; malaria transmission was heterogeneous and seasonal with most clinical cases occurred during the rainy season. A total of 1125 residents, 325 suspected malaria patients with clinical symptoms and 800 healthy individuals without any malaria symptom, were enrolled in this study. For the microscopic examination, thick and thin blood smears were prepared from a finger prick, and stained with 10% Giemsa for 15 min. The stained smears were observed with a light microscope at 1000× magnification to confirm the presence of malaria parasites. Microscopic examination of the smears was conducted independently by two experienced microscopists in the Department of Medical Research Pyin Oo Lwin Branch, Myanmar. Agreement between the independent readings of the two microscopists was 99.9% (p < 0.001). A slide was considered negative if no parasite was observed in a count of 500 leucocytes.
Blood collection for molecular analysis
Finger-prick blood samples were taken from 1125 residents. Written consent was obtained from each individual prior to blood collection. Two or three drops of blood (approximately 50 μl) from a finger prick were spotted on Whatman 3 MM filter paper (GE Healthcare, Maidstone, UK) and allowed to air dry for subsequent molecular analysis.
Ethics
This study was approved by the Ministry of Health, Myanmar, and by the Ethical Review Committee of Inha University School of Medicine, Korea. Informed written consent and permission (in the case of children under 18 years of age) were obtained from each patient or a parent/legal guardian.
Polymerase chain reaction
Genomic DNA was isolated from each spotted blood filter paper by using the QIAamp DNA Blood Mini Kit (Qiagen, Hilden, Germany). Genomic DNA was eluted with 100 μl of sterile H2O. Nested PCR specific to the five human malaria parasite species (P. falciparum, P. vivax, P. ovale curtisi, P. ovale wallikeri and P. malariae) was performed using a pair of primers targeted to the 18S rRNA gene described previously [5, 6]. P. knowlesi was detected by PCR using a set of primers specific to the 18S rRNA gene [28]. For the primary PCR reaction, 2 μl of purified genomic DNA (approximately 10 ng of DNA) were used in a 30 μl reaction. For the nested PCR, 1 μl of the primary PCR reactant were used as a template in a 30 μl reaction. The amplified PCR products were analysed using a 1.5% agarose gel, stained with ethidium bromide, and observed under ultraviolet light. To rule out false positives, the genomic DNA isolated from a healthy Korean individual residing in a region not considered endemic for malaria was included a negative control in all PCR reactions. The presence or absence of different Plasmodium species was analysed with species-specific amplicon sizes. Randomly selected PCR products for each Plasmodium species were also sequenced to confirm the accuracy of the amplified products.
Statistic analysis
The 95% confidence intervals (CI) were calculated as appropriate and the value of P < 0.05 was considered statistically significant.
Results
Microscopic examination
Thin and thick blood smears from a total of 1125 residents including 325 suspected malaria patients were examined for the presence of malaria parasites using microscope. The results of the microscopic examination identified 261 cases (23.2, 95% CI 20.7–25.7) as positive for malaria infection (Table 1). All positive results were obtained in blood samples from 261 suspected malaria patients. The remained 64 suspected malaria patients and 800 non-symptomatic healthy residents were determined to negatives. Of the 261 microscopically confirmed cases, 135 cases (51.7%) were P. falciparum mono-infections, 94 cases (36.0%) were P. vivax mono-infections, and 31 cases (11.9%) were P. falciparum and P. vivax mixed infections. There was a case (0.4%) confirmed as a P. malariae mono-infection.
Species-specific nested PCR analysis
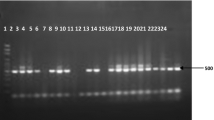
The 1125 blood samples were analysed for the presence of Plasmodium DNA by species-specific nested PCR. The representative species-specific nested PCR amplification results for the Plasmodium species are shown in Fig. 2. Correct amplifications for the sequences from each Plasmodium species were confirmed by nucleotide sequencing analysis of randomly selected PCR products. Plasmodium infection was confirmed in 299 cases (26.6, 95% CI 24.0–29.2%). The 261 positive cases were corresponded to the samples detected by microscopic examination, but the 38 positive cases were negative in the microscopic examination. The specific Plasmodium species were identified as follows: P. falciparum (140 cases), P. vivax (99 cases), P. malariae (1 case), P. ovale curtisi (2 cases), and mixed infection with P. falciparum and P. vivax (52 cases), mixed infection with P. falciparum and P. malariae (4 cases), and mixed infection with P. vivax and P. ovale curtisi (1 case) (Table 2). No case of P. knowlesi or P. ovale wallikeri infection was detected in the analysed samples.
