Abstract
Background
Enzymes from thermophiles are of great interest for research and bioengineering due to their stability and efficiency. Thermophilic expression hosts such as Thermus thermophilus [T. thermophilus] can overcome specific challenges experienced with protein production in mesophilic expression hosts, such as leading to better folding, increased protein stability, solubility, and enzymatic activity. However, available inducible promoters for efficient protein production in T. thermophilus HB27 are limited.
Results
In this study, we characterized the pilA4 promoter region and evaluated its potential as a tool for production of thermostable enzymes in T. thermophilus HB27. Reporter gene analysis using a promoterless β-glucosidase gene revealed that the pilA4 promoter is highly active under optimal growth conditions at 68 °C and downregulated during growth at 80 °C. Furthermore, growth in minimal medium led to significantly increased promoter activity in comparison to growth in complex medium. Finally, we proved the suitability of the pilA4 promoter for heterologous production of thermostable enzymes in T. thermophilus by producing a fully active soluble mannitol-1-phosphate dehydrogenase from Thermoanaerobacter kivui [T. kivui], which is used in degradation of brown algae that are rich in mannitol.
Conclusions
Our results show that the pilA4 promoter is an efficient tool for gene expression in T. thermophilus with a high potential for use in biotechnology and synthetic biology applications.
Similar content being viewed by others
Background
Enzymes from thermophiles have attracted significant interest for research and bioengineering application due to their high stability and high catalytic efficiency [1,2,3]. The production of thermostable enzymes used in these fields is most commonly performed in mesophilic expression hosts, for example Escherichia coli [E. coli], Bacillus subtilis [B. subtilis] or Pichia pastoris [P. pastoris] [4,5,6]. Although the heterologous production of thermostable enzymes in mesophilic expression hosts can be successful, often the folding of the proteins of thermophilic bacteria is much better at high production rate in thermophilic expression hosts. It was recently shown that thermostable enzymes can be folded very differently when produced in mesophilic expression hosts, which can influence activity and thermostability [7]. Some thermostable enzymes are not active at all, when produced in mesophilic expression hosts [8]. Moreover, problems arising from the absence of cofactors or chaperones in mesophilic hosts have to be overcome [9,10,11]. The latter might affect the incorporation of cofactors such heme or iron sulfur clusters. Furthermore, thermostable enzymes can form aggregates known as inclusion bodies when produced in mesophilic expression hosts [8].
The use of thermophilic expression hosts and the development of genetic tools for the overproduction of thermostable enzymes is a popular strategy to circumvent these problems [1, 12, 13]. Recent studies highlighted the potential of the fungus Chaetomium thermophilum [C. thermophilum] and the hyperthermophilic archaeon Sulfolobus solfatarius [S. solfatarius] as possible thermophilic expression hosts [14, 15]. In this study we focus on Thermus thermophilus [T. thermophilus] as thermophilic expression host. T. thermophilus is a thermophilic bacterium that grows optimally at high temperatures between 55 and 80 °C with high growth rates leading to high cell mass [16]. Another outstanding trait which makes T. thermophilus a highly suitable candidate as production platform for thermostable enzymes is its high frequency of natural transformation supporting the use of this organism as cell factory, but also as an expression host for directed evolution studies or for the construction of genetic libraries and genomic and metagenomic studies [17,18,19]. However, although T. thermophilus HB27 is a popular model organism, the pool of strong, regulable promoters for efficient protein production is still limited. Three inducible promoters, namely the Parg, PdnaK and Pscs−mdh promoters were characterized and two systems for efficient overproduction were reported [20,21,22]. One utilizes the Pnar-promoter from T. thermophilus HB8, a close relative to T. thermophilus HB27 which is induced by anaerobic conditions and the presence of nitrate; T. thermophilus HB27 does not perform nitrate respiration due to a lack of the genes involved [21, 23]. Another system uses a silica inducible promoter from T. thermophilus HB8 which is induced by addition of 10 mM silica. However, the latter led to a significant growth inhibition [22]. The problems encountered during expression of fully active enzymes from thermophiles in mesophilic expression hosts together with very limited number of thermophilic expression hosts prompted us to develop a novel expression system for T. thermophilus as thermophile expression host.
For the adhesion on solid surfaces and for the uptake of DNA from the environment T. thermophilus requires type IV pili (T4P) which consist of the major structural subunit PilA4 [24, 25]. For the formation of long pilus structures a high number of PilA4 subunits is required. This led to the suggestion that the pilA4 gene must be highly expressed under T4P-forming conditions. In previous studies we found that the growth temperature had a significant effect on T4P production and on pilA4 expression such as higher amounts of pilA4 transcript were detected during growth at 68 °C compared to 80 °C [26]. This indicates that the pilA4 promoter undergoes thermoregulation which can be mediated by many different factors in bacteria such as riboswitches, RNA-thermometers, temperature dependent transcription factors or heat shock proteins [27,28,29].
