Abstract
Background
Resistance to the carbamate insecticide bendiocarb is emerging in Anopheles gambiae populations from the city of Yaoundé in Cameroon. However, the molecular basis of this resistance remains uncharacterized. The present study objective is to investigate mechanisms promoting resistance to bendiocarb in An. gambiae populations from Yaoundé.
Methods
The level of susceptibility of An. gambiae s.l. to bendiocarb 0.1 % was assessed from 2010 to 2013 using bioassays. Mosquitoes resistant to bendiocarb, unexposed and susceptible mosquitoes were screened for the presence of the Ace-1R mutation using TaqMan assays. Microarray analyses were performed to assess the pattern of genes differentially expressed between resistant, unexposed and susceptible.
Results
Bendiocarb resistance was more prevalent in mosquitoes originating from cultivated sites compared to those from polluted and unpolluted sites. Both An. gambiae and Anopheles coluzzii were found to display resistance to bendiocarb. No G119S mutation was detected suggesting that resistance was mainly metabolic. Microarray analysis revealed the over-expression of several cytochrome P450 s genes including cyp6z3, cyp6z1, cyp12f2, cyp6m3 and cyp6p4. Gene ontology (GO) enrichment analysis supported the detoxification role of cytochrome P450 s with several GO terms associated with P450 activity significantly enriched in resistant samples. Other detoxification genes included UDP-glucosyl transferases, glutathione-S transferases and ABC transporters.
Conclusion
The study highlights the probable implication of metabolic mechanisms in bendiocarb resistance in An. gambiae populations from Yaoundé and stresses the need for further studies leading to functional validation of detoxification genes involved in this resistance.
Similar content being viewed by others
Background
Malaria prevention largely relies on the use of measures, such as long-lasting insecticidal nets (LLINs) and indoor residual spraying (IRS) [1]. Of the four insecticides classes used in public health, pyrethroids are by far the most widely used [1]. During the past decades, overreliance on pyrethroids in public health and agriculture, led to rapid expansion of pyrethroid resistance in malaria vectors populations which now threatens the continued effectiveness of current control efforts [2]. Resistance to pyrethroids is mainly due to mutations in the knock down genes (kdr) and metabolic detoxification mechanisms and is largely prevalent in all major malaria vectors [3–5]. Because of rapid spread rate of this resistance across sub-Saharan Africa, effective measures are needed to mitigate its impact. The World Health Organization (WHO) recommends application of insecticides having different mode of action or temporal replacement by different insecticide classes in case of resistance [6]. Carbamates and organophosphates due to their different mode of action, are actually considered as suitable alternative insecticides to pyrethroids for vector control such as IRS [7–10]. Field experiments conducted across West Africa showed the effectiveness of carbamates and organophosphates against pyrethroid resistant malaria vector populations [11, 12]. There is, an increasing number of countries which have started introducing the use of carbamate in their national vector control strategy [7, 9, 10]. However, increasing reports of carbamates resistance in the main malaria vectors across sub-Saharan Africa [13–17], could jeopardize current efforts to implement appropriate resistance management strategies against malaria vectors. Despite current expansion of bendiocarb resistance little is known on mechanisms promoting this resistance in Central African mosquito populations.
In Cameroon, despite efforts made over the past years to control malaria, the disease is still considered, a major threat [18, 19]. Major vectors in the country display high level of pyrethroid resistance [20–22]. Studies undertaken in the cities of Douala and Yaoundé, reported particularly high prevalence of pyrethroid and DDT resistance in both Anopheles gambiae and Anopheles coluzzii [23–25]. The use of insecticide tools for vector control in households, the selective pressure of pollutants in breeding habitats and uncontrolled use of pesticides in small scale urban vegetable farming are all considered to have caused, the fast evolution of insecticide resistance which is now also affecting bendiocarb [23, 25, 26]. However, the molecular basis of carbamate resistance remained uncharacterized in An. gambiae populations in Cameroon. Such information is crucial to guide the implementation of appropriate resistance management strategies to prolong the effectiveness of carbamates in Cameroon. The main resistance mechanisms to carbamates involved metabolic resistance and target-site resistance. Metabolic resistance to carbamates is often conferred by the up-regulation of detoxification genes such as cytochrome P450 s [27, 28] or carboxylesterases [29–31].
Target-site resistance to carbamates and organophosphates is conferred by a single point mutation causing acetylcholinesterase inhibition [32, 33]. The mutation encoded by the Ace-1 R gene induces a substitution from glycine to serine at position 119 (G119S). The G119S mutation has also been recorded in several species including Culex quinquefasciatus, Anopheles albimanus and An. gambiae [33–38]. Recent findings reported the duplication of this mutation in some An. gambiae individuals [39, 40]. In Cameroon, no G119S mutation has up to now been reported in An. gambiae population and the underlying molecular basis of the carbamate resistance in this major malaria vector remain to be established. The present study seeks to characterize mechanisms involved in the ongoing mosquito resistance to carbamate in the city of Yaoundé. The study also traces the dynamics of An. gambiae susceptibility to bendiocarb between 2010 and 2013.
Methods
Study site
Mosquito collections were conducted in districts of the city of Yaoundé (3°51′N 11°30′E). Yaoundé the capital city of Cameroon, is situated within the Congo-Guinean phytogeographic domain and display an equatorial climate consisting of four seasons: two rainy seasons (March–June and September–November; annual rainfall 1700 mm) and two dry seasons (December–February and July–August).
