Abstract
Background
Actinobacteria from freshwater habitats have been explored less than from other habitats in the search for compounds of pharmaceutical value. This study highlighted the abundance of actinobacteria from freshwater sediments of two rivers and one lake, and the isolates were studied for their ability to produce antimicrobial bioactive compounds.
Results
16S rRNA gene sequencing led to the identification of 84 actinobacterial isolates separated into a common genus (Streptomyces) and eight rare genera (Nocardiopsis, Saccharopolyspora, Rhodococcus, Prauserella, Amycolatopsis, Promicromonospora, Kocuria and Micrococcus). All strains that showed significant inhibition potentials were found against Gram-positive, Gram-negative and yeast pathogens. Further, three biosynthetic genes, polyketide synthases type II (PKS II), nonribosomal peptide synthetases (NRPS) and aminodeoxyisochorismate synthase (phzE), were detected in 38, 71 and 29% of the strains, respectively. Six isolates based on their antimicrobial potentials were selected for the detection and quantification of standard antibiotics using ultra performance liquid chromatography (UPLC–ESI–MS/MS) and volatile organic compounds (VOCs) using gas chromatography mass spectrometry (GC/MS). Four antibiotics (fluconazole, trimethoprim, ketoconazole and rifampicin) and 35 VOCs were quantified and determined from the methanolic crude extract of six selected Streptomyces strains.
Conclusion
Infectious diseases still remain one of the leading causes of death globally and bacterial infections caused millions of deaths annually. Culturable actinobacteria associated with freshwater lake and river sediments has the prospects for the production of bioactive secondary metabolites.
Similar content being viewed by others
Background
Actinobacteria are diverse group of Gram positive and filamentous bacteria that have high guanine–cytosine (GC) content ranging from 50 to 70 mol% in their genome [1]. They are considered excellent elaborators of pharmaceutical products such as antibiotics and industrial enzymes and are well known as a prominent source for finding novel biologically active secondary metabolites [2, 3].
Antibiotic resistance against available drugs is one of the primary reasons to seek new and novel drugs such as antibiotics from a natural source to fight against multidrug-resistant pathogens [4]. The infections caused by globally emerging Gram-negative multidrug-resistant pathogens are an important challenge. Vancomycin-resistant enterococci (VRE), Methicillin-resistant Staphylococcus aureus (MRSA), extended-spectrum β-lactamase (ESBLs) that produce Gram-negative bacteria, and Klebsiella pneumoniae carbapenemase (KPC) that produces Gram-negative bacteria are few of the most significant cases that are gradually increasing in ubiquity and virulence [5]. Due to this fact, there is an incessant requirement for the search for new bioactive compounds from unexplored/less explored environments [6]. As a result, it is important to target such environments that could be highly potent sources of novel and bioactive compounds. Among all living organisms, the actinobacteria phylum currently represents the most prospective group of microorganisms for the discovery of bioactive compounds such as antimicrobials, antitumor agents, antiparasitics, anticancer agents and enzymes [7, 8]. It has been shown that 45% of all reported bioactive compounds of microbial origin are produced by actinobacteria, more than 70% of which are produced by the largest genus in the phylum Streptomyces [9].
Since the discovery of the first antibiotic from actinobacteria in 1940, actinomycin, the exploration of these micro-organisms has resulted in the isolation of thousands of naturally occurring antibiotics to date [10]. Several novel species of Streptomyces have been reported worldwide as potential natural sources for the discovery of naturally occurring antibiotics [11,12,13]. Actinobacteria have been extensively reported from different ecosystems such as soil, freshwater, marine and as endophytes from plants [14, 15] and have been investigated for their potential contributions to the pharmaceutical industry by different researchers [16,17,18,19,20,21,22]. However, there has been a significant decline in the rate of discovery of novel actinobacteria in recent years [23, 24]. Therefore, the exploration of potential actinobacteria from unexplored habitats is an important approach to discovering novel antibiotics to meet the current needs [25, 26].
Northeast India is a large bioprospecting area that was identified as the Indo-Burma mega-biodiversity hotspot by Conservation International, and the area is well known for its rich biodiversity and unexplored biological resources [27, 28]. Bioprospection studies on the actinobacteria phylum have mainly focused on terrestrial and marine ecosystems, and few have focused on freshwater ecosystems [29]. There are several reports that have examined the diversity of actinobacteria in freshwater worldwide [29,30,31], but very few studies have reported on their biosynthetic potential. Therefore, it will be of great importance to characterize the various biologically active secondary metabolites produced by actinobacteria obtained from freshwater sediments. The present study intended to isolate actinobacterial cultures, screen them for in vitro antimicrobial inhibitory activity, detect their bioactive secondary metabolites and phylogenetically identify the potential antibiotic-producing actinobacteria from freshwater sediments of selected freshwater lakes and rivers in India.
Methods
Sediment sampling
Samples were collected from two rivers [Tlawng River (24°52′N; 92°36′E), Tuirial River (24°21′N 92°53′)] and one lake [Tamdil Lake (23°44′N; 92°57′E)] (Additional file 1: Fig. S8). Samples were randomly collected from five different stations of each river and lake at average depths of 2–5 m. The labeled samples were placed in sterile tubes (50 ml), transported to the laboratory and were processed immediately for the isolation of actinobacteria.
Isolation of freshwater actinobacterial strains
The collected samples were subjected to physical pretreatment (55 °C for 6 min) to hinder the growth of fast-growing bacteria and favor the growth of actinobacteria [7]. Actinobacteria were isolated using the serial dilution method and the spread plate technique. The stock solution of the sample was prepared with 1 ml of water sediment (water + sediment suspension) and 9 ml of sterile distilled water in a test tube, and the solution was mixed for 10 min. The suspension was serially diluted by transferring 1 ml aliquots to a series of test tubes; each containing 9 ml of sterile distilled water to prepare the final volumes of 10−1, 10−2 and 10−3, and the diluted suspension was spread over the surface of selected nutritional media. Seven selective media, starch casein agar (SCA), yeast extract-malt extract agar (ISP2), Actinomycetes isolation agar (AIA), Streptomyces agar (SA), glycerol–asparagine agar (ISP5), tyrosine agar medium (ISP7), and tap water yeast extract agar (TWYE), were supplemented with nalidixic acid (30 mg/ml) and cyclohexamide (30 mg/ml) to inhibit the growth of Gram-negative bacteria and fungi, respectively. The plates were incubated at 28 ± 1 °C for 7–30 days, and the colonies were observed periodically. Pure cultures were obtained after two to three successive sub-culturing rounds and transferred to fresh isolation media. The cultures were preserved in their respective slants at 4 °C and 30% glycerol at − 80 °C.