In this study we describe the use of the promoter of the pilA4 gene for temperature dependent production of thermostable β-glucosidase in T. thermophilus and fully active and soluble mannitol-1-phosphate dehydrogenase from the anaerobic thermophilic bacterium Thermoanaerobacter kivui [T. kivui]. Taken together this novel expression system can be used for the production of different enzymes from thermophilic and mesophilic bacteria and is a very promising system for advancing biotechnological applications.
Results and discussion
Construction of a PpilA4 expression vector
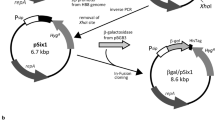
We previously reported that T. thermophilus produces two different forms of pili differing in structure and protein composition, wide T4P structures comprising of the pilin PilA4, and narrow pilus structures formed by PilA5. The latter is essential for twitching motility [24]. PilA4 was found to be essential for natural transformation and for assembly of both the wide and narrow pili [24]. To identify the potential promoter region of pilA4 we performed an in silico analysis of the DNA region upstream of pilA4 using the software phiSITE PromotorHunter [30]. A potential – 35 and – 10 region was detected 131–170 bp upstream of the pilA4 start codon (Fig. 1A). To examine the promoter activity of this region a 206 bp DNA-fragment spanning – 25 to – 231, relative to the translation start site of pilA4 was cloned into pMKE2-bglT-his (Fig. 1B). Agarose gel electrophoresis analysis of the plasmid construction and verification by restriction analysis is included in Additional file 1: Fig. S1. The detected DNA-fragments corresponded to the expected sizes and sequence analysis of the inserted fragments showed that they were inserted correctly. The temperature dependent pilA4 transcript levels detected in T. thermophilus in previous analyses suggested that the pilA4 expression undergoes temperature-dependent transcriptional regulation [26]. In order to include potential distantly located binding sites of proteins involved in temperature-dependent regulation of the pilA4 promoter a longer 499 bp DNA region spanning − 25 to -524 bp upstream of the pilA4 start codon was also cloned into pMKE2-bglT-his. The plasmid construction scheme is shown in Fig. 1B. The two plasmids, designated pMKE2-PpilA4-206 and pMKE2-PpilA4-499 were transformed into T. thermophilus ΔbglT via natural transformation.
Sequence of the intergenic region between comZ and pilA4 and plasmid construction. A The − 35 and − 10 regions of the predicted PpilA4-promoter, start of the transcript (+ 1) and the potential ribosome binding site (RBS) are indicated above the sequences. Arrows denote the transcriptional orientation of comZ and pilA4. B The plasmid pMKE2, which encodes for a kanamycin resistance gene (KanR), was digested using XbaI and NotI to eliminate the Pnar-fragment. The promoterless bglT gene amplified by PCR using genomic DNA of T. thermophilus HB27 was introduced into the XbaI and NotI restriction sites. The stop codon was omitted to generate a transcriptional fusion with the hexahistidin tag encoded by pMKE2 (HisTag). The two PpilA4 DNA fragments amplified from T. thermophilus genomic DNA by PCR were introduced via EcoRI and HindIII restriction sites
Effect of fragment length on β-glucosidase expression
To analyze the promoter activities reporter gene analyses were performed. Therefore, T. thermophilus (pMKE2-PpilA4-206) and T. thermophilus (pMKE2-PpilA4-499) were grown in TM+-medium at optimal growth temperature (68 °C) to mid exponential phase and the β-glucosidase activities were determined. Both recombinant strains exhibited β-glucosidase activities significantly higher than T. thermophilus ΔbglT and the control strain carrying a promoterless pMKE2-bglT-his plasmid (Fig. 2A, black bars). This leads to the conclusion that both, the 206 bp and the 499 bp region convey promoter activity. Interestingly, in T. thermophilus ΔbglT (pMKE2-PpilA4-499), β-glucosidase activities of ~ 270 MU were detected, whereas in T. thermophilus ΔbglT (pMKE2-PpilA4-206) a significantly lower level of activity of ~ 110 MU was detected (Fig. 2A). This suggests that the 499 bp DNA-fragment spanning the – 10 and – 35 region of the PpilA4 promoter contains regulatory regions important for optimal PpilA4 promoter activity.