Mosquito collection
Mosquito larvae were collected at all stages in water collections across the city of Yaoundé and reared separately according to their breeding habitats characteristics classified as cultivated, polluted or non polluted sites. Water collections with organic wastes were considered as polluted, non-polluted breeding sites were water collections without any sign of organic pollution, cultivated breeding sites were water collections associated with farming practices. In the laboratory, larvae were transferred into distilled water and reared separately at room temperature. During this period, they were fed using fish food until the pupa stage. Pupa were collected in cups and placed inside cages covered with netting for emergence.
Insecticide bioassays
Bioassays were conducted from October 2010 to December 2013 using 2–4 days old females emerging from larvae collected on the field. Morphological identification keys [41] were used to differentiate members of the An. gambiae complex to other mosquito species at both the larval and the adult stages. Unfed An. gambiae s.l. females aged 2–4 days were exposed to 0.1 % bendiocarb, 4 % DDT (dichloro-diphenyl-trichloroethane), 0.75 % permethrin, 0.05 % deltamethrin and 4 % malathion in susceptibility test kits from the WHO, following standardized procedures [42]. For bioassays using piperonyl butoxide (PBO) as synergist, unfed An. gambiae females were pre-exposed to 4 % PBO papers for 1 h before being immediately exposed for another 1 h to bendiocarb. Mortality was scored after 24 h but for mosquitoes surviving exposition to bendiocarb, they were maintained in observation for a total period of 48 h before storage in RNAlater. For each bioassay, exposition of mosquitoes to untreated papers was also undertaken as controls. Abbot formula [43] was used to adjust mortality rate in tested samples if the control group mortality rate was 5–20 %. WHO recommendations [42] were applied for classifying mosquitoes as resistant or susceptible.
Odd ratio calculations were undertaken to assess any association between phenotypes and genotypes [(resistants genotype A*susceptibles genotype B)/(susceptibles genotype A*resistants genotype B)]. Odd ratio estimates, mortality rates, the 95 % confidence intervals and p values were calculated with the software MedCalc V11.5.0.0.
Molecular identification of species and genotyping of Ace-1R G119S mutation
Genomic DNA utilized for the identification of An. gambiae s.l. species and the screening of the Ace-1R G119S mutation, was extracted from a leg or wing of adult mosquitoes by the Livak technique [44]. A polymerase chain reaction (PCR) was used for An. gambiae species identification [45]. The presence of the G119 mutation was screened using TaqMan assays as previously described [46]. TaqMan reactions were undertaken using the Agilent MX3005P machine. Each reaction was conducted in a 10 μl final volume with 1xSensiMix (Bioline), 800 nM of each primer and 200 nM of each probe.
Microarray experiments
Microarray experiments were conducted using only An. gambiae samples originating from cultivated sites where bendiocarb resistance was most prevalent. Differentially transcribed genes were compared between resistant, control (unexposed) and susceptible (Kisumu) samples.
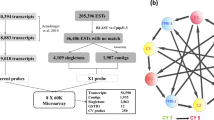
Pools of ten mosquitoes were used for total RNA extraction with the PicoPure RNA isolation Kit (Arcturus, Applied Biosystems, Mountain View CA USA). Each sample was constituted of three biological replicates. Total RNA extracted from mosquitoes was treated using DNase (RNase free DNase set, Qiagen Hilden Germany). A nanodrop spectrophotometer (Nanodrop Technologies UK) and a Bioanalyser (Agilent Technologies UK) were used to assess RNA concentration and quality. After amplification undertaken using 100 ng of total RNA, samples were labelled using Cy-3 or Cy-5 dye with the “Two colors low input Quick Amp labeling kit” (Agilent technologies, Santa Clara, CA, USA). This was immediately followed by samples purification undertaken using Qiagen purification kit. A spectrophotometer (NanoDrop Technologies) and Bioanalyzer (Agilent Technologies) was used to check for cRNA labelling and yield. Labelled cRNAs were hybridized to the ‘An. gambiae’ array Agilent 8x15 k chip (AGAM_15 K) (A-MEXP-2196) [46]. After 17 h hybridization at 65 °C and 10 rpm rotation, slides were washed according to the manufacturer instructions (Agilent Technologies). Microarray slides were then scanned with the Agilent G2565 Microarray Scanner System via the Agilent Feature Extraction Software (Agilent Technologies).
Five hybridizations per comparison including three independent biological replicates and two dye swaps were performed. Resistant samples were competitively hybridized against unexposed samples and the Kisumu laboratory strain.
Microarray data analysis
Genespring GX 11.1 software (Agilent Technologies) was used for microarray data analysis. Comparison of genes expression profiles between groups was undertaken after computing the mean transcription expression ratios to a one sample Student’s t test against zero. Benjamin and Hochberg calculation [47] was applied for multiple testing corrections. Transcripts significantly and differentially transcribed were those displaying both t test p values <0.05 and a fold change ≥twofold compared to the control or susceptible group. Gene ontology (GO) enrichment was performed using David functional 6.7 [48, 49] to determine GO significantly enriched using as background for comparison the totality of genes differentially transcribed for each group.