Morphological and microscopic characterization of actinobacterial strains
Pure cultures of the isolates were identified based on their morphological and cultural characteristics following the International Streptomyces Project (ISP) [32]; the nature of the colony, the color of aerial and substrate mycelium, the production of diffusible pigments and the utilization of carbon source were studied [33]. The spore chain morphologies of the isolates were studied using a scanning electron microscope (SEM). The mycelium structures were observed using a phase contrast microscope (Olympus), and the organisms were identified according to Bergey’s Manual of Determinative Bacteriology 9th edition.
Molecular identification and phylogenetic analysis
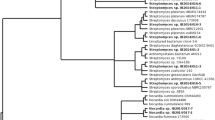
Genomic DNA was isolated and purified using a DNA extraction kit (Invitrogen) as described in previous studies [21]. Ribosomal RNA (16S rRNA) genes were amplified using universal bacterial primers [34]. The reactions and conditions of the PCR were performed exactly as reported in our previous studies [21], and sequencing was done commercially at Sci Genome Pvt. Ltd. Cochin, India. Sequences were compared with the reference strains of actinobacteria from the NCBI genomic database using a BLASTn search to determine similarity percentages. The strains with highest similarity percentages were retrieved from the EzTaxon database [35], and multiple sequence alignment was performed using Clustal W software packaged in MEGA 6.0 [36]. The evolutionary models were selected based on the lowest Bayesian information criterion (BIC) scores and the highest Akaike information criterion (AIC) values using MEGA 6.0 [37]. Phylogenetic analysis was performed using MEGA 6 software using the maximum-likelihood method and using the Tamura Nei parameters algorithm taking E. coli as the outgroup [38]. The significance of the branching order was determined by bootstrap analysis of 1000 alternative trees. The obtained nucleotide sequences of the 16S rRNA gene fragments were deposited, and accession numbers were acquired. Trees were viewed and edited using the FigTree 1.3.1 program.
Screening for antimicrobial activity
The antimicrobial activities of the actinobacterial isolates were tested against five bacterial pathogens [Gram-positive bacteria: Staphylococcus aureus MTCC-96, Bacillus subtilis NCIM-2097, and Micrococcus luteus NCIM-2170; Gram-negative bacteria: Pseudomonas aeruginosa MTCC-2453 and Escherichia coli MTCC-739 and yeast: Candida albicans MTCC-3017]. The pathogens were obtained from the Microbial Type Culture Collection (MTCC), Chandigarh and National Collection of Industrial Microorganisms (NCIM), Pune, India. The crude extracts were prepared by inoculating a single purified colony of actinobacteria in Tryptone yeast extract broth medium (ISP medium 1) and incubated at 28 °C, 150 rpm for 7–20 days. The grown cultures after centrifugation were used to assess antimicrobial activity by the agar well diffusion method [39]. The test pathogenic bacteria were spread on a nutrient agar plate, 6 mm diameter wells were prepared using a sterile cork borer, 70 µl of the clear supernatant of the actinobacteria was dispensed into individual wells, and the plates were incubated at 28 ± 2 °C for 24 h. The anti-microbial activity of the isolates was evaluated as described by Zothanpuia et al. [21].
Antimicrobial assay using crude extract
The actinobacteria isolates that were selected based on antimicrobial screening were grown in ISP1 broth using a 500 ml conical flask at 28 °C in a shaker incubator for 30 days. The filtrates of the grown cultures were used for the extraction using methanol 1:1 ratio (v/v). The methanolic crude extracts of the isolates were prepared in concentrations of 1, 2, 5, 20 mg and 40 mg/ml [21] with sterile water and used for antimicrobial activity by the agar well diffusion method and disk diffusion assay [39, 40].
Determination of MIC
The minimum inhibitory concentration (MIC) of the selected strains was determined using the broth micro dilution technique in a 96-well microtiter plate [41]. The methanolic extracts of the strains were dissolved and diluted in different concentrations (0.025, 0.05, 0.1, 0.2, 0.4, 0.8, 1.6 and 3.2 mg/ml) and were used to test the antimicrobial activity by growing them with bacterial culture in a 96-well microtiter plate. The ampicillin (1 mg/ml) amended bacterial culture was used as the positive control, and the bacterial cultures without treatment were used as the negative control. The plates were incubated at 37 °C for 36 h, and absorbance was taken at 700 nm in a UV–VIS spectrophotometer (MultiscanTM GO, Thermo Scientific, MA, USA). EC50 was expressed and calculated as previously described [21].
Amplifications of biosynthetic genes (PKS, phzE and NRPS)
The presence of biosynthetic genes [Polyketide synthase type II (PKS II) non-ribosomal peptide synthetase (NRPS) and aminodeoxyisochorismate synthase (phzE)] was evaluated using degenerate primers for highly conserved regions encoding enzymes associated with the biosynthesis of polyketides, peptides and phenazine, respectively. The primers that were employed and the PCR conditions for the amplification of PKS-II, phzE and NRPS gene fragments were described in previous studies [7, 21].
Phylogenetic analysis PKS II, NRPS and phzE gene
Biosynthetic gene sequences of PKS type II, NRPS and phzE from the selected seven strains were compared with the sequences from NCBI database using the BLASTn search tool [38] and were aligned by Clustal W software packaged in MEGA 6.0 [36]. The evolutionary model for PKS II, NRPS and phzE gene was selected based on lowest BIC value and highest AIC value using MEGA 6.0 [39]. Phylogenetic tree was constructed by the maximum likelihood method using MEGA 6.0 software with General Time Reversible (GTR + G) model for PKS II and Tamura 3-parameter (T92 + G) for NRPS and phzE gene [38, 39].
Detection of antibiotics using UPLC–ESI–MS/MS
Ultra-performance liquid chromatography (UPLC–ESI–MS/MS) was employed to detect antibiotics in the methanolic extracts of the selected strains. Four antibiotics (trimethoprim, fluconazole, ketoconazole and rifampicin) were selected, and a standard solution was prepared using methanol (Additional file 1: Table S4). The mixed standards were diluted in the ranges from 0.5 to 500 ng/ml, and a standard calibration curve was prepared. Instrumentation and analytical conditions were performed using the standardized methods as described in our previous paper (Fig. 4) [21].
GC–MS analysis
Gas chromatography–mass spectroscopy (GC–MS) was used to determine the volatile organic compounds (VOCs) present in the methanolic extracts of the selected strains. For GC–MS, the Clarus 680 GC was used in the analysis employed with a fused silica column packed with Elite-5MS (5% biphenyl 95% dimethylpolysiloxane, 30 m × 0.25 mm ID × 250 μm df), and the components were separated using helium as carrier gas at a constant flow of 1 ml/min. The injector temperature was set to 260 °C during the chromatographic run. A total of 1 μl of the extracted sample was injected into the instrument, and the oven temperatures were as follows: 60 °C (2 min); followed by 300 °C at a rate of 10 °C/min; and 300 °C for 6 min. The mass detector conditions were a transfer line temperature of 240 °C; an ion source temperature of 240 °C, an ionization mode electron impact at 70 eV, a scan time of 0.2 s and a scan interval of 0.1 s. The fragments from 40 to 600 Da were analyzed. The spectra of the detected compounds were compared with their mass spectra from the database of known components stored in the GC–MS NIST (2008) library.