P pilA4 promoter activities. β-glucosidase activities in T. thermophilus ΔbglT, T. thermophilus ΔbglT (pMKE2-bglT-his), T. thermophilus ΔbglT (pMKE2-PpilA4-206) and T. thermophilus ΔbglT (pMKE2-PpilA4-499) grown in TM+-medium at 68 °C (black bars) and 80 °C (grey bars) respectively A. β-glucosidase activities in T. thermophilus ΔbglT, T. thermophilus ΔbglT (pMKE2-bglT-his), T. thermophilus ΔbglT (pMKE2-PpilA4-206) and T. thermophilus ΔbglT (pMKE2-PpilA4-499) grown in minimal medium with 20 mM pyruvate as carbon sorce at 68 °C (black bars) and 80 °C (grey bars) B. The β-glucosidase activities are given in Miller units (MU). Values are expressed as means ± standard deviations (n = 3)
Effect of growth temperature on β-gucosidase expression
In former studies we found that elevated growth temperatures of 80 °C significantly decreased pilA4 transcript levels [26]. To analyze the PpilA4 promoter activities at 80 °C T. thermophilus ΔbglT (pMKE2-PpilA4-206) and T. thermophilus ΔbglT (pMKE2-PpilA4-499) were grown to mid-exponential growth phase in TM+-medium at 80 °C. The β-glucosidase activities in T. thermophilus ΔbglT (pMKE2-PpilA4-499) were ~ 30 MU which is significantly lower than those detected in cells grown at 68 °C (~ 270 MU) and in the range of the β-glucosidase activities in the control strain T. thermophilus ΔbglT (pMKE2-bglT-his) (~ 10 MU) (Fig. 2A). To ensure that the low β-glucosidase activities in T. thermophilus ΔbglT (pMKE2-PpilA4-499) were not due to plasmid loss at this high temperature, we analyzed the plasmid integrity by retransformation into E. coli TOP10 cells and found that the plasmid was successfully retransferred into E. coli. Analyses of the β-glucosidase activities in cells of T. thermophilus ΔbglT (pMKE2-PpilA4-206) grown at 80 °C also led to the detection of a decrease in activities (~ 73 MU) in comparison to cells grown at 68 °C (~ 110 MU) (Fig. 2A). However, the decrease in β-glucosidase activities was not as dramatic as in T. thermophilus ΔbglT (pMKE2-PpilA4-499). This suggests that the shorter ~ 200 bp PpilA4 region is also subject to temperature-dependent regulation but not as effectively as the longer fragment. This might be due to missing binding sites for bacterial enhancer binding proteins (bEBPs) which can be located quite distantly from the promoter sequence. Such bEBPs effect transcription of the pilin gene through topological changes in the DNA and interaction with the RNA-polymerase holoenzyme. Although so far only very little is known about regulation of T4P-related genes in T. thermophilus, bEBPs have been shown to be involved in the regulation of these genes in other T4P forming organisms such as Pseudomonas aeruginosa [P. aeruginosa] or Geobacter sulfurreducens [G. sulfurreducens] [26, 31, 32]. Complex regulatory mechanisms even including multiple promotors involving bEBPs have been previously identified for the regulation of major pilin subunit genes in Neisseria gonorrhoeae [N. gonorrhoeae] [33].
Effect of growth medium on β-glucosidase expression
In former studies we reported that growth in minimal medium led to significantly increased pilA4 transcript levels [26]. We next examined whether our expression system was affected by growth in minimal medium by analyzing the β-glucosidase activities in cells of T. thermophilus ΔbglT (pMKE2-PpilA4-499) throughout growth at 68 and 80 °C, respectively in minimal medium with pyruvate as carbon source (Fig. 2B). Growth in minimal medium at 68 °C led to significantly increased β-glucosidase activities of maximally ~ 700 MU (Fig. 2B, black bars) in comparison to ~ 270 MU detected in cells grown in TM+ medium. Growth in minimal medium at 80 °C also led to increased β-glucosidase activities of maximally ~ 350 MU in comparison to the activities of ~ 30 MU after growth at 80 °C in TM+ medium (Fig. 2B, grey bars). Taken together, these findings lead to the conclusion that both, growth temperature and medium have a significant effect on gene expression mediated by the PpilA4 promoter with maximal gene expression during growth at 68 °C in minimal medium.
Effect of growth phase on β-glucosidase expression
In former studies it was reported that T. thermophilus HB27 is transformable in all growth phases, with lowest transformation frequencies in stationary growth phase [34]. To address the effect of growth phase on BglT production we examined the activities throughout growth in TM+ medium and minimal medium at 68 or 80 °C by analyzing the β-glucosidase activities in T. thermophilus ΔbglT (pMKE2-PpilA4-499) (Fig. 3). As shown in Fig. 3A, growth at 68 °C in TM+ medium led to exponential growth (µ = 0.93) directly after inoculation of the culture. A final OD600 of 1.4 was reached in the stationary phase after 9 h. A maximal β-glucosidase expression of 300 MU was detected in late exponential growth phase. Growth at 80 °C in TM+ medium led to a prolonged lag phase of 3 h followed by an exponential phase with a doubling time of 0.30 h−1 (Fig. 3B). A final OD600 of 0.9 was reached in stationary phase 9 h after inoculation. During growth at 80 °C in TM+ medium very low β-glucosidase activities of 100 MU were detected in the lag phase which even decreased throughout growth down to 30 MU in stationary growth phase.
Growth-phase dependent β-glucosidase activities in cells. Shown are the optical density (•) and β-glucosidase activities (bars) of T. thermophilus ΔbglT (pMKE2-PpilA4-499) cells grown in TM+-medium at 68 °C (A), 80 °C (B) or in minimal medium with 20 mM pyruvate as carbon and energy source at 68 °C (C) or 80 °C (D)
Inoculation of fresh minimal medium at 68 °C with T. thermophilus ΔbglT (pMKE2-PpilA4-499) led to exponential growth directly after inoculation (Fig. 3C), with a growth rate of µ = 0.19 and a final optical density of 0.9 after 9 h. Maximal β-glucosidase activities of 700 MU were detected during exponential growth. Growth in minimal medium at 80 °C led to a reduced doubling time of 0.1 h−1 and a lower final optical density of 0.38. The measured β-glucosidase activities decreased throughout growth at 80 °C in minimal medium to a minimum of 150 MU in stationary phase (Fig. 3D). Taken together, highest β-glucosidase activities of 700 MU were detected during growth in minimal medium at 68 °C quite rapidly after only 5 h of cultivation, which underlines the suitability of this expression system for biotechnological applications, where time can be an important factor.
