Abstract
Metabolic syndrome includes glucose intolerance and dyslipidemia, both of which are strong risk factors for developing diabetes and atherosclerotic cardiovascular diseases. Recently, multiple groups independently studied a previously uncharacterized gene, officially named C19orf80 (human) and Gm6484 (mouse), but more commonly known as RIFL, Angptl8, betatrophin and lipasin. Both exciting and conflicting results have been obtained, and significant controversy is ongoing. Accumulating evidence from genome wide association studies and mouse genetic studies convincingly shows that lipasin is involved in lipid regulation. However, the mechanism of action, the identity of transcription factors mediating its nutritional regulation, circulating levels, and relationship among lipasin, Angptl3 and Angptl4, remain elusive. Betatrophin represents a promising drug target for replenishing β-cell mass, but current results have not been conclusive regarding its potency and specificity. Here, we summarize the consensus and controversy regarding functions of lipasin/betatrophin based on currently available evidence.
Similar content being viewed by others
Introduction
Metabolic syndrome represents common metabolic disorders that include glucose intolerance and dyslipidemia, and the prevalence of metabolic syndrome has increased dramatically in the past two decades [1]. Both glucose intolerance and dyslipidemia are strong risk factors for developing diabetes and atherosclerotic cardiovascular diseases [2],[3]. Clinically, in addition to hyperglycemia and hyperinsulinemia, type 2 diabetic patients commonly suffer from disturbances in production and clearance of plasma lipoproteins, known as diabetic dyslipidemia, characterized by increased triacylglycerides (TAG), reduced high-density lipoprotein cholesterol (HDL-C) and postprandial lipemia [4]-[6].
In the postprandial state, plasma TAGs are mainly associated with chylomicrons (CMs) that are predominately synthesized in the intestine, while in the fasted state, TAGs are mainly associated with very low-density lipoprotein (VLDL) that is synthesized in the liver. Lipoprotein lipase (LPL) is critical in determining plasma TAG levels and the partitioning of fatty acids that are taken up by peripheral tissues [7],[8]. In both mice and humans, loss-of-function mutations in LPL result in severe hypertriglyceridemia [9]-[11]. Similarly, loss-of-function mutations in GPIHBP1, which is required for proper targeting of LPL to the lumen of capillaries [12],[13], also cause hypertriglyceridemia. Therefore, LPL expression and activity are tightly controlled to meet the needs of various tissues under different physiological and pathological conditions [8].
Recently, multiple groups independently studied a previously uncharacterized gene, officially named C19orf80 (human) and Gm6484 (mouse) according to the HUGO Gene Nomenclature Committee [14]. Both exciting and conflicting results have been obtained, and significant controversy is ongoing. In this review we aim to summarize the consensus and controversy using the currently available evidence regarding this gene.
The first controversy is that many different names have been proposed for this protein, including RIFL [15], lipasin [16]-[18], Angptl8 [19],[20], betatrophin [21]-[24] and C19orf80 [25]. Here when mentioning an experiment or a finding, the name that the original authors used is adopted, so that all above names are used interchangeably. We define consensus as a result that has no published conflicting evidence or has been confirmed by independent studies.
Consensus on the role of lipasin in lipid metabolism
Evidence from human genome-wide association studies
Multiple studies have identified ANGPTL8 sequence variations that are associated with lipid profiles in human genome-wide association studies (GWAS) [19],[26]-[29]. The SNP rs2278426 represents a nucleotide transition (C vs. T, from CGG to TGG) that results in a non-synonymous amino acid change, from arginine (R) to tryptophan (W) at residue 59 (Table 1). The minor allele is T (W), and the minor allele frequency (MAF) is 15% in the 1000 Genome Project populations, which are mainly composed of whites [26]. The MAF is 26% in Hispanics, 18% in African Americans and 5% in European Americans [19].
Quagliarini et al. found that the 59W variant is associated with lower LDL-C in African Americans and Hispanics. In the Dallas Heart Study (DHS), African Americans and Hispanics with 59W homozygotes exhibited lower HDL-C than those with 59R homozygotes [19]. Consistently, in a study composed of 4361 Mexicans, Weissglas-Volkov et al. found that WW homozygotes had 14% lower HDL-C than RR homozygotes. African Americans in the DHS had 15% lower LDL-C in WW homozygotes than in RR homozygotes [29]. Of note, no association was found between R59W variant and triglyceride levels in both studies.
The SNP rs145464906 represents a nucleotide transition (C vs. T, from CAG to TAG) that results in a premature stop codon at residue 121, and therefore a truncated ANGPTL8 is generated by this SNP. The MAF of this SNP is extremely low, about 0.1% in European ancestry and 0.01% in African ancestry, and the carriers with European ancestry were 10 mg/dl higher in HDL-C and 15% lower in triglyceride levels [27].
The SNP rs737337 has also been found to be associated with HDL-C levels [28], and the SNP, although located in a region upstream of the ANGPTL8 transcription start site, represents a synonymous variant in the DOCK6 (dedicator of cytokinesis protein 6) gene. Therefore this SNP, although associated with lipid levels, may or may not be related to ANGPTL8 functions.
Taken together, multiple lines of evidence from GWAS suggest ANGPTL8 plays a role in lipid metabolism.