Statistical analysis
All experiments were conducted in triplicate, and the readings were taken as the mean ± the standard deviation of the mean of three replicates, which were calculated using Microsoft Excel XP 2010. One-way analysis of variance (ANOVA) was performed to analyzed significant difference (P = 0.05) between antimicrobial activities obtained isolates by using SPSS software version 20.0.
Results
Isolation and distribution of freshwater actinobacteria
A total of 68 isolates of actinobacteria were obtained from freshwater sediments; 30 strains from Tamdil Lake, 19 from Tlawng River, 19 from Tuirial River. From a total of seven different media employed for isolation, 31 isolates were recovered from the SCA medium, 24 from AIA, 3 from ISP5, 4 from SA, 1 from TWYE, 3 from ISP7, and 2 from ISP2. These results clearly indicated that SCA was the most suitable medium for the isolation of actinobacteria from freshwater sediments and yielded 45% of the total isolates followed by AIA (36%). After 30 days of incubation, the cultures matured and the actinobacteria colony was observed to exhibit white, black, yellow, orange, brownish white and pale yellow colors. Most of the actinobacterial strains analyzed by field emission gun-scanning electron microscopy (FEG-SEM) showed that the aerial mycelia produce spiral spore chains (Additional file 1: Fig. S1).
Molecular characterization and phylogenetic affiliation
According to the molecular identification using 16S rRNA gene sequencing, 68 actinobacteria isolates were classified into six families and nine genera with similarity percentages ranging from 98 to 100%. The majority of the isolates were grouped under Streptomycetaceae followed by Pseudonocardiaceae, Nocardiopsaceae, Nocardiaceae, Promicromonosporaceae and Micrococcaceae. Streptomyces was the most dominant genus (n = 49, 72%), and the other genera included Nocardiopsis (n = 6), Saccharopolyspora (n = 4), Rhodococcus (n = 2), Prauserella (n = 1), Amycolatopsis (n = 1), Promicromonospora (n = 1) Kocuria (n = 1) and Micrococcus (n = 3) (Additional file 1: Table S1). All sequences were deposited in the NCBI GenBank database, and accession numbers were given (KM243384, KM405296–KM405298, KM405300–KM405304, KM405306–KM405307, KM405310, KM406395, KM406397, KM406398, KR703473–KR703475, KR857285, KR857286, KR857288, KR857290–KR857296, KR857298–KR857318, KT232313–KT232316, KT429605–KT429610, KT429612, KT429614–KT429616, KY077681, MF536299–MF536302). The length of the sequences was used for the construction of phylogenetic tree ranges from 500 to 1000 bp. The phylogenetic tree was constructed using the maximum-likelihood and Tamura Nei parameters with the lowest BIC values (12,154.636) and highest AIC values (10,215.261) (Fig. 1). The topology of the tree that was generated differentiated the isolates into 3 major clades. All genera of Streptomyces formed major clade I with a bootstrap support value of 96%. Rare genera, such as Saccharopolyspora, Amycolatopsis and Prauserella, which fell under the family Pseudonocardiaceae, were clustered together with Rhodococcus and had a bootstrap value of 87%. Micrococcus and Kocuria under the family Micrococcaceae formed a separate clade II with Promicromonospora and had a bootstrap value of 83%. All strain types of Nocardiopsis formed a separate clade III with a bootstrap value of 100%.
Relative abundance
The relative abundance of actinobacteria at the genus level revealed that Streptomyces was the most dominant in Tamdil Lake (n = 20, 40.8%) followed by Tlawng River (n = 18, 36.7%) and Tuirial River (n = 11, 22.4%) from a total of 49 isolates. However, some rare actinobacteria, such as Promicromonospora sp., Prauserella sp., Rhodococcus sp., and Kocuria sp., were obtained from only Tamdil Lake, while Amycolatopsis sp. was found in only Tlawng River. Saccharopolyspora sp., Nocardiopsis sp. and Micrococcus sp. were obtained from Tamdil Lake and Tlawng River, whereas several different species of Streptomyces were obtained from all study sites (Fig. 2). These results showed that the freshwater actinobacteria population varies substantially between lakes and rivers. In comparison to the river ecosystem, the lake ecosystem was observed to be more favorable for actinobacterial growth, as indicated by the enhanced number of isolates obtained with greater diversity.
Evaluation of antimicrobial activity
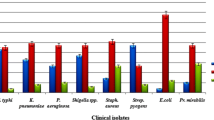
Initially, all isolates (n = 68) were subjected to preliminary screening against five bacterial pathogens (S. aureus, B. subtilis, M. luteus, P. aeruginosa, E. coli) and yeast (C. albicans). All isolates exhibited antagonistic activity against at least three of the six tested pathogens. E. coli was found to be the most susceptible pathogen and all isolates (100%) showed activity against it within the inhibition range of 7.4 mm to 15.5 mm diameter (Additional file 1: Table S2). This was followed by P. aeruginosa (95.65%), C. albicans (79.71%), B. subtilis (78.26%) and S. aureus (63.76%). Only 26 (37%) isolates showed positive activity against M. luteus. The maximum activity was recorded by Streptomyces flavogriseus strain DST30 (18.8 mm) followed by Streptomyces cyaneofuscatus strain DST57 (15.95 mm) and Streptomyces albidoflavus DST71 (15.9 mm) against M. luteus, C. albicans and B. subtilis, respectively. Six strains (Streptomyces sp. DST25, Streptomyces cellulosae DST28, Streptomyces flavogriseus DST52, Streptomyces albidoflavus DST71, Streptomyces sp. DST116 and Streptomyces sp. DST119) showed broad-spectrum antimicrobial activities (Table 1), inhibiting all six tested pathogens, and these strains were selected as potential candidates for further investigation. Mean (± SD) followed by the same letter(s) in each column are not significantly different at P < 0.05 using Duncan’s new multiple range test.
Antimicrobial activity using methanol crude extract
The methanolic crude extracts of the six selected strains that were tested for their antimicrobial activity showed adequate inhibition zones at 20 and 40 mg/ml (Additional file 1: Fig. S1) for all six samples, while all the isolates showed no activity in 1 and 2 mg/ml. The agar well diffusion assay showed better results compared to the filter paper disk diffusion assay.
MIC of selected strains
The methanolic crude extracts of the six strains were subjected to antimicrobial activity quantification by determining the MIC of each strain against six pathogens. Streptomyces sp. DST116 showed maximum activity against M. luteus (EC50 = 0.05103 mg/ml) among all tested pathogens. Streptomyces cellulosae DST28 (EC50 = 0.3371 mg/ml) and Streptomyces flavogriseus DST52 (EC50 = 0.003 mg/ml) also showed the highest antimicrobial activities against M. luteus. Streptomyces sp. DST25 showed the highest activity against B. subtilis (EC50 = 0.009 mg/ml), and Streptomyces albidoflavus DST71 showed the highest activity against P. aeruginosa (EC50 = 0.05042 mg/ml) (Table 2).