Identification of Plasmodium species by species-specific nested PCR. A total of 299 blood samples were confirmed to positive by species-specific nested PCR. Representative results are presented. Lane M, 100-bp ladder; lanes 1–4, Plasmodium falciparum (205 bp amplicon); lanes 5–8, Plasmodium vivax (120 bp amplicon); lanes 9–12, mixed with P. falciparum and P. vivax; lanes 13–14, mixed with P. falciparum and Plasmodium malariae (145 bp amplicon); lane 15, mixed with P. vivax and Plasmodium ovale curtisi (800 bp amplicon); lane NC, negative control. No positive case was detected for Plasmodium knowlesi
Comparison of microscopic examination and nested PCR for Plasmodium detection
The results of the nested PCR analysis were different from those of the microscopic examination, and an overall comparison of the results for these active malaria detection methods is summarized in Table 3. There was 87.3% (261/299) agreement between PCR and microscopy regarding the parasite species identification in the analysed blood samples. However, microscopic examination misdiagnosed the Plasmodium species, or did not detect mixed infections in 31 samples. Neither mono-infection nor mixed infections of P. ovale were detected by microscopic examination. However, 2 P. ovale curtisi mono-infections and a mixed infection of P. ovale curtisi and P. vivax, which were misdiagnosed to P. vivax infections in microscopic examination, were detected by nested PCR. Moreover, 4 cases of P. falciparum and P. malariae mixed infections were detected by nested PCR analysis, which were misdiagnosed as P. falciparum mono-infections or mixed infections with P. falciparum and P. vivax under microscopy. The number of P. falciparum and P. vivax mixed infections also increased from 31, based on the microscopic examination, to 52 in the nested PCR analysis. A total of 38 among the 864 microscopy negative samples were also identified to P. falciparum (23 cases) or P. vivax (15 cases) infections. These results suggest that the molecular approach is more sensitive and reliable than microscopic examination, particularly in the case of mixed infections and submicroscopic infections.
Discussion
Accurate diagnosis of the Plasmodium species is not only important for establishing a successful treatment regimen, but also for designing effective malaria control measures in endemic regions where multiple species of malaria parasites exist. Misidentification of the Plasmodium species could cause a severe public health concerns since it could extend the parasite clearance times and lead to recrudescence and drug resistance [29,30,31]. Conventional microscopic examination by qualified microscopists has been adopted as the primary method for malaria diagnosis and surveillance in Myanmar. Recently, RDTs were introduced as effective screening tools in field-based malaria surveys since they provide readily available results, and thereby enable rapid treatment. However, only limited information is available regarding the effectiveness of the parasite detection techniques that are widely used in Myanmar. As all common human malaria parasites, as well as P. knowlesi, have been shown to exist in Myanmar [25,26,27, 32], correct diagnosis of the infected Plasmodium species and prompt treatment are essential to malaria control and elimination in the country. Therefore, studies are needed to assess quality and impact of these diagnostic methods for optimal malaria diagnosis setting and nationwide malaria surveillance outcomes.