Evidence from mouse genetic studies
Mice that were deficient in Gm6484 were among the mouse knockout library created through collaboration between Genentech Inc. and Lexicon Pharmaceuticals Inc. This collaboration focused on genes that encoded secreted and trans-membrane proteins. A broad, unbiased phenotypic screen was performed on mouse lines in the library, and Gm6484-null mice clearly had lower triglyceride levels [30]. Wang et al. found that, based on an independent mouse line, Angptl8 knockout mice exhibited lower TAG levels in the fed state, but not in the fasted state. Furthermore, they showed that the knockout mice had a reduction in VLDL (very low density lipoprotein) secretion, an increase in LPL activity, and a markedly reduced uptake of fatty acids by white adipose tissue [20].
Overexpression of lipasin in mouse liver using adenovirus dramatically increased serum TAG levels [17]. Consistently, Quagliarini et al. found that adenovirus-mediated Angptl8 overexpression increased serum TAG levels, but in an Angptl3 dependent manner. Indeed, Angptl8 was co-immunoprecipitated with Angptl3 in mouse plasma and cultured hepatocytes, and promoted Angptl3 cleavage [19].
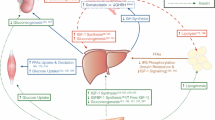
Therefore, both loss- and gain-of-function studies in mice clearly suggest that lipasin is involved in triglyceride metabolism (Figure 1).
Roles of lipasin (Angptl8) in regulating triglyceride metabolism and pancreatic beta-cell proliferation. Lipasin is secreted from the liver into the circulation, and is involved in triglyceride metabolism and in promoting pancreatic β -cell proliferation. Active as a dimmer, LPL binds to both HSPG and GPIHPB1 on the surface of capillary microvascular endothelial cells. LPL hydrolyzes TAG in chylomicrons and VLDL, yielding FFAs, which are then taken up by peripheral tissues, including fat, muscle and heart. Both Angptl3 and Angptl4 need to be cleaved to release functional N-termini to inhibit LPL, disrupting dimer formation either reversibly or irreversibly, respectively. Lipasin likely inhibits LPL directly or indirectly by promoting Angptl3 cleavage. Food intake dramatically induces the expression of lipasin, whereas fasting induces Angptl4. Dotted lines denote homologous regions. Angptl3, angiopoietin-like 3; Angptl4, angiopoietin-like 4; EC, endothelial cell; GPIHBP1, glycosylphosphatidylinositol anchored high density lipoprotein binding protein 1; FFA, free fatty acid; HSPG, heparan sulfate proteoglycans; LPL, lipoprotein lipase; TAG, triglyceride; VLDL, very low-density lipoprotein.
Expression pattern and transcriptional regulation
We found that in mice, lipasin is highly enriched in the liver, brown adipose tissue (BAT), and white adipose tissue (WAT), with liver being the tissue that has the highest expression, and in humans, it is predominantly in the liver among 48 tissues tested [17]. Ren et al. showed comparable expression of RIFL in mouse WAT, BAT and liver, and it is in human liver and WAT among the 4 tissues tested [15]. Quagliarini et al. showed comparable Angptl8 expression levels in liver, BAT and WAT, and also in the adrenal gland [19]. Because lipasin is sensitive to nutritional status, it is not surprising that there are discrepancies in expression patterns, especially in humans, when the nutritional status is not strictly controlled. Nevertheless, it is probably safe to conclude that lipasin is enriched in the liver and fat.
The mRNA levels of mouse lipasin in liver and fat are suppressed by fasting and elevated in mice with a high-fat diet treatment [17]. RIFL mRNA expression in fat and liver was found to be higher in ob/ob mice, which are obese due to the lack of leptin, and are highly induced by refeeding [15]. In mouse BAT, lipasin is induced by cold exposure [16]. C19orf80 is unregulated by T3 in the mouse liver [25]. In 3T3-L1 adipocytes, insulin increases, while cAMP decreases RIFL expression [15]. We found that that serum lipasin is increased 2 hours following a defined meal in humans [18]. Therefore, lipasin is clearly a liver-enriched nutritionally-regulated circulating factor.
Lipasin is a novel but atypical member of the ANGPTL protein family
The ANGPTL protein family contains 7 typical members, which are characterized by the presence of a coiled-coil domain at the N-terminus, a fibrinogen like domain at the C-terminus and a signal peptide for protein secretion [31]. N- and C-terminal domains of ANGPTLs have distinct functions. The N-terminal coiled-coil domain and the C-terminal fibrinogen-like domain of ANGPTL3 are involved in lipid regulation [32] and angiogenesis [33], respectively. Phylogenetic analysis using lipasin and N-terminal domains of the 7 ANGPTLs showed that lipasin and ANGPTL3 were most closely related, and shared a common ancestor with ANGPTL4 [16]. Quagliarini et al. suggested that ANGPTL8, which is in an intron of DOCK6, arose through gene duplication, because of a similar gene structure of ANGPTL3 and DOCK7 [19]. Of note, different from ANGPTL8 and ANGPTL3, ANGPTL4 is not in an intron of another gene. Lipasin, therefore, is a new but atypical member of the ANGPTL family, because it lacks the fibrinogen-like domain, but shares common ancestors with ANGPTL3 and ANGPTL4.
Role in adipogenesis and autophagy
RIFL is highly induced during adipocyte differentiation in 3T3-L1 cells, primary mouse and human pre-adipocytes. Indeed, knockdown using siRNA significantly suppressed adipocyte differentiation [15]. Tseng et al. found that C19orf80 is up-regulated in mouse liver by T3, a thyroid hormone that regulates liver lipid metabolism. Immunofluorescence analysis showed that C19orf80 is located around lipid droplets or within the lysosome-associated compartment. In hepatocyte cell lines, C19orf80 overexpression activates an autophagic response; conversely, shRNA mediated knockdown suppresses T3-activated autophagy and lipolysis. Therefore, C19orf80 is likely involved in an autophagic process that is activated by T3 in the liver [25].