Biosynthetic gene analysis
Out of the 68 isolates screened for a biosynthetic gene, NRPS was detected in 71% (n = 49) of the isolates, PKS type II was detected in 26 isolates (38%), and phzE was detected in 28% (n = 19) of the isolates (Additional file 1: Table S2). A total of 11 isolates (DST45, DST47, DST54, DST56, DST57, DST58, DST74, DST76, DST77, DST99, DST101) were found to have all three genes.
Phylogenetic analysis of biosynthetic genes
The nucleotide sequences of three biosynthetic genes (PKS II, NRPS and phzE) showed 82–92% similarity with the type strain from NCBI-BLASTn database. The transition and transversion bias ratio of PKSII, NRPS and phzE gene was 0.55, 0.33 and 0.17 respectively whereas the maximum log likelihood for the substitution computation was − 2765.453, − 501.484 and − 801.607 respectively. The phylogenetic tree constructed using PKS II sequences revealed that Streptomyces sp. DST29 formed separate clade with Streptomyces sp. MM48 Streptomyces gobitricini with bootstrap values of 99% while Streptomyces sp. DST116, Streptomyces sp. DST52 and Streptomyces sp. DST119 each formed a separate clade with a bootstrap support value of 99–100% (Fig. 3a). Similarly the NRPS gene sequences of Streptomyces sp. DST116 Streptomyces sp. DST25, Streptomyces sp. DST71 and Streptomyces sp. DST119 formed separate clade with Streptomyces sp. CAH29-18, Streptomyces albidus NBRC14052, Streptomyces cyaneofuscatus DST103, Streptomyces bamensis NBRC14727 and Streptomyces sp. BSH50-42 respectively with a bootstrap value of 84–89% (Fig. 3b). Similarly Streptomyces sp. DST119 and Streptomyces sp. DST71 were clustered separately in phzE gene sequences forming same clade with Streptomyces sp. HB291 and Streptomyces sp. 13–33–9 respectively with a bootstrap value of 100% (Fig. 3c).
GC–MS analysis
The methanolic crude extracts of the six selected strains were investigated to determine their volatile organic compounds using GC–MS, which revealed thirty-five VOCs (Additional file 1: Table S3). Fourteen compounds were detected from the extract of Streptomyces albidoflavus DST71 within the retention time of 15–29 min (Additional file 1: Fig. S2). Among the compounds, hexanal constituted the maximum amount, which accounted for 23.2% of the total volume. Six VOCs, valine, glutaraldehyde, d-leucine, 3,3-dimethyl-4-methylamino-butan-2-one, pentadecylamine, cyclopropane and 1-butyl-2-(2-methylpropyl)-, were detected from the extract of Streptomyces sp. DST25, and glutaraldehyde was the most abundant followed by an amino acid, valine (Additional file 1: Fig. S3). Only one compound (di-n-octyl phthalate) was detected in extracts of Streptomyces cellulosae DST28 (Additional file 1: Fig. S4). Seven compounds were determined from the extract of Streptomyces flavogriseus DST52, of which carbonic acid, 2, 2, 2-trichloroethyl undec-10-enyl ester alone constituted 49.78% (Additional file 1: Fig. S5). Only 2-methoxy-4,5-diphenyl-6-(2′-phenylethyl)pyrimidine was detected in the extract of Streptomyces sp. DST116 (Additional file 1: Fig. S6), while six compounds were detected in the extract of Streptomyces sp. DST119 in which 2-benzylthio-8-methyl-7-phenylpyrano [2,3-f]benzoxazol-6(h)-one constituted the maximum amount (42.66%) (Additional file 1: Fig. S7).
Detection and quantification of antibiotics using the UPLC-MRM method
The UPLC–ESI–MS/MS analysis for detection of certain standard antibiotics of the methanolic crude extracts of the selected isolates showed that rifamycin was present in the highest amount in all samples followed by ketoconazole, trimethoprim and fluconazole. Trimethoprim was found to be present in higher amounts (39 μg/g) in extracts of Streptomyces flavogriseus DST52 compared to the other samples. Extracts of Streptomyces cellulosae DST28 contained more fluconazole (17 μg/g), ketoconazole (50 μg/g) and rifamycin in (74 μg/g) compared to other samples (Table 3 and Figs. 4, 5). MS/MS Spectra of standard reference analytes i.e. trimethoprim, fluconazole, ketoconazole and rifampicin showed as Fig. 5 was used from our earlier publication [21].
MS/MS Spectra of reference analytes; a trimethoprim, b fluconazole, c ketoconazole, d rifampicin (as per [21])
Discussion
The bio-resources in freshwater ecosystems are largely unexplored, especially in the field of microbiology. Freshwater ecosystems are becoming a promising area for the isolation of bioactive compounds of pharmaceutical and biotechnological importance [21]. In the present investigation, 68 actinobacterial strains were isolated from three freshwater systems, and maximum strains were obtained from the lake sediment compared to the sediments from the two rivers. This could be because sediments containing actinobacteria in rivers are continuously removed by running water and get deposited in different areas throughout the river. At the same time, lake sediments are concentrated in particular areas that are not drastically affected by running water. Different nutritional media were employed to achieve maximum diversities of actinobacteria, since nutrient uptake differed between organisms. The results indicated that SCA was the best medium for the isolation of the maximum number of actinobacteria strains, which was in accordance with earlier studies [42,43,44,45].
Streptomyces represents the largest genus under the bacteria domain [46] and the actinobacteria phylum [21]. The present investigation also showed that Streptomyces was the most dominant genus in freshwater sediments, which is in accordance with the findings of Wohl and McArthur. [47]; Deshmukh and Sridhar. [42]; Ningthoujam et al. [48]; Sanasam et al. [49]; Jami et al. [50] and Zothanpuia et al. [45]. There are also several genera of actinobacteria other than Streptomyces that are called rare genera whose isolation frequencies were lower compared to Streptomyces [51]. Only 12% of the actinobacterial isolates that were recovered were rare genera, which included Kocuria, Nocardiopsis, Amycolatopsis, Saccharopolyspora, Rhodococcus, Prauserella, Promicromonospora and Micrococcus, and these genera have been previously reported from freshwater habitats [29, 49, 50, 52]. To the best of our knowledge, Amycolatopsis, Prauserella and Promicromonospora have not yet been reported from freshwater sediments and were isolated for the first time in the present study. However, halophilic actinobacteria, Amycolatopsis halophila [53], Prauserella salsuginis, Prauserella flava, Prauserella aidingensis, and Prauserella sediminis [54], were reported to be isolated from a saline lake in Xinjiang Province, northwest China, while Promicromonospora thailandica has also been reported from marine sediment [55].