Homologous production of BglT-his in T. thermophilus
Next, we analyzed whether our PpilA4-promoter based gene expression system can indeed be used to induce protein production in T. thermophilus. Therefore, T. thermophilus ΔbglT (pMKE2-PpilA4-499) was cultivated in TM+ medium or minimal medium at 68 or 80 °C and harvested in the exponential growth phase. Western blot analyses were performed using the penta-his-antibody. After growth in TM+ or minimal medium at 68 °C both conditions his-tagged BglT was detected in amounts visible in the Coomassie stained SDS-PAGE and also in the Western blot (Fig. 4). As expected no BglT was detected after growth in TM+ medium at 80 °C (Fig. 4A) whereas small amounts of BglT were detected after growth in minimal medium at 80 °C (Fig. 4B).
SDS-PAGE and Western blot analysis of homologous production of BglT-his in T. thermophilus. T. thermophilus ΔbglT (pMKE2-bglT-his) cells were grown in 50 mL TM+ or minimal medium at either 68 or 80 °C, harvested in the mid exponential growth phase followed by cell lysis and seperation of cell extracts via SDS-PAGE. Each lane contains 10 µL of cell extracts normalized to OD600 = 10. Proteins were detected by Coomassie brilliant blue staining (A). Western blot analyses were performed using the penta-his antibody (1:10000 dilution) (B). The white arrow indicates the BglT-his
Heterologous production of MtlD-his from T. kivui
To evaluate the suitability of our system for heterologous protein production, we used the mannitol-1-phosphate dehydrogenase MtlD from T. kivui. Therefore, we first generated pLK1, an expression vector utilizing the pilA4 promoter region as displayed in Fig. 5. The PpilA4 containing DNA fragment with a length of 0.56 kb and the vector amplified by PCR with a length of 6.64 kb were detected by agarose gel electrophoresis (Additional file 1: Fig. S2A, B). pLK1 was verified by restriction analysis using BsaI and PsiI. The length of the detected DNA-fragments (4.19 kb, 2.08 kb and 8.88 kb) corresponds to the expected length (Additional file 1: Fig. S2C).
The wildtype mtlD gene was cloned into pLK1 and transformed into T. thermophilus cells, but protein production was not observed (data not shown). This phenomenon has been observed with other proteins as well, likely due to the distinct codon usage of T. thermophilus, which has a high GC content of 68% compared to T. kivui with a GC content of 35%. To circumvent this problem we used a codon optimized mtlD gene. We cloned the codon-optimized gene into pLK1 to generate pLK1-mtlD-his which was transformed into T. thermophilus wildtype cells. pLK1-mtlD-his was verified via restriction analysis using NdeI and NotI. The lenght of the detected DNA-fragments (7.1 kb and 1.17 kb) corresponds to the expected length (Additional file 1: Fig. S2D).
Cells were grown until mid exponential growth phase (OD600 = 0.6) in minimal medium, harvested and MtlD-his was purified via Ni-NTA affinity chromatography (Fig. 6). A protein yield of 9 mg MtlD per liter of culture was obtained, which is comparable to the protein yields achieved for heterologous protein production in T. thermophilus using the Psip- system which depends on a silica inducible promoter [22]. In contrast to the production of MtlD in E. coli, which was previously reported by Moon et al. [35], no MtlD was detected in the cell debris (Fig. 6B). Which indicates that the enzyme is fully soluble in the expression host T. thermophilus.
Purification of MtlD from T. kivui produced in T. thermophilus. The enzyme was purified by Ni-NTA (elution with 300 mM imidazole) and analyzed on a 12% SDS gel. The protein was stained with Coomassie brilliant blue R-250 (A). Western blot analysis of the fractions obtained during purification of MtlD-his. Western blot analysis was performed using the penta-his antibody (1:1000 dilution) (B). The white arrow indicates MtlD-his.
To assess the functionality of the purified enzyme, we analyzed MtlD-dependent fructose-1-phosphate reduction. The enzyme displayed specific activities of ~ 800 µmol min−1 mg−1 which were comparable to the enzyme purified from E. coli BL21 (DE3). These results demonstrate our pilA4 promoter-based expression system can be used for heterologous production of thermostable, soluble and active proteins from other thermophilic bacteria in T. thermophilus. MtlD not only catalyzes the degradation of mannitol, as in T. kivui, but also the biosynthesis of mannitol-1-phosphate from fructose-6-phosphate. Mannitol has a wide range of applications in the food and pharmaceutical industry [36]. Our studies are the first step to establish a thermophilic production platform for mannitol.