Controversy on the role of lipasin in lipid metabolism
Despite the consensus on the involvement of lipasin in lipid regulation, there has been inconsistent evidence regarding the mechanism in this process. The proposed mechanism is that lipasin inhibits LPL activity, and therefore in mice with lipasin overexpression, the higher triglyceride phenotype is likely due to the reduced triglyceride clearance by the inhibition of LPL activity [17]. Consistently, a recombinant lipasin inhibits LPL activity in vitro. In vivo, Angptl8 increases the cleavage of Angptl3 [19], releasing its N-terminal domain, which has been shown to inhibit LPL activity [32]. Indeed, in mice with Angptl8 deficiency, post heparin plasma LPL activity was increased [20].
However, contradictory to the above hypothesis, incorporation of free fatty acids (FFA) into adipose tissue was dramatically suppressed in the Angptl8 KO mice [20]. If lipasin inhibits LPL, we would expect increased circulating FFA levels in the Angptl8 KO mice because more triglycerides are hydrolyzed due to increased LPL activity, but the KO mice exhibit reduced FFA levels. Also, lipasin has been shown to be significantly induced by feeding and suppressed by fasting. In white adipose tissue, fasting reduces LPL activity, and lipasin expression is reduced as well. Therefore, it is not in line with an inhibitory role on LPL activity by lipasin, which is also reduced. Lipasin has been shown to be induced in brown fat by cold exposure. However, cold exposure increases LPL activity in brown fat, in which, however, lipasin expression is increased. Therefore, no satisfactory mechanism has been obtained to explain the relation between lipasin and LPL activity. Lipasin is predominately expressed in the liver [17]; LPL, however, is mostly expressed in fat and muscle [34]. The expression pattern difference seems to suggest lipasin to act in an endocrine manner. Nevertheless, lipasin is also highly induced in fat upon feeding. Therefore, whether lipasin acts in an endocrine manner, a paracrine manner, or both, is still a question.
We previously pointed out that lipasin and Angptl4 show opposite changes in expression by various stimuli, such as fasting, refeeding, obesity and cold exposure [35]. Yi et al. showed increased liver betatrophin in the insulin resistance mouse model [21]. Using the deposited microarray data from the GEO database [21], we compared the expression levels of betatrophin, Angptl4 and Angptl3. In this insulin resistance model, likewise, betatrophin was induced but Angptl4 was suppressed in the liver (Figure 2). It is possible that the 3 factors are coordinated in regulating lipid levels, however, the relationship among the 3 factors and how they are coordinated are far from clear.
Expression levels of betatrophin, Angptl4 and Angptl3 in the mouse model of insulin resistance induced by the insulin receptor antagonist S961. A) Normalized and B) absolute expression levels of betatrophin, Angptl4 and Angptl3 in the liver of the mouse model of insulin resistance induced by the insulin receptor antagonist S961. The microarray data was obtained from the Gene Expression Omnibus with the accession number GSE45694. The gene symbol in the microarray dataset was Gm6484 (also known as Angptl8). Con, control; IR, insulin resistant.
In mice, TAG levels are increased by lipasin overexpression, and decreased by its deficiency, but cholesterol levels seem unaffected. In humans, however, most GWAS results show ANGPTL8 SNPs affect HDL-C or LDL-C levels, but the effect on triglycerides was very modest, compared to that on cholesterol (Table 1). The reasons for this discrepancy are unknown. Patients with type 2 diabetes are commonly associated with diabetic dyslipidemia [5],[6], and altered LPL activity was shown to be involved in linking HDL-C metabolism and insulin resistance [36],[37]. Because circulating lipasin levels are elevated in type 2 diabetes [18],[23],[38], it is possibly that lipasin plays a role in mediating lipid abnormality that is associated with insulin resistance.
RIFL is up-regulated during adipocyte differentiation, and PPARG was involved in the up-regulation [15]. In adipocytes, insulin stimulates expression of RIFL, which is down-regulated by cAMP [15]. C19orf80 is up-regulated by the thyroid hormone T3 [25]. We show that human lipasin and glucose levels are positively correlated, and there is a consensus ChREBP binding site in human lipasin promoter [18]. However, the exact transcription factors that coordinate lipasin transcription in response to food intake still remain elusive. It is important to identify the transcription factor, its in vivo binding sites, and the role it plays in response to feeding and insulin resistance, and to explain why the changes in lipasin and Angptl4 expressions are always opposite.
Consensus on circulating factor(s) to promote pancreatic proliferation
Replenishing functional pancreatic β-cell mass represents a promising therapeutic strategy for treating both type 1 and type 2 diabetes. There is no doubt that human β-cells have the capacity to proliferate to significantly increase the mass for compensation, in response to physiological and pathological stimuli, such as pregnancy, high blood sugar, pancreatic injury and insulin resistance [39]-[43].
In mouse models, insulin resistance, induced by distinct methods, robustly promotes pancreatic β-cell proliferation. In a mouse line with insulin receptor deletion specifically in the liver (LIRKO), pancreatic β-cell mass is dramatically increased, along with phenotypes including insulin resistance, severe glucose intolerance, and a failure of insulin to suppress hepatic glucose production [43]. Yi et al. induced insulin resistance with an insulin receptor antagonist. In this model, in addition to dramatically increased insulin resistance, β-cell mass and proliferation are also dramatically increased [21].