Actinobacteria are a potential candidate to fight against multidrug-resistant organisms and are well-known producers of antimicrobial compounds, and actinobacteria have been found in different habitats worldwide [4, 20, 21, 40]. The present study reports the antimicrobial activity of sixty-eight actinobacterial isolates that showed activity against at least three of the tested pathogens. Some rare genera of actinobacteria, such as Kocuria, Nocardiopsis, Amycolatopsis, Saccharopolyspora, Rhodococcus, Prauserella, Promicromonospora and Micrococcus, were also evaluated for their antimicrobial activities in the present study. Among them, Saccharopolyspora sp. DST31, Nocardiopsis DST32, Rhodococcus sp. DST38 and Nocardiopsis DST95 showed activity against five of the six tested pathogens. Sibanda et al. [29] isolated actinobacteria belonging to Saccharopolyspora and Actinosynnema from the Tyume River, South Africa, and found antibacterial activity against the tested pathogens, which supports the present investigation. Several other rare genera of actinobacteria from freshwater habitats, except for Amycolatopsis, Prauserella and Promicromonospora, have been previously reported for their antimicrobial activity, as discussed earlier.
Six potential Streptomyces strains that showed a broad spectrum of antimicrobial activities against all tested pathogens were further selected, and the methanolic extracts of the strains showed better activity using the agar well diffusion method compared to the filter paper disk diffusion assay, which was supported by the findings of Gebreyohannes et al. [40]. Recently, we recorded the potential microbial activity of Streptomyces cyaneofuscatus from freshwater sediments of Tamdil Lake [21]. Broad-spectrum antimicrobial activity was also measured in Streptomyces sp. AZ-NIOFD1 from the Nile River [56]. Various potential strains of Streptomyces have also been identified in freshwater habitats [21, 57,58,59]. The methanolic crude extract of Streptomyces flavogriseus DST52 showed antimicrobial activity with an MIC value of 0.003 mg/ml, which was lower than the actinobacterial strain SMS_SU21 from a mangrove ecosystem that showed antimicrobial activity with an MIC value of 0.05 mg/ml [60]. The MIC of Streptomyces sp. DST119 extract was 0.015 mg/ml against S. aureus, while that of Streptomyces flavogriseus DST52 was 0.056 mg/ml, which was far lower than those of the actinobacterial crude extract (1.65 and 1.84 mg/ml) against S. aureus and E. coli, respectively [40].
PKS-II, phzE and NRPS have been extensively described as responsible for the synthesis of a broad range of structurally diverse secondary metabolites in actinobacteria [7, 21, 61]. PKS and NRPS are responsible for the synthesis of bioactive polyketides and peptides, while phenazine is an antibiotic that has been reported to be derived from phzE, and all three genes are all renowned for playing vital roles in biological control [7, 21, 62]. The present study also correlated antimicrobial compounds with reference to their biosynthetic genes in some of the selected strains. Among the selected strains that showed antimicrobial activity against all tested pathogens, the biosynthetic genes PKS-II, phzE and NRPS were all detected and amplified with the expected size in Streptomyces sp. DST116 and DST119. However, none of the genes were detected in Streptomyces cellulosae DST28, which clearly showed that the strains that show antimicrobial activity do not necessarily contain PKS-II, phzE or NRPS genes, and these findings are in agreement with previous studies [63, 64]. The biosynthetic genes for six selected Streptomyces strains were sequenced and deposited in NCBI database and Genbank accession number were given as MG200184–MG200188 for NRPS; MG200189–MG200192 for PKSII; MG200193–MG200194 for phzE.
In the present study, four antibiotics were detected and quantified using the UPLC–ESI–MS/MS method. This method has been successfully employed to quantify bioactive compounds such as antibiotics and phenolic compounds [15, 21]. Antibiotics such as rifamycin and trimethoprim were detected and quantified from the crude methanol extract of six Streptomyces strains, which was supported by the findings of Passari et al. [19]. Fluconazole and ketoconazole were also quantified in all selected extracts of Streptomyces strains, which were also recently reported from Streptomyces cyaneofuscatus isolated from Tamdil Lake, Northeast India [21].
Secondary metabolite profiling based on GC–MS is becoming a foundation in the field of biological sciences and has been successfully employed to determine VOCs from various samples [20, 21]. The actinobacteria phylum has been reported as prolific producers of thousands of bioactive secondary metabolites. The present investigations measured 35 VOCs from six methanolic extracts of Streptomyces strains, of which maximum compounds were retrieved from Streptomyces albidoflavus DST71. Among the identified compounds from extracts of Streptomyces albidoflavus DST71, all except oxirane, 2-butyl-3-methyl-, cis, azacyclodecan-5-ol, N-(4-chlorobenzenesulfonyl)azetidin-3-one and 1,3,5-triazaadamantane detected compounds that have the antimicrobial activity, as reported by earlier researchers [65,66,67,68,69,70,71,72,73]. In the present study, the amount of hexanal in the methanol extract of Streptomyces albidoflavus DST71 was found to be maximum (23.2%), and this compound was reported to be one of the constituents of the crude extract of the roots of Leonurus sibiricus for its antibacterial, anti-inflammatory, antioxidant, and antiproliferative properties [70]. Antimicrobial activities of 2-thiophenecarboxylic acid, 5-(1, 1-dimethylethoxy)- and heptanal have also been observed in the extracts of Phormidium autumnale and Chlorella vulgaris, respectively [69]. The antimicrobial activity of glutaraldehyde was also discussed earlier [66, 74] and was also measured in the extract of Streptomyces sp. DST25. All compounds extracted from the crude extract of Streptomyces sp. DST25 except cyclopropane, 1-butyl-2-(2-methylpropyl)-were previously reported in antimicrobial studies [71, 75, 76]. The amino acid valine was also determined as a major compound next to glutaraldehyde in the present study, and this compound increases the production of the glycopeptide antibiotic, as reported by Beltrametti et al. [77] in the actinobacteria strain Nonomuraea sp. Only pyrrolo [1, 2-a] pyrazine-1, 4-dione, hexahydro-3-(2-methylpropyl) out of the six compounds detected from the extract of Streptomyces sp. DST119 has been previously reported for its antimicrobial activity [78, 79]. Two of the seven compounds, carbonic acid, 2,2,2-trichloroethyl undec-10-enyl ester and 1-butanol, 2-methyl-acetate from the extract of Streptomyces flavogriseus DST52 were reported earlier for their antimicrobial activities [80,81,82]. Only one compound was determined in the extracts of Streptomyces cellulosae DST28 and Streptomyces sp. DST116 with a single peak. Di-n-octyl phthalate obtained from Streptomyces cellulosae DST28 was reported earlier by various researchers for its antimicrobial activity [83, 84], while no activity was reported for 2-methoxy-4,5-diphenyl-6-(2′-phenylethyl)-pyrimidine obtained from the extract of Streptomyces sp. DST116 for its antimicrobial activity. Thus, from this study, we conclude that further investigation of the purification of these potent compounds will certainly explicate their efficacy in the pharmaceutical industry. Hence, the usage of freshwater bio-resources can be an ideal source for the isolation of actinobacterial cultures with rare and unique properties that could certainly add to the ever-growing pharmaceutical needs and other biotechnological applications.