Microarray validation by qRT-PCR (real-time quantitative reverse transcription polymerase chain reaction)
Quantitative RT-PCR analysis as described in Tene et al. [24] was used to confirm the overexpression of detoxification genes detected by microarray. Biological replicates consisting of two micrograms of total RNA per replicate were reverse transcribed into cDNA in a reaction mix containing superscript III (Invitrogen, Carlsbad, CA, USA) and oligo-dT20 primer as recommended by the manufacturer. A MX3005 Agilent system (Agilent) was used to perform quantitative PCR reactions. Each reaction was conducted in a final volume of 25 µl containing iQ SYBR Green supermix (Biorad), primers at the concentration of 0.3 µM each and 5 µl of 1:50 diluted cDNA. The specificity of PCR products generated was verified using melt curves analysis. Standard curves for each gene were generated using serial dilutions of cDNA. Selected transcripts fold changes were normalized to EFGM_ANOGA (AGAP009737_RA) and 40S ribosomal protein S7 (AGAP010592_RA). Fold changes differences of selected genes between test samples and susceptible (Kisumu), were estimated according to the 2−ΔΔCT method considering PCR efficiency [50, 51].
Results
Susceptibility to insecticides and species identification
The bendiocarb susceptibility of An. gambiae females aged 2–4 days was monitored regularly from October 2010 to December 2013. High variation of mosquito susceptibility according to breeding habitats characteristics was recorded. Mosquitoes originating from cultivated sites were two to five times more resistant to bendiocarb (mortality rate 77.1 %) compared to those originating from polluted (mortality rate 88.4 %) and unpolluted (mortality rate 94.7 %) sites. Mosquitoes originating from polluted sites, also appeared twice more resistant to bendiocarb compared to those originating from unpolluted sites (Table 1). Levels of susceptibility to bendiocarb of mosquitoes originating from cultivated sites apart of 2011 (when a 100 % mortality rate was recorded), were regularly low with mortality rates always below 80 % suggesting an established bendiocarb resistance in this An. gambiae population (Fig. 1). High prevalence of DDT, permethrin and deltamethrin resistance was also detected in mosquitoes originating from cultivated sites. However, these mosquitoes appeared highly susceptible to the organophosphate malathion (Table 2). When mosquitoes displaying high bendiocarb resistance (samples collected in 2013) were pre-exposed to PBO before being exposed to bendiocarb, a 100 % (n = 184) mortality rate was recorded. These data suggest the implication of P450 monooxygenase in mosquito resistance to bendiocarb.
Of the 233 mosquitoes recorded as resistant to bendiocarb and identified at the species level, 186 (80 %) were An. gambiae and 47 An. coluzzii. Anopheles coluzzii was the predominant species in polluted and unpolluted sites (43/47) whereas An. gambiae was the most abundant in cultivated sites (175/186) suggesting an ecological niche partitioning between both species in Yaoundé.
Screening of ACE-1R mutation
A total of 392 specimens including survivors after exposition to bendiocarb (resistant), dead (susceptible) and control (unexposed) were processed to search for Ace-1R mutation presence. None were detected carrying the Ace-1R mutation. Further supporting the full recovery of susceptibility observed after PBO exposure.
Genome-wide transcription analysis of bendiocarb resistance
Microarray analyses to detect detoxification genes overexpressed, were undertaken with An. gambiae samples originating from cultivated sites where mosquitoes display high level resistance to bendiocarb. Three pairwises comparisons were conducted: resistant vs control (unexposed) (Rb-C), resistant vs susceptible (Kisumu) (Rb-S), control vs susceptible (Kisumu) (C-S). The number of transcripts significantly and differentially transcribed (p < 0.05 and fold-change (FC) >2) varied from 30 between resistant and control (21 up-regulated and nine down-regulated), 423 between resistant and susceptible (Kisumu) (220 up-regulated and 205 down-regulated) and 609 between control (unexposed) and susceptible (Kisumu) (322 up-regulated and 287 down-regulated)(Fig. 2).
Differentially transcribed genes between resistant, unexposed and susceptible. The Venn diagram presents genes with a transcription ratio ≥twofold in either direction and a corrected p value <0.05 in bendiocarb resistant samples compared to unexposed and the Kisumu laboratory strain. Transcripts number are presented for each portion of the Venn diagram
Candidate detoxification genes
A hierarchical analysis was conducted to detect the most likely candidate genes involved in bendiocarb resistance with the assumption that these will likely be detected in more than one comparison. Because no gene was commonly overexpressed in the three comparisons Rb-C, Rb-S, C-S, more attention was focused on sets of genes commonly over-expressed between two comparisons.
Genes over-expressed in Rb-S/C-S
The number of detoxification genes commonly over-expressed in Rb-S/C-S and possibly connected with resistance to bendiocarb, included four cytochrome P450 genes (cyp6z3, cyp12f2, cyp6m3 and cyp6m4) and one Glutathione-S-transferase: gstms3. Four probes belonging to cyp6z3 gene were detected always over-expressed in Rb-S and only one cyp6z3 probe was detected significantly over-expressed in C-S. cyp6z3 is known to be associated with xenobiotic and insecticide detoxification in An. gambiae [52]. Four probes for cyp12f2 were also found overexpressed with fold changes exceeding 14 in both Rb-S and C-S comparisons. For cyp6m3 and cyp6m4, the number of probes detected significantly overexpressed varied from one and two for Rb-S to four and three for C-S comparisons respectively with no important variation of the fold change (Table 3). Both cyp6m3 and cyp6m4, are considered to be involved in xenobiotic detoxification [53]. Genes recorded commonly over-expressed in both Rb-C and Rb-S also included three probes for gstms3, one probe for each of the three glucosyl glucuronosyl transferases (AGAP005753-RA, AGAP007374-RA, AGAP005750-RA) as well as for xanthine dehydrogenase (Table 3).