Recently, El Ouaamari et al. showed that in the LIRKO mouse model, it is possible that liver-derived systemic factors contribute to the β-cell hyperplasia [44]. By performing parabiosis experiments, they first showed that circulating non-neuronal factors stimulate β-cell replication in the LIRKO mice. Next they showed that the serum from the LIRKO mice, when injected intra-peritoneally into the control mice, induced β-cell replication in vivo, indicating that circulating factors are responsible for the β-cell proliferation. Conditioned media from liver explant cultures or from primary hepatocytes showed β-cell stimulating effects, indicating that hepatocyte-derived circulating factors play a role in β-cell proliferation in the LIRKO mice [44]. Therefore, it is likely that one or more circulating factors can stimulate pancreatic β-cell proliferation. The question is what is (are) the circulating factor(s)?
Controversy on betatrophin in β-cell proliferation
Yi et al. induced insulin resistance in mice by infusing an insulin receptor antagonist, S961, which is a 43 amino acid inhibitory peptide with high affinity and selectivity for the insulin receptor. This model of insulin resistance invoked dramatic pancreatic β-cell proliferation. Because S961 had no effect on β-cells ex vivo, a circulating factor was hypothesized to have this effect, and betatrophin expression was found to be induced in liver. Indeed, adenovirus mediated expression of betatrophin stimulated β-cell expansion and enhanced glucose clearance [21]. These results suggest betatrophin to be a liver-derived circulating factor that triggers compensatory β-cell proliferation upon insulin resistance.
Despite the initial excitement, the work triggered ongoing controversy, because subsequent work from other laboratories did not confirm a physiologic role for betatrophin in β-cell mass expansion. The first report showed that mice lacking Angptl8 had normal glucose homeostasis [20]. The second report showed that elevated hepatic betatrophin does not increase human β-cell replication in a transplant setting [45]. The two reports provide contradictory evidence for a physiological role of betatrophin in the mouse β-cell function or a pharmacological role for betatrophin on human β-cells; the conclusions, however, are far from conclusive.
The normal glucose tolerance of the betatrophin KO mice suggests that betatrophin is not required physiologically for the maintenance of the β-cell mass. However, a compensatory increase in homologous genes, such as Angptl3 and Angptl4, may have obscured the phenotype of the KO mice. The result that the human β-cells were completely unresponsive to betatrophin argues for lack of a physiological role for betatrophin in man. These experiments, however, as correctly pointed out by Dr. Andrew Stewart [46], lacked critical positive controls. For instance, it is possible that 1) the transplanted human β-cells were not capable of proliferation; 2) the overexpressed mouse betatrophin could not activate human receptors in the transplanted cells; and 3) despite the increase in betatrophin mRNA, the supposedly functional form, circulating betatrophin, was not increased. Therefore, the results would be more conclusive if a known human and/or mouse β-cell mitogen was overexpressed to show the transplanted human β-cells were responsive, and circulating betatrophin was measured to make sure it was increased. Taken together, it is hard to draw a conclusion on the role of betatrophin in β-cell function based on currently available evidence. Irisin is an exercise-induced hormone that stimulates thermogenesis through browning of adipocytes [47]. Irisin was found to promote betatrophin expression [48], and therefore an intriguing hypothesis of the p38-PGC-1 α -irisin-betatrophin axis was proposed to connect these pathways [49].
Controversy on circulating lipasin/betatrophin levels
To understand roles of betatrophin in human disease, there has been a surge in interest in examining circulating betatrophin levels in patients [18],[22]-[24],[38],[50],[51]. These studies show that betatrophin levels were altered in various physiologic conditions, such as the postprandial state [18], and pathological conditions, such as type 2 diabetes [18],[23],[38],[50], type 1 diabetes [22], obesity [18],[50], and were associated with metabolic parameters, such as BMI [18],[50], glucose [18],[38], insulin resistance [38],[50], LDL-C [24], HDL-C and triglycerides [50].
However, results from these studies show a wide range of variations (Table 2). For instance, circulating levels of betatrophin in lean and non-diabetic subjects ranged from 0.3 ng/ml [22] to 45 ng/ml [50], and the levels were either increased [18],[23],[38] or decreased [50] in type 2 diabetes, either increased [18] or decreased [50] in obesity, either positively [18],[38] or negatively [50] correlated with insulin, and either correlated with atherogenic lipid profiles [24] or with HDL-C [50] (Table 2). Indeed, we showed that for the same 30 human subjects, correlations between betatrophin and BMI can be either positive or negative based on ELISA kits that rely on antibodies recognizing the C-terminal or N-terminal betatrophin, respectively [51].
To resolve these discrepancies, we proposed that different betatrophin species are measured by ELISA kits that rely on either the N- or the C-terminal antibodies [51]. When human sera were analyzed by Western blot, the C-terminal antibody recognized both full-length protein (band position corresponding to 22kD) and C-terminal fragments (band position corresponding to small molecular weight) [52]. The small fragments were identified as amino acids 118–198 and 133–198 by HPLC and MALDI-TOF, and therefore, betatrophin is likely cleaved in vivo to release the C-terminal fragments, while the N-terminal fragments are degraded [52]. Therefore, the N-terminal kit likely measures full-length betatrophin and the C-terminal kit measures total betatrophin, including both full-length protein and the C-terminal fragments. Indeed, the betatrophin levels in lean and non-diabetic subjects determined by the C-terminal kit are higher than those determined by the N-terminal kit (Table 2).