Change history
05 June 2018
Upon publication of this article [1], it was brought to our attention that Figs. 3, 4 and 5 are incorrectly presented in the original version of the article. The figures were inadvertently swapped in the original submission and published. Figure 3 should be treated as Fig. 5; Fig. 4 should be 3 and Fig. 5 should be Fig. 4.
References
Ventura M, Canchaya C, Tauch A, Chandra G, Fitzgerald GF, Chater KF, van Sinderen D. Genomics of actinobacteria: tracing the evolutionary history of an ancient phylum. Microbiol Mol Biol Rev. 2007;71:495–548.
Goodfellow M, Fiedler HP. A guide to successful bioprospecting: informed by actinobacterial systematics. Anton Van Leeuwen. 2010;98:119–42.
Bae KS, Kim MS, Lee JH, Kang JW, Kim DI, Lee JH, et al. Korean indigenous bacterial species with valid names belonging to the phylum actinobacteria. J Microbiol. 2016;54:789–95.
Claverías FP, Undabarrena A, González M, Seeger M, Cámara B. Culturable diversity and antimicrobial activity of actinobacteria from marine sediments in Valparaíso bay, Chile. Front Microbiol. 2015;6:737.
Chambers HF, DeLeo FR. Waves of resistance: Staphylococcus aureus in the antibiotic era. Nat Rev Microbiol. 2009;7:629–41.
Bull AT, Stach JEM. Marine actinobacteria: new opportunities for natural product search and discovery. Trends Microbiol. 2007;15:491–9.
Yuan M, Yu Y, Li HR, Dong N, Zhang XH. Phylogenetic diversity and biological activity of actinobacteria isolated from the Chukchi. Self marine sediments in the Arctic Ocean. Mar Drugs. 2014;12:1281.
Chaudhary HS, Soni B, Shrivastava AR, Shrivastava S. Diversity and versatility of actinomycetes and its role in antibiotic production. J App Pharm Sci. 2013;3(8 Suppl 1):S83–94.
Berdy J. Bioactive microbial metabolites. J Antibiot. 2005;58:1–26.
Trujillo ME. Actinobacteria. Chichester: Wiley; 2008. https://doi.org/10.1002/9780470015902.a0020366.
Le Roes-Hill M, Meyers PR. Streptomyces polyantibioticus sp. nov., isolated from the banks of a river. Int J Syst Evol Microbiol. 2009;59:1302–9.
Ray L, Mishra SR, Panda AN, Rastogi G, Pattanaik AK, Adhya TK, et al. Streptomyces barkulensis sp. nov., isolated from an estuarine lake. Int J Syst Evol Microbiol. 2014;64:1365–72.
Biswas K, Choudhury JC, Mahansaria R, Saha M, Mukherjee J. Streptomyces euryhalinus sp. nov., a new actinomycete isolated from a mangrove forest. J Antibiot. 2017;70(6):747–53.
Goodfellow M, Williams ST. Ecology of actinomycetes. Annu Rev Microbiol. 1983;37:189–216.
Passari AK, Mishra VK, Singh G, Singh P, Kumar B, Gupta VK, et al. Insights into the functionality of endophytic actinobacteria with a focus on their biosynthetic potential and secondary metabolites production. Sci Rep. 2017;7:11809.
Lam KS. Discovery of novel metabolites from marine actinomycetes. Curr Opin Microbiol. 2006;9:245–51.
Kavitha A, Prabhakar P, Narasimhulu M, Vijayalakshmi M, Venkateswarlu Y, Rao KV, et al. Isolation, characterization and biological evaluation of bioactive metabolites from Nocardia levis MK-VL-113. Microbiol Res. 2010;165:199–210.
Wang C, Wang Z, Qiao XLZ, Li F, Chen M, et al. Antifungal activity of volatile organic compounds from Streptomyces alboflavus TD-1. FEMS Microbiol Lett. 2013;341:45–51.
Passari AK, Chandra P, Zothanpuia Mishra VK, Leo VV, Gupta VK, et al. Detection of biosynthetic gene and phytohormone production by endophytic actinobacteria associated with Solanum lycopersicum and their plant growth-promoting effect. Res Microbiol. 2016;167:692–705.
Sharma P, Kalita MC, Thakur D. Broad spectrum antimicrobial activity of forest derived soil actinomycete, Nocardia sp. PB-52. Front Microbiol. 2016;7:347.
Zothanpuia, Passari AK, Chandra P, Leo VV, Mishra VK, Kumar B, et al. Production of potent antimicrobial compounds from Streptomyces cyaneofuscatus associated with fresh water sediment. Front Microbiol. 2017;8:68.
Wang Q, Zhang Y, Wang M, Tan Y, Hu X, He H, et al. Neo-actinomycins A and B, natural actinomycins bearing the 5H-oxazolo[4,5-b]phenoxazine chromophore, from the marinederived Streptomyces sp. IMB094. Sci Rep. 2017;7:3591.
Alvan G, Edlund C, Heddini A. The global need for effective antibiotics—summary of plenary presentations. Drug Resis Updat. 2011;14:70–6.
Zotchev SB. Marine actinomycetes as an emerging resource for the drug development pipelines. J Biotechnol. 2012;158:168–75.
Lazzarini A, Cavaletti L, Toppo G, Marinelli F. Rare genera of actinomycetes as potential producers of new antibiotics. Anton Van Leeuwen. 2000;79:399–405.
Poulsen M, Oh DC, Clardy J, Currie CR. Chemical analyses of wasp-associated Streptomyces bacteria reveal a prolific potential for natural products discovery. PLoS ONE. 2011;6:e16763.
Myers N, Mittermeier RA, Mittermeier CG, da Fonseca GAB, Kent J. Biodiversity hotspots for conservation priorities. Nature. 2000;403:853–8.
Zothanpuia, Passari AK, Gupta VK, Singh BP. Detection of antibiotic-resistant bacteria endowed with antimicrobial activity from a freshwater lake and their phylogenetic affiliation. Peer J. 2016;4:e2103.
Sibanda T, Mabinya LV, Mazomba N, Akinpelu DA, Bernard K, Olaniran AO. Antibiotic producing potentials of three freshwater actinomycetes isolated from the eastern Cape province of South Africa. Int J Mol Sci. 2010;11:2612–23.
Radhika S, Bharathi S, Radhakrishnan M, Balagurunathan V. Bioprospecting of fresh water actinobacteria: isolation, antagonistic potential and characterization of selected isolates. J Pharm Res. 2011;4:2584–6.