Genes overexpressed in Rb-C
Further attention was paid to Rb-C as this compares mosquitoes having similar genetic background and which are only different in the resistance phenotype. Two cytochrome P450 genes cyp12f1 and cyp4c36 are over-expressed but with low fold-change of around two including four probes for cyp12f1 and one for cyp4c36. However, the expression level of these two P450 s is low <twofold in Rb-S. Three genes with no recognized role in insecticide detoxification were also over-expressed: Cytosol aminopeptidase, Zinc carboxypeptidase a1 and Chymotrypsin 1 (Table 3).
Genes over-expressed only in Rb-S or C-S
Several cytochrome P450 genes, including four probes for cyp6z1, three probes for cyp6p4, and three probes for cyp6ag1 were over-expressed only in Rb-S. Other genes overexpressed in this comparison included four probes for gstms1 and gstd1-3, one probe for gstd7, as well as for an ABC transporter and a glucosyl glucuronosyl transferase. For C-S comparison, four probes for cyp6m2, three probes for gstms2, one probe for gsts1-1 and one for a thioredoxin dependent peroxidase (tpx2) were detected overexpressed (Table 3).
Annotation and gene ontology analysis
Enrichment analysis using DAVID Functional program was conducted to assess GO terms frequent in the group of transcripts up-regulated in resistant vs control (unexposed), resistant or control vs Kisumu. Three GO terms were detected significantly enriched with an enrichment fold of over 20 % when transcripts up-regulated between resistant vs unexposed were analysed. All the terms were associated with proteolysis activity. None of the terms remained significant when the Benjamin and Hochberg multiple testing correction was applied. When the enrichment analysis was conducted with transcripts upregulated between resistant vs Kisumu, an enrichment fold varying from 3.2 to 4.5 % was detected for Cytochrome P450 genes (Table 4). Monooxygenase activity remained significant when the Benjamin and Hochberg multiple testing correction was applied (p < 0.01). When transcripts recorded as up-regulated between unexposed vs Kisumu were analysed three were found associated with cytochrome P450 monooxygenase activity with an enrichment fold varying from 2.5 to 2.9 %. However, no activity was scored significantly enriched when the Benjamin and Hochberg multiple testing correction was applied.
Validation of microarray data by RT-PCR
Eleven transcripts overexpressed in resistant samples including six cytochrome P450 (cyp6z3, cyp12f2, cyp12f1, cyp4c36, cyp6p4, cyp6ag1), two GST (gstd1-4, gstsm3), two aminopeptidase (cytosol aminopeptidase, chymotrypsin1) and one UDPGT (AGAP005750-RA) were selected to validate microarray data using qRT-PCR. A positive but non-significant correlation (R2 = 0.44; p = 0.24) was recorded between qRT-PCR and microarray fold change measurements (Fig. 3).
Discussion
Despite fast evolution of insecticide resistance in vector populations across Cameroon, molecular mechanisms conferring resistance are still poorly studied. The present study was conducted to characterize molecular mechanisms promoting bendiocarb resistance in An. gambiae populations in the city of Yaoundé. Both An. gambiae and An. coluzzii were found resistant to bendiocarb. Mosquitoes originating from cultivated sites were found to be more resistant to bendiocarb than those collected from polluted or unpolluted sites and could be related to their frequent exposition to xenobiotics including insecticides.
No mosquito was found carrying the G119S mutation conferring target site resistance to carbamate and organophosphate. The increase mortality after the use of piperonyl butoxide (PBO) as synergist suggested the likely implication of cytochrome P450 s in bendiocarb resistance. Our data was similar to previous investigations conducted across West Africa supporting the implication of metabolic mechanisms in carbamate resistance [27]. Although G119S mutation is recognized as the primary resistance mechanism against carbamates and organophosphates it remains less expanded across Central Africa [54]. Its distribution might be constrained by its high fitness cost [39]. However, possession of both G119S mutation and metabolic resistance could lead to extremely resistant phenotypes [27, 40].
Microarray analysis identified several cytochrome P450 genes with the most important being cyp6z3, cyp6z1, cyp12f2, cyp6p4 and cyp6ag1, which were overexpressed when resistant or unexposed samples were compared to the Kisumu susceptible strain (Rb-S and C-S). However, in addition to their potential implication in insecticide resistance, the high fold change difference detected for some of the genes could likely results from the different genetic background between Kisumu strain originating from Kenya and local An. gambiae populations from Cameroon. Similar observations have been reported from previous studies [24]. The over-expression of the two P450 genes cyp12f1 and cyp4c36 in the comparison between bendiocarb resistant and control non exposed mosquitoes (Rb-C) was low and not observed in the Rb-S comparison suggesting that these genes may not be the main bendiocarb resistance genes. Although further functional characterization studies will help to establish the exact role of these candidate genes. Cyp12f1 gene was already reported overexpressed in mosquitoes resistant to DDT [53] while no role for cyp4c36 in insecticide resistance have so far been reported. Nevertheless cytochrome P450 are known to metabolise a large number of xenobiotics including pyrethroids and carbamates [55, 56]. For the set of genes detected only overexpressed in comparison between control and susceptible (C-S), despite a probable absence of role in bendiocarb resistance, it is likely that these detoxification genes (cyp6m2, gstms2, tpx2, gsts1-1) as well as many others detected over-expressed, might be implicated in the metabolism of an important number of compounds since mosquito populations screened during the study were also recorded resistant to DDT and pyrethroids.