In this study, 1125 blood samples collected from residents in malaria endemic areas in Upper Myanmar were analysed using microscopic examination and species-specific nested PCR. Overall malaria incidence as determined by microscopic examination and nested PCR method were relatively similar, with a coincidence of 87.3%, but important discrepancies between the microscopic examination and nested PCR analysis were also identified. The role of microscopic examination as the gold standard for malaria diagnosis has been questioned due to false negative results at low parasitaemiae levels (less than 20–30 parasites/μl of blood) and frequent errors in species identification in mixed infections [33]. The low diagnosis reliability of microscopy for species-specific and mixed-infections in endemic areas has been reported previously [12, 34,35,36,37,38,39]. Consistent with these previous reports, mixed infections that were missed in the microscopic examination were identified in 27 samples by molecular detection method. In particular, the prevalence of P. falciparum and P. vivax mixed infections highly increased with nested PCR analysis. Misdiagnosis using microscopic examination technique can easily occur, particularly in the case of mixed infections where one parasite species is dominant over the others [40]. It has been postulated that co-existence of P. falciparum and P. vivax in a single human host results in parasitic suppression through interspecies inhibition [41,42,43]. The clinical symptoms in mixed infections can also vary depending on whether it is a P. falciparum or P. vivax super-infection. Although it still remains controversial whether mixed infections involving P. falciparum or P. vivax and other Plasmodium species are beneficial or harmful [44, 45], acute diagnosis of malaria parasites in mixed infections followed by prompt treatment is essential to the prevention of relapses or recrudescences, and the associated severe complications.
Asymptomatic infection, particularly in the cases of P. falciparum and P. vivax, has been recognized an important obstacle to controlling malaria since asymptomatic carriers do not seek treatment for the infection, so that they constitute a reservoir available for new infections. A significant proportion of P. falciparum and P. vivax infections are asymptomatic or subclinical in malaria endemic countries [46,48,49,50,51,52]. Repeated exposure and acquisition of partial immunity can result in persistent low-grade infection, even though infected individuals may not have typical malaria symptoms such as fever and anemia. Consistent with these previous reports, 38 asymptomatic cases (22 cases of P. falciparum and 15 cases of P. vivax), which were not detected by microscopic examination, were identified by molecular detection method. These results suggest that the importance of detection of asymptomatic infections with submicroscopic level in Myanmar for successful malaria control and elimination in the country. Aggressive approach to detect asymptomatic carriers followed by prompt treatment would be required.
The results of this study also provide partial epidemiological information regarding the prevalence of Plasmodium species in the regions studied. Based on the results from the nested PCR analysis, overall malaria infection rate in the residents in the studied endemic areas was 26.6% (299/1125). It has been estimated that total malaria cases confirmed either by microscopy examination or by RDT in Myanmar were 420,808 in 2010 [53]. Most of the confirmed cases were P. falciparum and P. vivax infections (420,462 cases) and the ratio of P. falciparum/P. vivax infections was 70% [53]. Consistent with the previous results, P. falciparum and P. vivax accounted for most of the PCR-confirmed cases in this study. But the ratio of P. falciparum/P. vivax was not as high as previously reported and a high proportion of mixed P. falciparum and P. vivax infections (17.4%) were identified. It is assumed that P. vivax is becoming predominant species in the studied areas with the recent decrease of falciparum malaria cases. Asymptomatic and submicroscopic infections by the two Plasmodium species were also a great concern. The prevalence of P. malariae and P. ovale was low, but single as well as mixed infections involving either P. falciparum or P. vivax were identified. Distribution of P. malariae and P. ovale in Myanmar has been reported previously [25, 26, 54]. In particular, 8 confirmed P. ovale cases (mixed infections with P. vivax and P. malariae) were reported in Pyin Oo Lwin area [25]. These observations, along with the present study, suggest that P. malariae and P. ovale are endemic in Upper Myanmar, though the prevalence is not high. P. malariae and P. ovale infections can be easily missed to diagnose by microscopic examination, primarily due to quite low parasitaemia of the parasites [55, 56]. Moreover, in mixed infections with other Plasmodium species, it is more difficult to differentiate the parasites from a background with large numbers of parasites of other species [55, 56]. In fact, the P. malariae and P. ovale infections that were not detected or misread in microscopic examination were successfully identified by nested PCR methods in this study. High rates of P. knowlesi co-infection with other malaria parasites also has been reported in North-Eastern Myanmar [27], but no cases of P. knowlesi mono- or co-infection were detected in this study. It is worthy to realize the limitation of PCR-based molecular diagnosis methods to detect parasite DNA, especially in very low level of parasitaemia. A routine extraction process of parasite DNA from dried blood filters could result in loss or dilution of parasite DNA for successful amplification [36]. Nevertheless, as confirmed in this study and other previous studies [12, 34,35,36,37,38,39], PCR-based methods showed greater diagnostic performance than conventional microscopic examination to detect Plasmodium species with high specificity and sensitivity. Molecular diagnostic approach is highly recommended for the accurate diagnosis of Plasmodium species as a part of the malaria surveillance and elimination programs in Myanmar.