Conclusions
We reported the development of a temperature inducible PpilA4-based-expression system for homologous and heterologous production of thermostable proteins in T. thermophilus. We demonstrate the successful expression of soluble, fully active T. thermophilus β-glucosidase and T. kivui mannitol-1-phosphate dehydrogenase in T. thermophilus. We showed that the pilA4-promoter-dependent expression system is highly efficient yielding protein levels of 9 mg/L of culture. The high induction of the pilA4 promoter at optimal growth temperature in exponential growth phase makes this novel expression system very well suitable for the fast production of high amounts of thermostable proteins for biochemical protein analysis and also for industrial applications. The promoter is regulated by an easy to perform temperature switch.
Materials and methods
Organisms and cultivation
E. coli TOP 10 was cultivated in LB “Luria-Bertani” complex medium (10 g/L tryptone, 10 g/L NaCl, 5 g/L yeast extract) at 37 °C and, when appropriate, 20 µg/mL kanamycin were added. T. thermophilus strains were grown at either 68 or 80 °C in TM+ medium (8 g/L tryptone, 4 g/L yeast extract, 3 g/L NaCl, 0.6 mM MgCl2 and 0.17 mM CaCl2) or Thermus minimal medium [37] with 20 mM pyruvate as carbon source. 20 or 40 µg/mL kanamycin in liquid or solid medium were added when appropriate.
Plasmid construction
The primers and used in this study are presented in Additional file 1: Tables S1 and S2. Plasmid construction was carried out as displayed in Figs. 1B and 5. To generate a pilA4-promoter-dependent expression vector the plasmid pMKE2 [21] was used as backbone. First, the Pnar promoter was deleted from pMKE2 by XbaI and NotI digestion and replaced by the promoterless β-glucosidase gene bglT (TT_P0042, GenBank accession number AF135400.2), which was amplified from chromosomal DNA of T. thermophilus HB27 (GenBank accession number AE017221) with Phusion DNA polymerase (New England Biolabs, Ipswitch MA, USA) using the primers bglT_for and reverse primer bglT_rev (Fig. 1B). The DNA was denatured at 98 °C for 3 min, followed by 35 cycles of denaturation at 98 °C for 30 s, annealing at 60–80 °C for 30 s, and extension at 72 °C for the appropriate time depending on the desired product length assuming an optimal polymerase speed of 0.5 kb per minute. A final extension step was performed at 72 °C for 5 min. The resulting DNA fragment encodes a promoterless bglT gene devoid of the stop codon flanked by XbaI and NotI restriction sites. A 499 bp and 206 bp DNA fragment spanning the putative promoter region upstream of the pilin gene pilA4 (TT_C0858, GenBank accession number AAS81202.1) from T. thermophilus was amplified via PCR using the two forward primers pA4_500_for and pA4_200_for, and the reverse primer pA4_rev, respectively. The PCR products were purified via the PCR CleanUP Kit (Merck, Darmstadt, Germany) digested with HindIII and EcoRI, and inserted into the pMKE2-bglT-his plasmid leading to the two recombinant plasmids pMKE2-PpilA4-206 and pMKE2-PpilA4-499, respectively (Fig. 1B). The plasmids were verified by restriction (Additional file 1: Fig. S1) and sequencing. The PpilA4-dependent expression was performed in a markerless ΔbglT mutant kindly provided by Prof. Dr. José Berenguer.
To produce MtlD from T. kivui in T. thermophilus HB27 using the PpilA4 promoter, the expression vector pLK1 was constructed by introducing a 499 bp DNA fragment of the pilA4 promoter region into pMKE2, thereby replacing the Pnar fragment in front of the multiple cloning site (Fig. 5). For heterologous production of MtlD of T. kivui (TKV_c02860, GenBank accession number CP009170.1) in T. thermophilus a codon optimized mtlD gene was used. The codon optimization was performed by Genscript (Rijswijk, Netherlands) using their GeneSmart™ algorithm. The codon-optimized sequence is provided in Additional file 1: Table. S3. From Genscript we received the codon-optimized mtlD on the plasmid pUC57 (GenBank accession number Y14837.1). The mtlD gene was flanked by NdeI and NotI restriction sites which were used to insert it into pLK1, leading to the expression plasmid pLK1-mtlD-his. We verified pLK1 and pLK-mtlD-his by restriction analyses (Additional file 1: Fig. S2).