Among potential candidate genes conferring bendiocarb resistance, cyp12f2 was reported over-expressed in response to bacterial challenge or during malaria parasite invasion in mosquitoes [57] and in permethrin-resistant An. arabiensis in South Africa [58]. cyp6ag1, cyp6z3 and cyp6p4 were reported over-expressed in DDT and pyrethroid resistant An. gambiae and/or An. arabiensis populations [53, 58–60]. Ortholog of cyp6p4 and cyp6z3 have been connected to pyrethroid resistance in the malaria vector Anopheles funestus [3, 61]. Whereas, cyp6z1 in addition to its confirm involvement in DDT and pyrethroid resistance in An. gambiae [62, 63], was recently reported as the main gene conferring metabolic resistance to bendiocarb to An. funestus the other major African malaria vector [28].
Previous investigations from Yaoundé identified several candidates genes including cyp6m2, cyp6p3, cyp6z3, gstd1-6, involved in DDT or pyrethroid resistance [24]. Cyp6m2 and cyp6p3 also emerged as main candidate genes conferring bendiocarb resistance in a study conducted in Côte d’Ivoire [27]. However, none of these two genes emerged as potential candidate for bendiocarb resistance. The fact that during the present study only An. gambiae individuals were screened for microarray analysis while in Côte d’Ivoire mosquito population screened consisted exclusively of An. coluzzii might somewhere explain the difference recorded. Different detoxification gene expression pattern have been recorded for An. gambiae, An. coluzzii or An. arabiensis [53, 59, 64]. Several Glutathione S transferase genes including gstms3, gstms1, gstd1-3 and gstd7 were also detected overexpressed in Rb-S and/or C-S comparisons. GSTs are known to metabolize several xenobiotics including pyrethroids, organochlorines and organophosphates and to catalyse the secondary metabolism process of a large number of compounds oxidized by cytochrome P450 [30, 65, 66]. In pyrethroid resistant strains, the overexpression of GSTs attenuates lipid peroxidation induced by pyrethroid and reduce mortality [67].
In the city of Yaoundé, mosquito tolerance to DDT and pyrethroids and the prevalence of the kdr allele, have been increasing with time [25, 68]. It remains to be established whether increase resistance to DDT and pyrethroids could also have promoted cross-resistance to carbamates. Yet the increase prevalence of bendiocarb resistance poses serious challenges for malaria control since carbamates are considered as a main alternative to pyrethroids.
Conclusion
Insecticide resistance is considered as a key challenge for malaria vector control. In this study, we revealed increase tolerance of mosquito to bendiocarb (carbamate). The use of carbamates in IRS are considered as one of the main alternatives to the use of pyrethroid-treated nets particularly in geographical settings with high pyrethroid resistance. Elucidating mechanisms involved in carbamate resistance will enable the monitoring of this resistance in field populations. The data support the implication of cytochrome P450 monooxygenase in mosquito resistance to carbamates however there is a need to conduct further analysis to assess the role of candidate detoxification genes detected during this study.
Abbreviations
- LLINs:
-
long-lasting insecticidal nets
- IRS:
-
indoor residual spraying
- Kdr :
-
knock down resistance
- WHO:
-
World Health Organization
- PBO:
-
piperonyl butoxide
- PCR:
-
polymerase chain reaction
- GO:
-
gene ontology
- qRT-PCR:
-
real-time quantitative reverse transcription polymerase chain reaction
- DDT:
-
dichloro-diphenyl-trichloroethane
References
WHO. Malaria vector control and personal protection. WHO Technical Report Series 936. Geneva: World Health Organization; 2006.
WHO. Global Plan for insecticide resistance management (GPIRM). Geneva: World Health Organization; 2012.
Wondji C, Irving H, Morgan J, Lobo N, Collins F, Hunt R, et al. Two duplicated P450 genes are associated with pyrethroid resistance in Anopheles funestus, a major malaria vector. Genome Res. 2009;19:452–9.
Ranson H, Abdalla H, Badolo A, Guelbeogo W, Kerah-Hinzoumbe C, Yangalbe-Kalnone E, et al. Insecticide resistance in Anopheles gambiae: data from the first year of a multi-country study highlight the extent of the problem. Malar J. 2009;8:299.
Nkya T, Akhouayri I, Kisinza W, David J. Impact of environment on mosquito response to pyrethroid insecticides: facts, evidences and prospects. Insect Biochem Mol Biol. 2013;43:407–16.
WHO. Global plan for insecticide resistance management in malria vectors (GPIRM). Geneva: World Health Organization; 2012.
Bradley J, Matias A, Schwabe C, Vargas D, Monti F, Nseng G, et al. Increased risks of malaria due to limited residual life of insecticide and outdoor biting versus protection by combined use of nets and indoor residual spraying on Bioko Island Equatorial Guinea. Malar J. 2012;11:242.
Osse R, Aikpon R, Padonou G, Oussou O, Yadouleton A, Akogbeto M. Evaluation of the efficacy of bendiocarb in indoor residual spraying against pyrethroid resistant malaria vectors in Benin: results of the third campaign. Parasit Vectors. 2012;5:163.