Conclusions
This study has shown that microscopic examination does not reliably distinguish Plasmodium species in the studied areas co-endemic for multiple Plasmodium species. Nested PCR not only detected all microscopy-positive samples, but also detected single or mixed infections and submicroscopic infections that were missed or misread by microscopy. Given the observed higher sensitivity and specificity associated with the nested PCR technique, molecular detection method holds enormous promise for malaria diagnosis and can be used as effective monitoring tools for malaria surveillance, control and elimination in Myanmar. Submicroscopic and asymptomatic infections also highlight the urgent need to develop new public health strategies and appropriate community-based interventions for the effective control and elimination of malaria in Myanmar.
References
Wongsrichanalai C, Barcus MJ, Muth S, Sutamihardja A, Wernsdorfer WH. A review of malaria diagnostic tools: microscopy and rapid diagnostic test (RDT). Am J Trop Med Hyg. 2007;77:119–27.
Fançony C, Sebastião YV, Pires JE, Gamboa D, Nery SV. Performance of microscopy and RDTs in the context of a malaria prevalence survey in Angola: a comparison using PCR as the gold standard. Malar J. 2013;12:284.
WHO. World malaria report 2008. Geneva: World Health Organization; 2008. p. 2008.
Payne D. Use and limitation of light microscopy for diagnosing malaria at the primary health care level. Bull World Health Organ. 1988;66:621–6.
Snounou G, Viriyakosol S, Jarra W, Thaithong S, Brown KN. Identification of the four human malaria parasite species in field samples by the polymerase chain reaction and detection of a high prevalence of mixed infections. Mol Biochem Parasitol. 1993;58:283–92.
Snounou G, Singh B. Nested PCR analysis of Plasmodium parasites. Methods Mol Med. 2002;72:189–203.
Roper C, Elhassan IM, Hviid L, Giha H, Richardson W, Babiker H, et al. Detection of very low level Plasmodium falciparum infections using the nested polymerase chain reaction and a reassessment of the epidemiology of unstable malaria in Sudan. Am J Trop Med Hyg. 1996;54:325–31.
Kimura M, Kaneko O, Liu Q, Zhou M, Kawamoto F, Wataya Y, et al. Identification of the four species of human malaria parasites by nested PCR that targets variant sequences in the small subunit rRNA gene. Parasitol Int. 1997;46:91–5.
Perandin F, Manca N, Calderaro A, Piccolo G, Galati L, Ricci L, et al. Development of a real-time PCR assay for detection of Plasmodium falciparum, Plasmodium vivax, and Plasmodium ovale for routine clinical diagnosis. J Clin Microbiol. 2004;42:1214–9.
Ebrahimzadeh A, Fouladi B, Fazaeli A. High rate of detection of mixed infections of Plasmodium vivax and Plasmodium falciparum in South-East of Iran, using nested PCR. Parasitol Int. 2007;56:61–4.
Krishna S, Bharti PK, Chandel HS, Ahmad A, Kumar R, Singh PP, et al. Detectionof mixed infections with Plasmodium spp. by PCR, India, 2014. Emerg Infect Dis. 2015;21:1853–7.
Mekonnen SK, Aseffa A, Medhin G, Berhe N, Velavan TP. Re-evaluation of microscopy confirmed Plasmodium falciparum and Plasmodium vivax malaria by nested PCR detection in southern Ethiopia. Malar J. 2014;13:48.