Saravanan S, Sivakami R, Prem GK. Actinomycetes diversity in five fresh water systems of Pudukkottai, Tamilnadu and their antimicrobial activity. Int J Curr Microbiol App Sci. 2015;4:672.
Shirling EB, Gottlieb D. Methods for characterization of Streptomyces species. Int J Syst Bacteriol. 1966;16:313–40.
Goodfellow M, Haynes JA. Actinomycetes in marine sediments. In: Ortiz-Ortiz L, Bojalil LF, Yakoleff V, editors. Biological, biochemical, and biomedical aspects of actinomycetes. New York: Academic Press; 1984. p. 453–72.
Weisburg WG, Barns SM, Pelletier DA, Lane DJ. 16S ribosomal DNA amplification for phylogenetic study. J Bacteriol. 1991;173:697–703.
Kim TU, Cho SH, Han JH, Shin YM, Lee HB, Kim SB. Diversity and physiological properties of root endophytic actinobacteria in native herbaceous plants of Korea. J Microbiol. 2012;50:50–7.
Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. The Clustal X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997;24:4876–82.
Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–25.
Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 2011;28:2731–9.
Saadoun I, Muhana A. Optimal production conditions, extraction, partial purification and characterization of inhibitory compound(s) produced by Streptomyces Ds-104 isolate against multi-drug resistant Candida albicans. Curr Trends Biotechnol Pharm. 2008;2:402–20.
Gebreyohannes G, Moges F, Sahile S, Raja N. Isolation and characterization of potential antibiotic producing actinomycetes from water and sediments of Lake Tana4 Ethiopia. Asian Pac J Trop Biomed. 2013;3:426–35.
Eloff JN. Which extractant should be used for the screening and isolation of antimicrobial components from plants. J Ethnopharmacol. 1998;60:1–8.
Deshmukh MB, Sridhar KR. Distribution and antimicrobial activity of actinomycetes of a freshwater coastal stream. Asian Jr Microbiol Biotech Env Sc. 2002;4:335–40.
Rifaat HM. The biodiversity of actinomycetes in the river Nile exhibiting antifungal activity. J Mediterr Ecol. 2003;4:5–7.
Rizvi R, Kamble L, Kadam A. Searching the submerged: a report on prevalence of actinomycetes in sediments of river Godavari and optimized strategy for their isolation. Trends Biotechnol Res. 2012;1:2.
Zothanpuia P, Passari A, Singh BP. Molecular characterization of actinomycetes isolated from Tuichang river and their biosynthetic potential. Sci Vis. 2015;15:136.
Subramani R, Aalbersberg W. Marine actinomycetes: an ongoing source of novel bioactive metabolites. Microbiol Res. 2012;167:571–80.
Wohl DL, McArthur JV. Actinomycete–Lora associated with submersed freshwater macrophytes. FEMS Microbiol Ecol. 1998;26:135–40.
Ningthoujam DS, Sanasam S, Nimaichand S. Studies on bioactive actinomycetes in a Niche Biotope, Nambul River in Manipur, India. J Microbial Biochem Technol. 2011;S6:001. https://doi.org/10.4172/1948-5948.S6-001.
Sanasam S, Nimaichand S, Ningthoujam D. Novel bioactive actinomycetes from a niche biotope, Loktak Lake, in Manipur, India. J Pharm Res. 2011;4:1707–10.
Jami M, Ghanbaria M, Kneifela W, Domig KJ. Phylogenetic diversity and biological activity of culturable actinobacteria isolated from freshwater fish gut microbiota. Microbiol Res. 2015;175:6–15.
Tiwari K, Gupta RK. Rare actinomycetes: a potential storehouse for novel antibiotics. Crit Rev Biotechnol. 2012;32:108–32.
Jiang CL, Xu LH. Diversity of aquatic actinomycetes in lakes middle plateau, Yunnan, China. Appl Environ Micobiol. 1996;62:249–53.
Tang SK, Wang Y, Guan TW, Lee JC, Kim CJ, Li WJ. Amycolatopsis halophila sp. nov., a halophilic actinomycete isolated from a salt lake. Int J Syst Evol Microbiol. 2010;60:1073–8.
Li Y, Tang SK, Chen YG, Wu JY, Zhi XY, Zhang YQ, et al. Prauserella salsuginis sp. nov., Prauserella flava sp. nov., Prauserella aidingensis sp. nov. and Prauserella sediminis sp. nov., isolated from a salt lake. Int J Syst Evol Microbiol. 2009;59:2923–8.
Thawai C, Kudo T. Promicromonospora thailandica sp. nov., isolated from marine sediment. Int J Syst Evol Microbiol. 2012;62:2140–4.
Atta HM, Dabour SM, Desoukey SG. Sparsomycin antibiotic production by Streptomyces sp.-NIOFD1: taxonomy, fermentation, purification and biological activities. Am-Eur J Agric Environ Sci. 2009;5:368–77.
Nwodo UU, Agunbiade MO, Green E, Mabinya LV, Okoh AI. A freshwater Streptomyces, isolated from Tyume river, produces a predominantly extracellular glycoprotein bioflocculant. Int J Mol Sci. 2012;13:8679–95.
Singh LS, Sharma H, Talukdar NC. Production of potent antimicrobial agent by actinomycete, Streptomyces sannanensis strain SU118 isolated from phoomdi in Loktak Lake of Manipur, India. BMC Microbiol. 2014;14:278.
Zhao J, Guo L, Liu C, Bai L, Han C, Li J, et al. Streptomyces tyrosinilyticus sp. nov., a novel actinomycete isolated from river sediment. Int J Syst Evol Microbiol. 2015;65:3091–6.
Sengupta S, Pramanik A, Ghosh A, Bhattacharyya M. Antimicrobial activities of actinomycetes isolated from unexplored regions of Sundarbans mangrove ecosystem. BMC Microbiol. 2015;15:170.
Schwarzer D, Finking R, Marahiel MA. Nonribosomal peptides: from genes to products. Nat Prod Rep. 2003;20:275–87.
Ayuso-Sacido A, Genilloud O. New PCR primers for the screening of NRPS and PKS-I systems in actinomycetes: detection and distribution of these biosynthetic gene sequences in major taxonomic groups. Microb Ecol. 2005;49:10–24.
Qin S, Li J, Chen HH, Zhao GZ, Zhu WY. Isolation, diversity and antimicrobial activity of rare actinobacteria from medicinal plants of tropical rain forests in Xishuangbanna, China. Appl Environ Microbiol. 2009;75:6176–86.
Passari AK, Mishra VK, Saikia R, Gupta VK, Singh BP. Isolation, abundance and phylogenetic affiliation of endophytic actinomycetes associated with medicinal plants and screening for their in vitro antimicrobial biosynthetic potential. Front Microbiol. 2015;6:273.