There are at least two alternative explanations. The first is alternative splicing of the betatrophin transcript. Indeed, 2 additional transcripts are reported in the Ensemble database, in addition to the full length transcript ENSP00000252453 which encodes 198 residues: isoforms ENSP00000464941 and ENSP00000465378 encode 99 and 58 residues, respectively (lacking 99 and 140 N-terminal amino acid residues, respectively). However, the finding that only full-length transcript was detected in human liver [19] does not support the alternative splicing hypothesis.
The discrepancies could also be due to sample degradation. If the C-terminal fragments are more stable than the N-terminal ones, the C-terminal fragments accumulate in the circulation, resulting in higher total betatrophin. However, because of the presence of proteinase cleavage sites and the corresponding C-terminal fragments identified by mass spec in vivo[52], the explanation of proteolytic regulation is more likely. Regardless of the explanation, caution will have to be exercised in interpreting betatrophin ELISA results by considering which antibodies are used, and which betatrophin species is measured.
Conclusion
Accumulating evidence from GWAS and mouse genetic studies convincingly shows that lipasin/Angptl8/RIFL is involved in lipid regulation. However, the mechanism of action, discrepancy between human and mouse studies and the relationship among lipasin, Angptl3 and Angptl4, remain elusive. Betatrophin represents a promising drug target in replenishing β-cell mass, but further evidence is needed to support this result.
Authors’ contributions
Conceived study and wrote the manuscript: RZ. Revised manuscript: RZ and ABA-S. Overall responsibility: RZ. Both authors read and approved the final manuscript.
Abbreviations
- Angptl:
-
Angiopoietin-like protein
- BAT:
-
Brown adipose tissue
- BMI:
-
Body mass index
- cAMP:
-
Cyclic adenosine monophosphate
- ChREBP:
-
Carbohydrate-responsive element-binding protein
- CM:
-
Chylomicrons
- C-terminus:
-
Carboxyl-terminus
- DHS:
-
Dallas Heart Study
- Dock6:
-
Dedicator of cytokinesis 6
- ELISA:
-
The enzyme-linked immunosorbent assay
- FFA:
-
Free fatty acids
- GEO:
-
Gene Expression Omnibus
- GPIHBP1:
-
Glycosylphosphatidylinositol anchored high density lipoprotein binding protein 1
- GWAS:
-
Genome wide association study
- HDL-C:
-
High-density lipoprotein cholesterol
- HPLC:
-
High-performance liquid chromatography
- KO:
-
Knockout
- LDL-C:
-
Low-density lipoprotein cholesterol
- LIRKO:
-
Liver-specific insulin receptor knockout
- LPL:
-
Lipoprotein lipase
- N-terminus:
-
Amino-terminus
- MAF:
-
Minor allele frequency
- MALDI-TOF:
-
Matrix assisted laser desorption ionization time-of-flight
- PAGE:
-
Polyacrylamide gel electrophoresis
- PGC-1 α:
-
Peroxisome proliferator-activated receptor gamma coactivator 1-alpha
- Poly(A):
-
Polyadenylation
- RIFL:
-
Refeeding induced in fat and liver
- shRNA:
-
Short hairpin RNA
- SNP:
-
Single-nucleotide polymorphism
- T3:
-
Triiodothyronine
- TAG:
-
Triacylglyceride
- VLDL:
-
Very low-density lipoprotein
- WAT:
-
White adipose tissue
References
Eckel RH, Grundy SM, Zimmet PZ: The metabolic syndrome. Lancet. 2005, 365: 1415-1428. 10.1016/S0140-6736(05)66378-7.
Grundy SM, Benjamin IJ, Burke GL, Chait A, Eckel RH, Howard BV, Mitch W, Smith SC, Sowers JR: Diabetes and cardiovascular disease: a statement for healthcare professionals from the American Heart Association. Circulation. 1999, 100: 1134-1146. 10.1161/01.CIR.100.10.1134.
Klein S, Burke LE, Bray GA, Blair S, Allison DB, Pi-Sunyer X, Hong Y, Eckel RH: Clinical implications of obesity with specific focus on cardiovascular disease: a statement for professionals from the American Heart Association Council on Nutrition, Physical Activity, and Metabolism: endorsed by the American College of Cardiology Foundation. Circulation. 2004, 110: 2952-2967. 10.1161/01.CIR.0000145546.97738.1E.
Miller M, Stone NJ, Ballantyne C, Bittner V, Criqui MH, Ginsberg HN, Goldberg AC, Howard WJ, Jacobson MS, Kris-Etherton PM, Lennie TA, Levi M, Mazzone T, Pennathur S: Triglycerides and cardiovascular disease: a scientific statement from the American Heart Association. Circulation. 2011, 123: 2292-2333. 10.1161/CIR.0b013e3182160726.
Haffner SM: Management of dyslipidemia in adults with diabetes. Diabetes Care. 2003, 26 (Suppl 1): S83-S86.
Mooradian AD: Dyslipidemia in type 2 diabetes mellitus. Nat Clin Pract Endocrinol Metab. 2009, 5: 150-159. 10.1038/ncpendmet1066.