Sitali L, Chipeta J, Miller JM, Moonga HB, Kumar N, Moss WJ, et al. Patterns of mixed Plasmodium species infections among children six years and under in selected malaria hyper-endemic communities of Zambia: population-based survey observations. BMC Infect Dis. 2015;15:204.
Makler MT, Palmer CJ, Ager AL. A review of practical techniques for the diagnosis of malaria. Ann Trop Med Parasitol. 1998;92:419–33.
Moges B, Amare B, Belyhun Y, Tekeste Z, Gizachew M, Workineh M, et al. Comparison of CareStart™ HRP2/pLDH COMBO rapid malaria test with light microscopy in north-west Ethiopia. Malar J. 2012;11:234.
Nyunt MH, Kyaw MP, Win KK, Myint KM, Nyunt KM. Field evaluation of HRP2 and pan pLDH-based immunochromatographic assay in therapeutic monitoring of uncomplicated falciparum malaria in Myanmar. Malar J. 2013;12:123.
Chong CK, Cho PY, Na BK, Ahn SK, Kim JS, Lee JS, et al. Evaluation of the accuracy of the EasyTest™ malaria Pf/Pan Ag, a rapid diagnostic test, in Uganda. Korean J Parasitol. 2014;52:501–5.
Maltha J, Gamboa D, Bendezu J, Sanchez L, Cnops L, Gillet P, et al. Rapid diagnostic tests for malaria diagnosis in the Peruvian Amazon: impact of pfhrp2 gene deletions and cross-reactions. PLoS ONE. 2012;7:e43094.
Cheng Q, Gatton ML, Barnwell J, Chiodini P, McCarthy J, Bell D, et al. Plasmodium falciparum parasites lacking histidine-rich protein 2 and 3: a review and recommendations for accurate reporting. Malar J. 2014;13:283.
WHO. Strategy for malaria elimination in the Greater Mekong Subregion (2015–2030). Geneva: World Health Organization; 2015.
Kyaw MP, Nyunt MH, Chit K, Aye MM, Aye KH, Aye MM, et al. Reduced susceptibility of Plasmodium falciparum to artesunate in southern Myanmar. PLoS ONE. 2013;8:e57689.
Zhou G, Sun L, Xia R, Duan Y, Xu J, Yang H, et al. Clinical malaria along the China–Myanmar border, Yunnan Province, China, January 2011–August 2012. Emerg Infect Dis. 2014;20:675–8.
Sonkong K, Chaiklieng S, Neave P, Suggaravetsiri P. Factors affecting delay in seeking treatment among malaria patients along Thailand–Myanmar border in Tak Province, Thailand. Malar J. 2015;14:3.
Tun KM, Imwong M, Lwin KM, Win AA, Hlaing TM, Hlaing T, et al. Spread of artemisinin-resistant Plasmodium falciparum in Myanmar: a cross-sectional survey of the K13 molecular marker. Lancet Infect Dis. 2015;15:415–21.
Win TT, Lin K, Mizuno S, Zhou M, Liu Q, Ferreira MU, et al. Wide distribution of Plasmodium ovale in Myanmar. Trop Med Int Health. 2002;7:231–9.
Lin K. Malaria control in Myanmar. In: Kano S, Tongol-Rivera P, editors. Asian parasitology malaria in Asia. Chiba: Federation of Asian Parasitologists; 2005. p. 123–34.
Jiang N, Chang Q, Sun X, Lu H, Yin J, Zhang Z, et al. Co-infections with Plasmodium knowlesi and other malaria parasites, Myanmar. Emerg Infect Dis. 2010;16:1476–8.
Lucchi NW, Poorak M, Oberstaller J, DeBarry J, Srinivasamoorthy G, Goldman I, et al. A new single-step PCR assay for the detection of the zoonotic malaria parasite Plasmodium knowlesi. PLoS ONE. 2012;7:e31848.
de Roode JC, Culleton R, Bell AS, Read AF. Competitive release of drug resistance following drug treatment of mixed Plasmodium chabaudi infections. Malar J. 2004;3:33.
Mayxay M, Pukrittayakamee S, Newton PN, White NJ. Mixed-species malaria infections in humans. Trends Parasitol. 2004;20:233–40.