Padonou G, Gbedjissi G, Yadouleton A, Azondekon R, Razack O, Oussou O, et al. Decreased proportions of indoor feeding and endophily in Anopheles gambiae s.l. populations following the indoor residual spraying and insecticide-treated net interventions in Benin (West Africa). Parasit Vectors. 2012;5:262.
Akogbeto M, Padonou G, Bankole H, Gazard D, Gbedjissi G. Dramatic decrease in malaria transmission after large-scale indoor residual spraying with Bendiocarb in Benin, an area of high resistance of Anopheles gambiae to pyrethroids. Am J Trop Med Hyg. 2011;85:586–93.
Asidi A, N’Guessan R, Koffi A, Curtis C, Hougard J, Chandre F, et al. Experimental hut evaluation of bednets treated with an organophosphate (chlorpyrifos-methyl) or a pyrethroid (lambdacyhalothrin) alone and in combination against insecticide-resistant Anopheles gambiae and Culex quinquefasciatus mosquitoes. Malar J. 2005;4:25.
Akogbeto M, Padonou G, Gbenou D, Irish S, Yadouleton A. Bendiocarb, a potential alternative against pyrethroid resistant Anopheles gambiae in Benin. West Afr Malar J. 2010;9:204.
AhouaAlou L, Koffi A, Adja M, Tia E, Kouassi P, Kone M, et al. Distribution of ace-1R and resistance to carbamates and organophosphates in Anopheles gambiae s.s. populations from Côte d’Ivoire. Malar J. 2010;9:167.
Edi C, Koudou B, Jones C, Weetman D, Ranson H. Multiple-insecticide resistance in Anopheles gambiae mosquitoes Southern Cote d’Ivoire. Emerg Infect Dis. 2012;18:1508–11.
Oduola A, Idowu E, Oyebola M, Adeogun A, Olojede J, Otubanjo O, et al. Evidence of carbamate resistance in urban populations of Anopheles gambiae s.s. mosquitoes resistant to DDT and deltamethrin insecticides in Lagos, South-Western Nigeria. Parasit Vectors. 2012;5:116.
Dabire K, Diabate A, Namontougou M, Djogbenou L, Kengne P, Simard F, et al. Distribution of insensitive acetylcholinesterase (ace-1R) in Anopheles gambiae s.l. populations from Burkina Faso (West Africa). Trop Med Int Health. 2009;14:396.
Essandoh J, Yawson A, Weetman D. Acetylcholinesterase (Ace-1) target site mutation 119S is strongly diagnostic of carbamate and organophosphate resistance in Anopheles gambiae s.s. and Anopheles coluzzii across southern Ghana. Malar J. 2013;12:404.
PNLP. Plan Stratégique nationale de lutte contre le paludisme 2011–2015. Rapport Miniatere de la Santé, Cameroun; 2012.
WHO Global Malaria Programme. World Malaria Report 2012. Geneva: World Health Organization; 2012.
Nwane P, Etang J, Chouaibou M, Toto J, Mimpfoundi R, Simard F. Kdr-based insecticide resistance in Anopheles gambiae s.s. populations in Cameroon: spread of the L1014F and L1014S mutations. BMC Res Notes. 2011;4:463.
Chouaibou M, Etang J, Brevault T, Nwane P, Hinzoumbe C, Mimpfoundi R, et al. Dynamics of insecticide resistance in the malaria vector Anopheles gambiae s.l. from an area of extensive cotton cultivation in northern Cameroon. Trop Med Int Health. 2008;13:476–86.
Ndjemai H, Patchoke S, Atangana J, Etang J, Simard F, Bilong Bilong C, et al. The distribution of insecticide resistance in Anopheles gambiae s.l. populations from Cameroon: an update. Trans R Soc Trop Med Hyg. 2008;103:1127–38.
Antonio-Nkondjio C, Fossog B, Ndo C, Djantio B, Togouet S, Awono-Ambene P, et al. Anopheles gambiae distribution and insecticide resistance in the cities of Douala and Yaounde (Cameroon): influence of urban agriculture and pollution. Malar J. 2011;10:154.
TeneFossog B, Poupardin R, Costantini C, Awono-Ambene H, Wondji C, Ranson H, et al. Resistance to DDT in an urban setting: Common mechanisms implicated in both M and S forms of Anopheles gambiae in the city of Yaoundé Cameroon. PLoS One. 2013;8:e61408.
Nwane P, Etang J, Chouaibou M, Toto J, Koffi A, Mimpfoundi R, et al. Multiple insecticide resistance mechanisms in Anopheles gambiae s.l. populations from Cameroon, Central Africa. Parasit Vectors. 2013;6:41.
Bigoga J, Manga L, Titanji VP, Etang J, Coetzee M, Leke RG. Susceptibility of Anopheles gambiae Giles (Diptera: Culicidae) to pyrethroids, DDT and carbosulfan in coastal Cameroon. Afr Entomol. 2007;15:1–7.
Edi CV, Djogbénou L, Jenkins AM, Regna K, Muskavitch MAT, Poupardin R, et al. CYP6 P450 enzymes and duplication produce extreme and multiple insecticide resistance in the malaria mosquito Anopheles gambiae. PLoS Genet. 2014;10:e1004236.