Patsch JR, Miesenbock G, Hopferwieser T, Muhlberger V, Knapp E, Dunn JK, Gotto AM, Patsch W: Relation of triglyceride metabolism and coronary artery disease. Studies in the postprandial state. Arterioscler Thromb. 1992, 12: 1336-1345. 10.1161/01.ATV.12.11.1336.
Kersten S: Physiological regulation of lipoprotein lipase. Biochim Biophys Acta. 1841, 2014: 919-933.
Santamarina-Fojo S, Brewer HB: The familial hyperchylomicronemia syndrome. New insights into underlying genetic defects. JAMA. 1991, 265: 904-908. 10.1001/jama.1991.03460070086049.
Coleman T, Seip RL, Gimble JM, Lee D, Maeda N, Semenkovich CF: COOH-terminal disruption of lipoprotein lipase in mice is lethal in homozygotes, but heterozygotes have elevated triglycerides and impaired enzyme activity. J Biol Chem. 1995, 270: 12518-12525. 10.1074/jbc.270.21.12518.
Weinstock PH, Bisgaier CL, Aalto-Setala K, Radner H, Ramakrishnan R, Levak-Frank S, Essenburg AD, Zechner R, Breslow JL: Severe hypertriglyceridemia, reduced high density lipoprotein, and neonatal death in lipoprotein lipase knockout mice. Mild hypertriglyceridemia with impaired very low density lipoprotein clearance in heterozygotes. J Clin Invest. 1995, 96: 2555-2568. 10.1172/JCI118319.
Beigneux AP, Davies BS, Gin P, Weinstein MM, Farber E, Qiao X, Peale F, Bunting S, Walzem RL, Wong JS, Blaner WS, Ding ZM, Melford K, Wongsiriroj N, Shu X, de Sauvage F, Ryan RO, Fong LG, Bensadoun A, Young SG: Glycosylphosphatidylinositol-anchored high-density lipoprotein-binding protein 1 plays a critical role in the lipolytic processing of chylomicrons. Cell Metab. 2007, 5: 279-291. 10.1016/j.cmet.2007.02.002.
Beigneux AP, Franssen R, Bensadoun A, Gin P, Melford K, Peter J, Walzem RL, Weinstein MM, Davies BS, Kuivenhoven JA, Kastelein JJ, Fong LG, Dallinga-Thie GM, Young SG: Chylomicronemia with a mutant GPIHBP1 (Q115P) that cannot bind lipoprotein lipase. Arterioscler Thromb Vasc Biol. 2009, 29: 956-962. 10.1161/ATVBAHA.109.186577.
Gray KA, Daugherty LC, Gordon SM, Seal RL, Wright MW, Bruford EA: Genenames.org: the HGNC resources in 2013. Nucleic Acids Res. 2013, 41: D545-D552. 10.1093/nar/gks1066.
Ren G, Kim JY, Smas CM: Identification of RIFL, a novel adipocyte-enriched insulin target gene with a role in lipid metabolism. Am J Physiol Endocrinol Metab. 2012, 303: E334-E351. 10.1152/ajpendo.00084.2012.
Fu Z, Yao F, Abou-Samra AB, Zhang R: Lipasin, thermoregulated in brown fat, is a novel but atypical member of the angiopoietin-like protein family. Biochem Biophys Res Commun. 2013, 430: 1126-1131. 10.1016/j.bbrc.2012.12.025.
Zhang R: Lipasin, a novel nutritionally-regulated liver-enriched factor that regulates serum triglyceride levels. Biochem Biophys Res Commun. 2012, 424: 786-792. 10.1016/j.bbrc.2012.07.038.
Fu Z, Berhane F, Fite A, Seyoum B, Abou-Samra AB, Zhang R: Elevated circulating lipasin/betatrophin in human type 2 diabetes and obesity. Sci Rep. 2014, 4: 5013-
Quagliarini F, Wang Y, Kozlitina J, Grishin NV, Hyde R, Boerwinkle E, Valenzuela DM, Murphy AJ, Cohen JC, Hobbs HH: Atypical angiopoietin-like protein that regulates ANGPTL3. Proc Natl Acad Sci U S A. 2012, 109: 19751-19756. 10.1073/pnas.1217552109.
Wang Y, Quagliarini F, Gusarova V, Gromada J, Valenzuela DM, Cohen JC, Hobbs HH: Mice lacking ANGPTL8 (Betatrophin) manifest disrupted triglyceride metabolism without impaired glucose homeostasis. Proc Natl Acad Sci U S A. 2013, 110: 16109-16114. 10.1073/pnas.1315292110.
Yi P, Park JS, Melton DA: Betatrophin: a hormone that controls pancreatic beta cell proliferation. Cell. 2013, 153: 747-758. 10.1016/j.cell.2013.04.008.
Espes D, Lau J, Carlsson PO: Increased circulating levels of betatrophin in individuals with long-standing type 1 diabetes. Diabetologia. 2014, 57: 50-53. 10.1007/s00125-013-3071-1.
Espes D, Martinell M, Carlsson PO: Increased circulating betatrophin concentrations in patients with type 2 diabetes. Int J Endocrinol. 2014, 2014: 323407-10.1155/2014/323407.
Fenzl A, Itariu BK, Kosi L, Fritzer-Szekeres M, Kautzky-Willer A, Stulnig TM, Kiefer FW: Circulating betatrophin correlates with atherogenic lipid profiles but not with glucose and insulin levels in insulin-resistant individuals. Diabetologia. 2014, 57: 1204-1208. 10.1007/s00125-014-3208-x.