Lee G, Jeon E, Le Tien D, Kim T, Yoo J, Yong Kim H, et al. Development and evaluation of a rapid diagnostic test for P. falciparum, P. vivax and mixed-species malaria antigens. Am J Trop Med Hyg. 2011;85:989–93.
Wang RB, Zhang J, Zhang QF. Malaria baseline survey in four special regions of northern Myanmar near China: a cross-sectional study. Malar J. 2014;13:302.
Kimura M, Miyak H, Kim HS, Tanabe M, Arai M, Kawai S, et al. Species-specific PCR detection of malaria parasites by microtiter plate hybridization: clinical study with malaria patients. J Clin Microbiol. 1995;33:2342–6.
Barber BE, William T, Grigg MJ, Yeo TW, Anstey NM. Limitations of microscopy to differentiate Plasmodium species in a region co-endemic for Plasmodium falciparum, Plasmodium vivax and Plasmodium knowlesi. Malar J. 2013;12:8.
Alemu A, Fuehrer HP, Getnet G, Kassu A, Getie S, Noedl H. Comparison of Giemsa microscopy with nested PCR for the diagnosis of malaria in North Gondar, north-west Ethiopia. Malar J. 2014;13:174.
Li P, Zhao Z, Wang Y, Xing H, Parker DM, Yang Z, et al. Nested PCR detection of malaria directly using blood filter paper samples from epidemiological surveys. Malar J. 2014;13:175.
Mwingira F, Genton B, Kabanywanyi AN, Felger I. Comparison of detection methods to estimate asexual Plasmodium falciparum parasite prevalence and gametocyte carriage in a community survey in Tanzania. Malar J. 2014;13:433.
Tajebe A, Magoma G, Aemero M, Kimani F. Detection of mixed infection level of Plasmodium falciparum and Plasmodium vivax by SYBR Green I-based real-time PCR in North Gondar, north-west Ethiopia. Malar J. 2014;13:411.
Mahende C, Ngasala B, Lusingu J, Yong TS, Lushino P, Lemnge M, et al. Performance of rapid diagnostic test, blood-film microscopy and PCR for the diagnosis of malaria infection among febrile children from Korogwe District, Tanzania. Malar J. 2016;15:391.
Sethabutr O, Brown A, Panyim S, Kain KC, Webster HK, Echeverria P. Detection of Plasmodium falciparum by polymerase chain reaction in a field study. J Infect Dis. 1992;166:145–8.
Maitland K, Williams T, Newbold C. Plasmodium vivax and P. falciparum: biological interactions and the possibility of cross-species immunity. Parasitol Today. 1997;13:227–31.
Mayxay M, Pukritrayakamee S, Chotivanich K, Imwong M, Looareesuwan S, White NJ. Identification of cryptic coinfection with Plasmodium falciparum in patients presenting with vivax malaria. Am J Trop Med Hyg. 2001;65:588–92.
Mohapatra MK, Dash LK, Bariha PK, Karua PC. Profile of mixed species (Plasmodium vivax and falciparum) malaria in adults. J Assoc Physicians India. 2012;60:20–4.
Black J, Hommel M, Snounou G, Pinder M. Mixed infections with Plasmodium falciparum and P. malariae and fever in malaria. Lancet. 1994;343:1095.
Smith T, Genton B, Baea K, Gibson N, Narara A, Alpers MP. Prospective risk of morbidity in relation to malaria infection in an area of high endemicity of multiple species of Plasmodium. Am J Trop Med Hyg. 2001;64:262–7.
Dunyo S, Milligan P, Edwards T, Sutherland C, Targett G, Pinder M. Gametocytaemia after drug treatment of asymptomatic Plasmodium falciparum. PLoS Clin Trials. 2006;1:e20.
Vafa M, Troye-Blomberg M, Anchang J, Garcia A, Migot-Nabias F. Multiplicity of Plasmodium falciparum infection in asymptomatic children in Senegal: relation to transmission, age and erythrocyte variants. Malar J. 2008;7:17.