Ibrahim SS, Riveron JM, Stott R, Irving H, Wondji CS. The cytochrome P450 CYP6P4 is responsible for the high pyrethroid resistance in knockdown resistance-free Anopheles arabiensis. Insect Biochem Mol Biol. 2016;68:23–32.
Hemingway J, Hawkes N, Mc Carroll L, Ranson H. The molecular basis of insecticides resistance in mosquitoes. Insect Biochem Molec Biol. 2004;34:653–65.
Li X, Schuler M, Berenbaum M. Molecular mechanisms of metabolic resistance to synthetic and natural xenobiotics. Annu Rev Entomol. 2007;52:231–53.
Silva AX, Jander G, Samaniego H, Ramsey JS, Figueroa CC. Insecticide resistance mechanisms in the green peach aphid Myzus persicae (Hemiptera: Aphididae) I: a transcriptomic survey. PLoS One. 2012;7:e36366.
Weill M, Lutfalla G, Mogensen K, Chandre F, Berthomieu A, Berticat C, et al. Comparative genomics: insecticide resistance in mosquito vectors. Nature. 2003;423:136–7.
Weill M, Malcolm C, Chandre F, Mogensen K, Berthomieu A, Marquine M, et al. The unique mutation in ace-1 giving high insecticide resistance is easily detectable in mosquito vectors. Insect Mol Biol. 2004;13:1–7.
Penilla R, Rodriguez A, Hemingway J, Torres J, Arredondo-Jimenez J, Rodriguez M. Resistance management strategies in malaria vector mosquito control. Baseline data for a large-scale field trial against Anopheles albimanus in Mexico. Med Vet Entomol. 1998;12:217–33.
Scott M, McAllister J. Comparison of biochemical and molecular tests for detecting insecticide resistance due to insensitive acetylcholinesterase in Culex quinquefasciatus. J Am Mosq Contr Assoc. 2012;28:323–6.
Labbe P, Berthomieu A, Berticat C, Alout H, Raymond M, Lenormand T, et al. Independent duplications of the acetylcholinesterase gene conferring insecticide resistance in the mosquito Culex pipiens. Mol Biol Evol. 2007;24:1056–67.
N’Guessan R, Darriet F, Guillet P, Carnevale P, Traore-Lamizana M, Corbel V, et al. Resistance to carbosulfan in Anopheles gambiae from Ivory Coast, based on reduced sensitivity of acetylcholinesterase. Med Vet Entomol. 2003;17:19–25.
Djogbenou L, Dabire K, Diabate A, Kengne P, Akogbeto M, Hougard J, et al. Identification and geographical distribution of ace-1R mutation in the malaria vector Anopheles gambiae in south-western Burkina Faso, West Africa. Am J Trop Med Hyg. 2008;78:298.
Djogbenou L, Chandre F, Berthomieu A, Dabire R, Koffi A, Alout H, et al. Evidence of introgression of the ace-1(R) mutation and of the ace-1 duplication in West African Anopheles gambiae s.s. PLoS One. 2008;3:e2172.
Weetman D, Mitchell S, Wilding C, Birks D, Yawson A, Essandoh J, et al. Contemporary evolution of resistance at the major insecticide target site Ace-1 by mutation and copy number variation in the malaria mosquito Anopheles gambiae. Mol Ecol. 2015;24:2656–72.
Gillies M, De Meillon B. The Anophelinae of Africa South of the Sahara. Johannesburg: South Africa Institute of Medical Research; 1968.
World Health Organization. Test procedures for insecticide resistance monitoring in malaria vectors. Bio-efficacy and persistence of insecticides on treated surfaces. WHO/MAL/9812 Report of the WHO Informal Consultation. Geneva: World Health Organization; 1998.
Abbott WS. A method of computing the effectiveness of an insecticide. J Am Mosq Control Assoc. 1987;3:302–3.
Livak K. Organization and mapping of a sequence on the Drosophila melanogaster X and Y chromosomes that is transcribed during spermatogenesis. Genetics. 1984;107:611–34.
Fanello C, Petrarca V, Della Torre A, Santolamazza F, Dolo G, Coulibaly M, et al. The pyrethroid knock-down resistance gene in the Anopheles gambiae complex in Mali and further indication of incipient speciation within Anopheles gambiae s.s. Insect Mol Biol. 2003;12:241–5.
Bass C, Nikou D, Donnelly M, Williamson M, Ranson H, Ball A, et al. Detection of knockdown resistance (kdr) mutations in Anopheles gambiae: a comparison of two new high-throughput assays with existing methods. Malar J. 2007;6:111.
Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc B. 1995;57:289–300.
Huang DW, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: path toward the comprehensive functional analysis of large gene lists. Nucl Acids Res. 2009;37:1–13.
Huang D, Sherman B, Lempicki R. Systematic and integrative analysis of large gene lists using DAVID Bioinformatics Resources. Nat Protoc. 2009;4:44–57.
Pfaffl M. A new mathematical model for relative quantification in real-time RT-PCR. Nucl Acids Res. 2001;29:6.
Schmittgen T, Livak K. Analyzing real-time PCR data by the comparative C(T) method. Nat Protoc. 2008;3:1101–8.
David J, Strode C, Vontas J, Nikou D, Vaughan A, Pignalelli P, et al. The Anopheles gambiae detoxification chip: a highly specific microarray to study metabolic based insecticide resistance in malaria vectors. Proc Natl Acad Sci USA. 2005;102:4080–4.