Kabara JJ, Conley AJ, Truant JP. Relationship of chemical structure and antimicrobial activity of alkyl amides and amines. Antimicrob Agents Chemother. 1972;2:492–8.
Lerones C, Mariscal A, Carnero M, Garcia-Rodriguez A, Fernandez-Crehuet J. Assessing the residual antibacterial activity of clinical materials disinfected with glutaraldehyde, o-phthalaldehyde, hydrogen peroxide or 2-bromo-2-nitro-1,3-propanediol by means of a bacterial toxicity assay. Clin Microbiol Infect. 2004;10:984–9.
Dehpour AA, Babakhani B, Khazaei S, Asadi M. Chemical composition of essential oil and antibacterial activity of extracts from flower of Allium atroviolaceum. J Med Plants Res. 2011;5(16):3667–72.
Ramakrishnan S, Venkataraman R. Screening of antioxidant activity, total phenolics and gas chromatography-mass spectrophotometer (GC-MS) study of ethanolic extract of Aporosa lindleyana Baill. Af J Biochem Res. 2011;5(14):360–4.
Al-Wathnani H, Ara I, Tahmaz RR, Al-Dayel TH, Bakir MA. Bioactivity of natural compounds isolated from cyanobacteria and green algae against human pathogenic bacteria and yeast. J Med Plants Res. 2012;6(18):3425–33.
Sitarek P, Rijo P, Garcia C, Skała E, Kalemba D, Białas AJ, et al. Antibacterial, antiinflammatory, antioxidant, and antiproliferative properties of essential oils from hairy and normal roots of Leonurus sibiricus L. and their chemical composition. Oxid Med Cell Longev. 2017;2017:7384061.
Dineshkumar M, Kannappan S, Sivakumar K. Effect of mangrove plant (Sesuvium portulacastrum) extract against Vibrio harveyi during shrimp larviculture. J Environ Biol. 2017;38:47–53.
Sepahi M, Jalal R, Mashreghi M. Antibacterial activity of poly-l-arginine under different conditions. Iran J Microbiol. 2017;9:103–11.
Foo LW, Salleh E, Hana SN. Green extraction of antimicrobial bioactive compound from piper betle leaves: probe type ultrasound-assisted extraction vs supercritical carbon dioxide extraction. Chem Eng Trans. 2017;56:109–14.
Hill SD, Berry CW, Seale NS, Kaga M. Comparison of antimicrobial and cytotoxic effects of glutaraldehyde and formocresol. Oral Surg Oral Med Oral Pathol. 1991;71:89–95.
Lee E, Shin A, Jeong KW, Jin B, Jnawali HN, Shin S, et al. Role of phenylalanine and valine10 residues in the antimicrobial activity and cytotoxicity of piscidin-1. PLoS ONE. 2014;9(12):e114453.
Ahmad A, Azmi S, Srivastava S, Kumar A, Tripathi JK, Mishra NN, et al. Design and characterization of short antimicrobial peptides using leucine zipper templates with selectivity towards microorganisms. Amino Acids. 2014. https://doi.org/10.1007/s00726-014-1802-3.
Beltrametti F, Jovetic S, Feroggio M, Gastaldo L, Selva E, Marinelli F. Valine influences production and complex composition of glycopeptide antibiotic A40926 in fermentations of Nonomuraea sp. ATCC 39727. J Antibiot (Tokyo). 2004;57:37–44.
Sheoran N, Nadakkakath AV, Munjal V, Kunduc A, Subaharan K, Venugopal V, Rajamma S, et al. Genetic analysis of plant endophytic Pseudomonas putida BP25 and chemo-profiling of its antimicrobial volatile organic compounds Sheoran. Microbiol Res. 2015;173:66–78.
Jinfeng EC, Rafi MIM, Hoon KC, Lian HK, Kqueen CY. Analysis of chemical constituents, antimicrobial and anticancer activities of dichloromethane extracts of Sordariomycetes sp. endophytic fungi isolated from Strobilanthes crispus. World J Microbiol Biotechnol. 2017;33(1):5.
Ezra D, Strobel GA. Effect of substrate on the bioactivity of volatile antimicrobials produced by Muscodor albus. Plant Sci. 2003;165:1229–38.
Ezra D, Hess WH, Strobel GA. New endophytic isolates of M. albus, a volatile antibiotic-producing fungus. Microbiology. 2004;150:4023–31.
Musini A, Rao MJP, Giri A. Phytochemical investigations and antibacterial activity of Salacia oblonga Wall ethanolic extract. Ann Phytomed. 2013;2(1):102–7.
Philip D, Kaleena PK, Valivittan K. GC–MS analysis and antibacterial activity of chromatographically separated pure fractions of leaves of Sansevieria roxburghiana. Asian J Pharm Clin Res. 2011;4:30.
Shafaghat A, Farshid S, Vahidmani-Hooshyar A. A phytochemical and antimicrobial activity of Lavandula officinalis leaves and stems against some pathogenic micro organisms. J Med Plants Res. 2012;6:455–60.
Authors’ contributions
Conceptualization: Z; BPS. Data analysis: Z; AKP; VVL. UPLC-MS/MS analysis: BK. GC–MS analysis: CN. Resources: BPS. Methodology: Z; AKP; VVL; BPS. Software: Z; AKP; VVL. Supervision: BPS. Validation: BPS; CN. Visualization: Z; AKP; VVL. Writing ± original draft: Z; BPS. Writing ± review and editing: AKP; VVL; AH; EFAA; AAA; CN. All authors read and approved the final manuscript.
Acknowledgements
B.P.S. is thankful to the Department of Biotechnology, Government of India, New Delhi for financial support under DBT’s Unit of Excellence programme for NE (102/IFD/SAN/4290-4291/2016-2017). The authors are also thankful to the Department of Biotechnology for the establishment of the DBT-BIF Center and the DBT State Biotech Hub in the Department, which were used for the present study. The authors are thankful to SAIF, NEHU for SEM and VIT-SIF Lab, SAS, Chemistry Division for NMR and GC–MS Analysis for GC–MS analysis of the samples. The authors would like to extend their sincere appreciation to the Deanship of Scientific Research at King Saud University for its funding to the Research Group Number (RGP -271).
Competing interests
The authors declare that they have no competing interests.
Additional information
All authors give consent to publish the research in microbial cell factories.
Availability of data and materials
All data generated or analysed during this study are included in this published article (and its additional files).
Ethics approval and consent to participate
Not applicable.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Author information
Authors and Affiliations
Corresponding author
Additional file
Additional file 1.
Additional tables and figures.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Zothanpuia, Passari, A.K., Leo, V.V. et al. Bioprospection of actinobacteria derived from freshwater sediments for their potential to produce antimicrobial compounds. Microb Cell Fact 17, 68 (2018). https://doi.org/10.1186/s12934-018-0912-0
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12934-018-0912-0