Kritsiriwuthinan K, Ngrenngarmlert W. Asymptomatic malaria infections among foreign migrant workers in Thailand. Asian Pac J Trop Med. 2011;4:560–3.
Ganguly S, Saha P, Guha SK, Biswas A, Das S, Kundu PK, et al. High prevalence of asymptomatic malaria in a tribal population in eastern India. J Clin Microbiol. 2013;51:1439–44.
Pava Z, Burdam FH, Handayuni I, Trianty L, Utami RA, Tirta YK, et al. Submicroscopic and asymptomatic Plasmodium parasitaemia associated with significant risk of anaemia in Papua, Indonesia. PLoS ONE. 2016;11:e0165340.
Baum E, Sattabongkot J, Sirichaisinthop J, Kiattibutr K, Jain A, Taghavian O, et al. Common asymptomatic and submicroscopic malaria infections in Western Thailand revealed in longitudinal molecular and serological studies: a challenge to malaria elimination. Malar J. 2016;15:333.
Imwong M, Nguyen TN, Tripura R, Peto TJ, Lee SJ, Lwin KM, et al. The epidemiology of subclinical malaria infections in South-East Asia: findings from cross-sectional surveys in Thailand–Myanmar border areas, Cambodia, and Vietnam. Malar J. 2015;14:381.
Hewitt S, Delacollette C, Chavez I. Malaria situation in the Greater Mekong Subregion. Southeast Asian J Trop Med Public Health. 2013;44(Suppl 1):46–72.
Zhou M, Liu Q, Wongsrichanalai C, Suwonkerd W, Panart K, Prajakwong S, et al. High prevalence of Plasmodium malariae and P. ovale in malaria patients along the Thai–Myanmar border, as revealed by acridine orange staining and PCR-based diagnoses. Trop Med Int Health. 1998;3:304–12.
Obare P, Ogutu B, Adams M, Odera JS, Lilley K, Dosoo D, et al. Misclassification of Plasmodium infections by conventional microscopy and the impact of remedial training on the proficiency of laboratory technicians in species identification. Malar J. 2013;12:113.
Chavatte J-M, Tan SBH, Snounou G, Lin RTPV. Molecular characterization of misidentified Plasmodium ovale imported cases in Singapore. Malar J. 2015;14:454.
Authors’ contributions
JMK carried out nested PCR analysis. PYC, MM, JL, HJ, HWL, SKA, MKM and KL contributed the blood sample collection and microscopic diagnosis of malaria slides. JMK, JL, TIK, JHP, KL, TSK and BKN analysed and interpreted the data. KL, TSK and BKN designed and supervised the experiments. BKN wrote the draft of the manuscript. All authors read and approved the final manuscript.
Acknowledgements
We thank the staffs in Department of Medical Research Pyin Oo Lwin Branch and the health professionals in Naung Cho, Pyin Oo Lwin and Tha Beik Kyin townships for their contribution and technical support in field study.
Competing interests
The authors declare that they have no competing interests.
Availability of data and materials
The original datasets analysed during the current study are available from the corresponding author upon request.
Consent for publication
All authors have given their consent for publication.
Ethics approval and consent to participate
This study was approved by the Ethics Review Committee, Department of Medical Research, Myanmar (97/Ethics 2015), and by the Ethical Review Committee of Inha University School of Medicine, Korea (INHA 15-013). Informed written consent and permission (in the case of children under 18 years of age) were obtained from each patient or a parent/legal guardian.
Funding
This work was supported by the National Research Foundation of Korea (NRF) grants funded by the Korea government (MEST) (NRF-2015K1A3A9A01034893) and the Korea government (MSIP) (NRF-2015R1A2A2A01004310).
Author information
Authors and Affiliations
Corresponding authors
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Kang, JM., Cho, PY., Moe, M. et al. Comparison of the diagnostic performance of microscopic examination with nested polymerase chain reaction for optimum malaria diagnosis in Upper Myanmar. Malar J 16, 119 (2017). https://doi.org/10.1186/s12936-017-1765-4
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12936-017-1765-4