Tseng YH, Ke PY, Liao CJ, Wu SM, Chi HC, Tsai CY, Chen CY, Lin YH, Lin KH: Chromosome 19 open reading frame 80 is upregulated by thyroid hormone and modulates autophagy and lipid metabolism. Autophagy. 2014, 10: 20-31. 10.4161/auto.26126.
Abecasis GR, Auton A, Brooks LD, DePristo MA, Durbin RM, Handsaker RE, Kang HM, Marth GT, McVean GA: An integrated map of genetic variation from 1,092 human genomes. Nature. 2012, 491: 56-65. 10.1038/nature11632.
Peloso GM, Auer PL, Bis JC, Voorman A, Morrison AC, Stitziel NO, Brody JA, Khetarpal SA, Crosby JR, Fornage M, Isaacs A, Jakobsdottir J, Feitosa MF, Davies G, Huffman JE, Manichaikul A, Davis B, Lohman K, Joon AY, Smith AV, Grove ML, Zanoni P, Redon V, Demissie S, Lawson K, Peters U, Carlson C, Jackson RD, Ryckman KK, Mackey RH, et al: Association of low-frequency and rare coding-sequence variants with blood lipids and coronary heart disease in 56,000 whites and blacks. Am J Hum Genet. 2014, 94: 223-232. 10.1016/j.ajhg.2014.01.009.
Teslovich TM, Musunuru K, Smith AV, Edmondson AC, Stylianou IM, Koseki M, Pirruccello JP, Ripatti S, Chasman DI, Willer CJ, Johansen CT, Fouchier SW, Isaacs A, Peloso GM, Barbalic M, Ricketts SL, Bis JC, Aulchenko YS, Thorleifsson G, Feitosa MF, Chambers J, Orho-Melander M, Melander O, Johnson T, Li X, Guo X, Li M, Shin Cho Y, Jin Go M, Jin Kim Y, et al: Biological, clinical and population relevance of 95 loci for blood lipids. Nature. 2010, 466: 707-713. 10.1038/nature09270.
Weissglas-Volkov D, Aguilar-Salinas CA, Nikkola E, Deere KA, Cruz-Bautista I, Arellano-Campos O, Munoz-Hernandez LL, Gomez-Munguia L, Ordonez-Sanchez ML, Reddy PM, Lusis AJ, Matikainen N, Taskinen MR, Riba L, Cantor RM, Sinsheimer JS, Tusie-Luna T, Pajukanta P: Genomic study in Mexicans identifies a new locus for triglycerides and refines European lipid loci. J Med Genet. 2013, 50: 298-308. 10.1136/jmedgenet-2012-101461.
Tang T, Li L, Tang J, Li Y, Lin WY, Martin F, Grant D, Solloway M, Parker L, Ye W, Forrest W, Ghilardi N, Oravecz T, Platt KA, Rice DS, Hansen GM, Abuin A, Eberhart DE, Godowski P, Holt KH, Peterson A, Zambrowicz BP, de Sauvage FJ: A mouse knockout library for secreted and transmembrane proteins. Nat Biotechnol. 2010, 28: 749-755. 10.1038/nbt.1644.
Mattijssen F, Kersten S: Regulation of triglyceride metabolism by Angiopoietin-like proteins. Biochim Biophys Acta. 1821, 2012: 782-789.
Ono M, Shimizugawa T, Shimamura M, Yoshida K, Noji-Sakikawa C, Ando Y, Koishi R, Furukawa H: Protein region important for regulation of lipid metabolism in angiopoietin-like 3 (ANGPTL3): ANGPTL3 is cleaved and activated in vivo. J Biol Chem. 2003, 278: 41804-41809. 10.1074/jbc.M302861200.
Camenisch G, Pisabarro MT, Sherman D, Kowalski J, Nagel M, Hass P, Xie MH, Gurney A, Bodary S, Liang XH, Clark K, Beresini M, Ferrara N, Gerber HP: ANGPTL3 stimulates endothelial cell adhesion and migration via integrin alpha vbeta 3 and induces blood vessel formation in vivo. J Biol Chem. 2002, 277: 17281-17290. 10.1074/jbc.M109768200.
Eckel RH: Lipoprotein lipase. A multifunctional enzyme relevant to common metabolic diseases. N Engl J Med. 1989, 320: 1060-1068. 10.1056/NEJM198904203201607.
Zhang R, Abou-Samra AB: Emerging roles of Lipasin as a critical lipid regulator. Biochem Biophys Res Commun. 2013, 432: 401-405. 10.1016/j.bbrc.2013.01.129.
Leanca CC, Nunes VS, Panzoldo NB, Zago VS, Parra ES, Cazita PM, Jauhiainen M, Passarelli M, Nakandakare ER, de Faria EC, Quintao EC: Metabolism of plasma cholesterol and lipoprotein parameters are related to a higher degree of insulin sensitivity in high HDL-C healthy normal weight subjects. Cardiovasc Diabetol. 2013, 12: 173-10.1186/1475-2840-12-173.
Huang Y, Li X, Wang M, Ning H, Li Y, Sun C: Lipoprotein lipase links vitamin D, insulin resistance, and type 2 diabetes: a cross-sectional epidemiological study. Cardiovasc Diabetol. 2013, 12: 17-10.1186/1475-2840-12-17.