β-Glucosidase reporter-gene assay
To analyze bglT production under the control of the T. thermophilus pilA4 promoter the plasmids pMKE2-PpilA4-206 and pMKE2-PpilA4-499 were transferred into T. thermophilus ΔbglT by natural transformation. Therefore, exponentially grown cells were incubated with 500 ng of plasmid DNA for 3 h at 68 °C and then plated on TM+-agar containing 40 µg/mL kanamycin. The β-glucosidase activities were analyzed as previously described with slight modifications [38]. Cells containing either pMKE2-PpilA4-206 or pMKE2-P-pilA4-499 were grown in either TM+ medium or minimal medium at either 68 or 80 °C and harvested in different growth phases by centrifugation. After removal of the supernatant the cells were resuspended in 1 mL Z-buffer (8.8 mM Na2HPO4, 4.5 mM NaH2PO4, 100 mM KCl, 10 mM MgSO4 × 7H2O, 3 mM β-mercaptoethanol, pH 7) and treated with 10 µL toluol for 1 min at 80 °C. 200 µL of the permeabilized cells were transferred into 800 µL preheated (80 °C) Z-buffer. The β-glucosidase-reaction was started by addition of 50 µl 2-nitrophenyl-ß-D-glucopyranosid (2NPGlc) from initial 10 mg/mL stock solution (Carl Roth GmbH, Karlsruhe, Germany) and terminated by addition of 200 µL of 1 M Na2CO3 after a sufficient yellow color had developed. After centrifugation for 5 min at 16,000×g, absorption of the supernatant was recorded at 420 and 550 nm respectively. Miller units [MU] = 1000 × OD420 / reaction time (min) × reaction volume (ml) × OD550 were calculated according to Miller [39].
Heterologous production and purification of MtlD-his
To produce MtlD of T. kivui in T. thermophilus HB27, 1 L of minimal medium was inoculated with T. thermophilus (pLK1-mtlD-his) to OD600 = 0.1. The cells were grown to mid exponential phase (OD600 = 0.6) at 68 °C and harvested by centrifugation. The cells were resuspended in 25 mL buffer 1 (50 mM Tris-HCl, 300 mM NaCl, pH = 7), disrupted via French press and cell debris was removed by centrifugation (14,000×g for 15 min). MtlD-his was purified via immobilized nickel ion chromatography using 3 mL of Ni-NTA resin in a 14 ml protino column (Macherey-Nagel, Düren, Germany). The cell-free extract was incubated with Ni-NTA for 1 h at 4 °C followed by washing with 60 ml buffer 1 containing 50 mM imidazole. MtlD-his protein was eluted with 6 mL buffer 1 containing 300 mM imidazole and concentrated in a Vivaspin® protein concentrator spin column (Sartorius, Göttingen, Germany; 10,000 Da cutoff). The protein concentration was determined according to Bradford [40]. Bovine serum albumin (BSA) was used as the standard protein. Standard solutions with known concentrations (0–90 µg/mL) were prepared. Samples were diluted 1:100 with buffer 1. 100 µL of the sample were mixed with 1 mL Bradford reagent (0.01% (w/v) Coomassie Brilliant Blue G-250, 4.7% (w/v) ethanol, 8.5% (w/v) H3PO4) in 1 mm plastic cuvettes (Sarstedt, Nümbrecht, Germany). The mixture was gently vortexed and incubated at room temperature for 10 min. Absorption was measured at 595 nm after calibrating the spectral photometer with plain Bradford solution. A calibration curve was generated by plotting the known BSA concentrations against their corresponding absorbance values. The equation of the standard curve was used to calculate the protein concentration of each unknown sample. All measurements were taken in triplicate.
Western-blot analysis
T. thermophilus ΔbglT (pMKE2-PpilA4-499) and T. thermophilus (pLK1-mtlD-his) were grown in 50 mL TM+ or minimal medium at 68 or 80 °C. Cells were harvested in mid exponential growth phase (OD600 = 0.8) by centrifugation for 2 min at 16,000×g followed by lysis in SDS-sample buffer (4% (w/v) SDS, 12% (v/v) glycerol, 50 mM Tris-HCl, pH 6.8, 2% (v/v) mercaptoethanol, 0.01% (w/v) Serva Blue G, OD600 = 10) by boiling for 20 min. The samples (10 µg protein) were separated by SDS-PAGE (12%) [41]. The proteins were blotted onto a nitrocellulose membrane and Western blot analyses were performed using a penta-his antibody (1:1000) (Qiagen, Venlo, Netherlands) as described previously [42].
MtlD activity assay
To determine the activity of the heterologously produced MtlD from T. kivui, the NADH dependent fructose-6-phosphate reduction was followed by measuring NADH oxidation at 340 nm (ε = 6,22 mM−1 cm−1) at 65 °C [35]. Specific activity = ((ΔA340 / Δt [min]) × Dilution Factor) / (ε × cm × mg).
Statistical analysis
The β-glucosidase and MtlD activity analyses were performed in technical and biological triplicates and are presented as the mean ± standard deviation. Statistical analysis was performed using GraphPad Prism 5.0 software (Graphpad Software, Inc., Boston, MA, USA).
Availability of data and materials
All data are available.
Abbreviations
- T4P:
-
Type IV pili
- BP:
-
Base pairs
- RBS:
-
Ribosome binding site
- SDS:
-
Sodium dodecyl sulfate
References
Cava F, Hidalgo A, Berenguer J. Thermus thermophilus as biological model. Extremophiles. 2009;13(2):213–31.
Niehaus F, Bertoldo C, Kahler M, Antranikian G. Extremophiles as a source of novel enzymes for industrial application. Appl Microbiol Biotechnol. 1999;51(6):711–29.
Taylor MP, van Zyl L, Tuffin IM, Leak DJ, Cowan DA. Genetic tool development underpins recent advances in thermophilic whole-cell biocatalysts. Microb Biotechnol. 2011;4(4):438–48.