Müller P, Donnelly M, Ranson H. Transcription profiling of a recently colonised pyrethroid resistant Anopheles gambiae strain from Ghana. BMC Genom. 2007;8:36.
Reddy M, Godoy A, Dion K, Matias A, Callender K, Kiszewski A, et al. Insecticide resistance allele frequencies in Anopheles gambiae before and after anti-vector interventions in continental Equatorial Guinea. Am J Trop Med Hyg. 2013;88:897–907.
Guzov V, Unnithan G, Chermogolov A, Feyereisen R. CYP12A1, a mitochondrial cytochrome P450 from the house fly. Arch Insect Biochem Physiol. 1998;359:231–40.
Ranson H, Nikou D, Hutchinson M, Wang X, Roth C, Hemingway J, et al. Molecular analysis of multiple cytochrome P450 genes from the malaria vector Anopheles gambiae. Insect Mol Biol. 2002;11:409–18.
Felix R, Silveira H. The role of Anopheles gambiae P450 cytochrome in insecticide resistance and infection. Insecticides—pest Engineering. Rijeka: Intech; 2012.
Nardini L, Christian R, Coetzer N, Koekemoer L. DDT and pyrethroid resistance in Anopheles arabiensis from South Africa. Parasit Vectors. 2013;6:229.
Müller P, Chouaibou M, Pignatelli P, Etang J, Walker E, Donnelly M, et al. Pyrethroid tolerance is associated with elevated expression of antioxidants and agricultural practice in Anopheles arabiensis sampled from an area of cotton fields in Northern Cameroon. Mol Ecol. 2008;17:1145–55.
Witzig C, Parry M, Morgan J, Irving H, Steven A, Cuamba N, et al. Genetic mapping identifies a major locus spanning P450 clusters associated with pyrethroid resistance in kdr-free Anopheles arabiensis from Chad. Heredity (Edinb). 2013;110:389–97.
Irving H, Riveron J, Ibrahim S, Lobo NF, Wondji C. Positional cloning of rp2 QTL associates the P450 genes CYP6Z1, CYP6Z3 and CYP6M7 with pyrethroid resistance in the malaria vector Anopheles funestus. Heredity (Edinb). 2012;109:383–92.
David J, Ismail H, Chandor-Proust A, Paine M. Role of cytochrome P450 s in insecticide resistance: impact on the control of mosquito-borne diseases and use of insecticides on Earth. Phil Trans R Soc Lond B Biol Sci. 2013;368:20120429.
Chiu T, Wen Z, Rupasinghe S, Schuler M. Comparative molecular modeling of Anopheles gambiae CYP6Z1, a mosquito P450 capable of metabolizing DDT. Proc Natl Acad Sci USA. 2008;105:8855–60.
Djouaka R, Bakare A, Coulibaly O, Akogbeto M, Ranson H, Hemingway J, et al. Expression of the cytochrome P450 s, CYP6P3 and CYP6M2 are significantly elevated in multiple pyrethroid resistant populations of Anopheles gambiae s.s. from Southern Benin and Nigeria. BMC Genom. 2008;9:538.
Ranson H, Jensen B, Wang X, Prapanthadara L, Hemingway J, Collins F. Genetic mapping of two loci affecting DDT resistance in the malaria vector Anopheles gambiae. Insect Mol Biol. 2000;9:499–507.
Felix R, Müller P, Ribeiro V, Ranson H, Silveira H. Plasmodium infection alters Anopheles gambiae detoxification gene expression. BMC Genom. 2010;11:312.
Vontas J, Small G, Hemingway J. Glutathione S-transferases as antioxidant defence agents confer pyrethroid resistance in Nilaparvata lugens. Biochem J. 2001;357:65–72.
Antonio-Nkondjio C, Tene Fossog B, Kopya E, Poumachu Y, Menze-Djantio B, Ndo C, et al. Rapid evolution of pyrethroid resistance prevalence in Anopheles gambiae populations from the cities of Douala and Yaoundé (Cameroon). Malar J. 2015;14:155.
Authors’ contributions
Conceived and designed the study protocol: CAN, CSW, Participated in field and laboratory analyses: EK, RP, BTF, PAA, CAN, CSW. Critically revised the manuscript: CSW, CC, BTF, RP, PAA Interpreted, analysed data and wrote the paper: CAN with contribution of other authors. All the authors read and approved the final manuscript.
Acknowledgements
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Availability of data and material
The data have been deposited in ArrayExpress under the accession number E-MTAB-1563.
Ethics approval and consent to participate
The study was conducted under the ethical clearance N° 216/CNE/SE/09 delivered by the Cameroon National Ethics (CNE) Committee Ref N° IORG0006538-IRB00007847-FWA00016054. The study did not used any human subject.
Funding
This work received financial support from Wellcome Trust Intermediate Fellowship in Public Health and Tropical Medicine (WTO86423MA) to CAN. The funding body did not had any role in the design, collection of data, analysis and interpretation of data and in writing of the manuscript.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Antonio-Nkondjio, C., Poupardin, R., Tene, B.F. et al. Investigation of mechanisms of bendiocarb resistance in Anopheles gambiae populations from the city of Yaoundé, Cameroon. Malar J 15, 424 (2016). https://doi.org/10.1186/s12936-016-1483-3
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12936-016-1483-3