Hu H, Sun W, Yu S, Hong X, Qian W, Tang B, Wang D, Yang L, Wang J, Mao C, Zhou L, Yuan G: Increased circulating levels of betatrophin in newly diagnosed type 2 diabetic patients.Diabetes Care 2014. in press.,
Parsons JA, Brelje TC, Sorenson RL: Adaptation of islets of Langerhans to pregnancy: increased islet cell proliferation and insulin secretion correlates with the onset of placental lactogen secretion. Endocrinology. 1992, 130: 1459-1466.
Alonso LC, Yokoe T, Zhang P, Scott DK, Kim SK, O'Donnell CP, Garcia-Ocana A: Glucose infusion in mice: a new model to induce beta-cell replication. Diabetes. 2007, 56: 1792-1801. 10.2337/db06-1513.
Bruning JC, Winnay J, Bonner-Weir S, Taylor SI, Accili D, Kahn CR: Development of a novel polygenic model of NIDDM in mice heterozygous for IR and IRS-1 null alleles. Cell. 1997, 88: 561-572. 10.1016/S0092-8674(00)81896-6.
Kulkarni RN, Jhala US, Winnay JN, Krajewski S, Montminy M, Kahn CR: PDX-1 haploinsufficiency limits the compensatory islet hyperplasia that occurs in response to insulin resistance. J Clin Invest. 2004, 114: 828-836. 10.1172/JCI21845.
Michael MD, Kulkarni RN, Postic C, Previs SF, Shulman GI, Magnuson MA, Kahn CR: Loss of insulin signaling in hepatocytes leads to severe insulin resistance and progressive hepatic dysfunction. Mol Cell. 2000, 6: 87-97. 10.1016/S1097-2765(00)00010-1.
El Ouaamari A, Kawamori D, Dirice E, Liew CW, Shadrach JL, Hu J, Katsuta H, Hollister-Lock J, Qian WJ, Wagers AJ, Kulkarni RN: Liver-derived systemic factors drive beta cell hyperplasia in insulin-resistant states. Cell Rep. 2013, 3: 401-410. 10.1016/j.celrep.2013.01.007.
Jiao Y, Le Lay J, Yu M, Naji A, Kaestner KH: Elevated mouse hepatic betatrophin expression does not increase human beta-cell replication in the transplant setting. Diabetes. 2014, 63: 1283-1288. 10.2337/db13-1435.
Stewart AF: Betatrophin versus bitter-trophin and the elephant in the room: time for a new normal in beta-cell regeneration research. Diabetes. 2014, 63: 1198-1199. 10.2337/db14-0009.
Bostrom P, Wu J, Jedrychowski MP, Korde A, Ye L, Lo JC, Rasbach KA, Bostrom EA, Choi JH, Long JZ, Kajimura S, Zingaretti MC, Vind BF, Tu H, Cinti S, Hojlund K, Gygi SP, Spiegelman BM: A PGC1-alpha-dependent myokine that drives brown-fat-like development of white fat and thermogenesis. Nature. 2012, 481: 463-468. 10.1038/nature10777.
Zhang Y, Li R, Meng Y, Li S, Donelan W, Zhao Y, Qi L, Zhang M, Wang X, Cui T, Yang LJ, Tang D: Irisin stimulates browning of white adipocytes through mitogen-activated protein kinase p38 MAP kinase and ERK MAP kinase signaling. Diabetes. 2014, 63: 514-525. 10.2337/db13-1106.
Sanchis-Gomar F, Perez-Quilis C: The p38-PGC-1alpha-irisin-betatrophin axis: Exploring new pathways in insulin resistance. Adipocyte. 2014, 3: 67-68. 10.4161/adip.27370.
Gomez-Ambrosi J, Pascual E, Catalan V, Rodriguez A, Ramirez B, Silva C, Gil MJ, Salvador J, Fruhbeck G: Circulating betatrophin concentrations are decreased in human obesity and type 2 diabetes.J Clin Endocrinol Metab 2014. in press.,
Fu Z, Abou-Samra AB, Zhang R: An explanation for recent discrepancies in levels of human circulating betatrophin. Diabetologia. 2014, 57: 2232-2234. 10.1007/s00125-014-3346-1.
Chang JK, Lyu RM, Chen XQ, Tian Q, Sabirov A, Dun SL, Dun NJ: Detection of C-terminal betatrophin peptides in human plasma and rodent liver. Detection of C-terminal betatrophin peptides in human plasma and rodent liver. The 4th Asia-Pacific International Peptide Symposium. Peptide Science, Japan. Edited by: Nishiuchi Y, Teshima T. 2013, 243-246.
Grant support
The study was supported in part by funding from Wayne State University to RZ, and by funding from Medical Research Center of Hamad Medical Corporation and the Qatar National Research Fund to ABA-S.
Author information
Authors and Affiliations
Corresponding author
Additional information
Competing interests
The authors declare that they have no competing interests.
Authors’ original submitted files for images
Below are the links to the authors’ original submitted files for images.
Rights and permissions
This article is published under an open access license. Please check the 'Copyright Information' section either on this page or in the PDF for details of this license and what re-use is permitted. If your intended use exceeds what is permitted by the license or if you are unable to locate the licence and re-use information, please contact the Rights and Permissions team.
About this article
Cite this article
Zhang, R., Abou-Samra, A.B. A dual role of lipasin (betatrophin) in lipid metabolism and glucose homeostasis: consensus and controversy. Cardiovasc Diabetol 13, 133 (2014). https://doi.org/10.1186/s12933-014-0133-8
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12933-014-0133-8