Adams MW, Kelly RM. Finding and using hyperthermophilic enzymes. Trends Biotechnol. 1998;16(8):329–32.
Liu H, Cheng Y, Du B, Tong C, Liang S, Han S, et al. Overexpression of a novel thermostable and chloride-tolerant laccase from Thermus thermophilus SG0.5JP17-16 in Pichia pastoris and its application in synthetic dye decolorization. PLoS ONE. 2015;10(3):e0119833.
Lan Thanh Bien T, Tsuji S, Tanaka K, Takenaka S, Yoshida K. Secretion of heterologous thermostable cellulases in Bacillus subtilis. J Gen Appl Microbiol. 2014;60(5):175–82.
Kruglikov A, Wei Y, Xia X. Proteins from thermophilic Thermus thermophilus often do not fold correctly in a mesophilic expression system such as Escherichia coli. ACS Omega. 2022;7(42):37797–806.
Hidalgo A, Betancor L, Moreno R, Zafra O, Cava F, Fernandez-Lafuente R, et al. Thermus thermophilus as a cell factory for the production of a thermophilic Mn-dependent catalase which fails to be synthesized in an active form in Escherichia coli. Appl Environ Microbiol. 2004;70(7):3839–44.
Sudhamsu J, Kabir M, Airola MV, Patel BA, Yeh SR, Rousseau DL, Crane BR. Co-expression of ferrochelatase allows for complete heme incorporation into recombinant proteins produced in E. coli. Protein Expr Purif. 2010;73(1):78–82.
Zheng L, Cash VL, Flint DH, Dean DR. Assembly of iron-sulfur clusters. Identification of an iscSUA-hscBA-fdx gene cluster from Azotobacter vinelandii. J Biol Chem. 1998;273(21):13264–72.
Blasco F, Dos Santos JP, Magalon A, Frixon C, Guigliarelli B, Santini CL, Giordano G. NarJ is a specific chaperone required for molybdenum cofactor assembly in nitrate reductase a of Escherichia coli. Mol Microbiol. 1998;28(3):435–47.
Zeldes BM, Keller MW, Loder AJ, Straub CT, Adams MW, Kelly RM. Extremely thermophilic microorganisms as metabolic engineering platforms for production of fuels and industrial chemicals. Front Microbiol. 2015;6:1209.
Kim K, Choe D, Lee DH, Cho BK. Engineering biology to construct microbial chassis for the production of difficult-to-express proteins. Int J Mol Sci. 2020;21(3):990.
Kellner N, Griesel S, Hurt E. A homologous recombination system to generate epitope-tagged target genes in Chaetomium thermophilum: a genetic approach to investigate native thermostable proteins. Int J Mol Sci. 2022;23(6):3198.
Albers SV, Jonuscheit M, Dinkelaker S, Urich T, Kletzin A, Tampe R, et al. Production of recombinant and tagged proteins in the hyperthermophilic archaeon Sulfolobus solfataricus. Appl Environ Microbiol. 2006;72(1):102–11.
Oshima T, Imahori K. Description of Thermus thermophilus (Yoshida and Oshima) comb. nov., a nonsporulating thermophilic bacterium from a japanese thermal spa. Int J Syst Bacteriol. 1974;24:102–12.
Mate DM, Rivera NR, Sanchez-Freire E, Ayala JA, Berenguer J, Hidalgo A. Thermostability enhancement of the Pseudomonas fluorescens esterase I by in vivo folding selection in Thermus thermophilus. Biotechnol Bioeng. 2020;117(1):30–8.
Koyama Y, Hoshino T, Tomizuka N, Furukawa K. Genetic transformation of the extreme thermophile Thermus thermophilus and of other Thermus spp. J Bacteriol. 1986;166(1):338–40.
Liebl W, Angelov A, Jürgensen J, Chow J, Löschcke A, Drepper T, et al. Alternative hosts for functional (meta)genome analysis. Appl Microbiol Biotechnol. 2014;98(19):8099–109.
Park HS, Kilbane JJn. Gene expression studies of Thermus thermophilus promoters PdnaK, Parg and Pscs-mdh. Lett Appl Microbiol. 2004;38(5):415–22.
Moreno R, Zafra O, Cava F, Berenguer J. Development of a gene expression vector for Thermus thermophilus based on the promoter of the respiratory nitrate reductase. Plasmid. 2003;49(1):2–8.
Fujino Y, Goda S, Suematsu Y, Doi K. Development of a new gene expression vector for Thermus thermophilus using a silica-inducible promoter. Microb Cell Fact. 2020;19(1):126.
Moreno R, Haro A, Castellanos A, Berenguer J. High-level overproduction of his-tagged tth DNA polymerase in Thermus thermophilus. Appl Environ Microbiol. 2005;71(1):591–93.
Neuhaus A, Selvaraj M, Salzer R, Langer JD, Kruse K, Kirchner L, et al. Cryo-electron microscopy reveals two distinct type IV pili assembled by the same bacterium. Nat Commun. 2020;11(1):2231.
Rumszauer J, Schwarzenlander C, Averhoff B. Identification, subcellular localization, and functional interactions of PilMNOWQ and PilA4 involved in transformation competency and pilus biogenesis in the thermophilic bacterium Thermus thermophilus HB27. FEBS J. 2006;273:3261–72.
Salzer R, Kern T, Joos F, Averhoff B. Environmental factors affecting the expression of type IV pilus genes as well as piliation of Thermus thermophilus. FEMS Microbiol Lett. 2014;357(1):56–62.
Sharma A, Alajangi HK, Pisignano G, Sood V, Singh G, Barnwal RP. RNA thermometers and other regulatory elements: diversity and importance in bacterial pathogenesis. Wiley Interdiscip Rev RNA. 2022;13(5):e1711.
Maleki F, Khosravi A, Nasser A, Taghinejad H, Azizian M. Bacterial heat shock protein activity. J Clin Diagn Res. 2016;10(3):BE01–3.
Liberek K, Galitski TP, Zylicz M, Georgopoulos C. The DnaK chaperone modulates the heat shock response of Escherichia coli by binding to the σ32 transcription factor. Proc Natl Acad Sci U S A. 1992;89(8):3516–20.
Klucar L, Stano M, Hajduk M. phiSITE: database of gene regulation in bacteriophages. Nucleic Acids Res. 2010;38:D366-70.
Juarez K, Kim BC, Nevin K, Olvera L, Reguera G, Lovley DR, Methe BA. PilR, a transcriptional regulator for pilin and other genes required for Fe(III) reduction in Geobacter sulfurreducens. J Mol Microbiol Biotechnol. 2009;16(3–4):146–58.
Ishimoto KS, Lory S. Formation of pilin in Pseudomonas aeruginosa requires the alternative sigma factor (RpoN) of RNA polymerase. Proc Natl Acad Sci U S A. 1989;86(6):1954–7.
Fyfe JA, Carrick CS, Davies JK. The pilE gene of Neisseria gonorrhoeae MS11 is transcribed from a σ70 promoter during growth in vitro. J Bacteriol. 1995;177(13):3781–7.
Hidaka Y, Hasegawa M, Nakahara T, Hoshino T. The entire population of Thermus thermophilus cells is always competent at any growth phase. Biosci Biotechnol Biochem. 1994;58(7):1338–39.
Moon J, Henke L, Merz N, Basen M. A thermostable mannitol-1-phosphate dehydrogenase is required in mannitol metabolism of the thermophilic acetogenic bacterium Thermoanaerobacter kivui. Environ Microbiol. 2019;21(10):3728–36.
Song SH, Vieille C. Recent advances in the biological production of mannitol. Appl Microbiol Biotechnol. 2009;84(1):55–62.
Schwarzenlander C, Haase W, Averhoff B. The role of single subunits of the DNA transport machinery of Thermus thermophilus HB27 in DNA binding and transport. Environ Microbiol. 2009;11(4):801–08.
Ohta T, Tokishita S, Imazuka R, Mori I, Okamura J, Yamagata H. β-glucosidase as a reporter for the gene expression studies in Thermus thermophilus and constitutive expression of DNA repair genes. Mutagenesis. 2006;21(4):255–60.
Miller JH. Assay of β-galactosidase. In: Platt T, Miller-Hill B, Miller JH, editors. Experiments in molecular genetics. New York, USA: Cold Spring Harbour Laboratory Press; 1972. pp. 319–53.
Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–54.
Lämmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–5.
Towbin H, Staehelin T, Gordon J. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc Natl Acad Sci U S A. 1979;76(9):4350–54.
Acknowledgements
The markerless T. thermophilus ΔbglT mutant was kindly provided by Prof. Dr. José Berenguer, Universida Autónoma de Madrid. This study was supported by a grant from the Deutsche Forschungsgemeinschaft (AV 9/6 − 2) and the Federal Ministry of Education and Research, Germany (ThermoSynCon).
Funding
Open Access funding enabled and organized by Projekt DEAL. Deutsche Forschungsgemeinschaft (DFG) grant AV 9/6 − 2. Federal Ministry of Education and Research (BMBF) grant ThermoSynCon.
Author information
Authors and Affiliations
Contributions
BA and LK designed the experiments, LK performed the experiments. BA and LK analyzed the data. BA, VM and LK wrote the paper.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
All authors agreed.
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Additional file 1
: Table S1.: Plasmids used in this study; Table S2.: Primers used in this study; Table S3.. Alignment of mtlD from Thermoanaerobacter kivui with the codon optimized mtlD for Thermus thermophilus; Figure S1.. Verification of the PCR-products and the generated pMKE2-PpilA4-206 and pMKE2-PpilA4-499 plasmids; Figure S2.. Verification of the PCR-products and restriction analyses of the generated pLK1-plasmids.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Kirchner, L., Müller, V. & Averhoff, B. A temperature dependent pilin promoter for production of thermostable enzymes in Thermus thermophilus. Microb Cell Fact 22, 187 (2023). https://doi.org/10.1186/s12934-023-02192-1
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12934-023-02192-1