Abstract
Background
Preimplantation genetic testing (PGT) has already been applied in chromosomally balanced translocation carriers to improve the clinical outcome of assisted reproduction. However, traditional methods could not further distinguish embryos carrying a translocation from those with a normal karyotype prior to implantation.
Methods
To solve this problem, we developed a method named “Chromosomal Phasing on Base level” (BasePhasing), which based on Infinium Asian Screening Array-24 v1.0 (ASA) and a specially phasing pipeline. Firstly, by comparing the number of single nucleotide polymorphism (SNP) loci in different minor allele frequencies (MAFs) and in 2Mbp continuous windows of ASA chip and karyomap-12 chip, we verified whether ASA could be adopted for genome-wide haplotype linkage analysis. Besides, the whole gene amplification (WGA) of 3–10 cells of GM16457 cell line was used to verify whether ASA chip could be used for testing of WGA products. Finally, two balanced translocation families were utilized to carry out BasePhasing and to validate the feasibility of its clinical application.
Results
The average number of SNP loci in each window of ASA (473.2) was twice of that of Karyomap-12 (201.2). The coincidence rate of SNP loci in genomic DNA and WGA products was about 97%. The 5.3Mbp deletion was detected positively in cell line GM16457 of both genomic DNA and WGA products, and haplotype linkage analysis was performed in genome wide successfully. In the two balanced translocation families, 18 blastocysts were analyzed, in which 8 were unbalanced and the other 10 were balanced or normal chromosomes. Two embryos were transferred back to the patients successfully, and prenatal cytogenetic analysis of amniotic fluid was performed in the second trimester. The results predicted by BasePhasing and prenatal diagnosis were totally consistent.
Conclusions
Infinium ASA bead chip based BasePhasing pipeline shows good performance in balanced translocation carrier testing. With the characteristics of simple operation procedure and accurate results, we demonstrate that BasePhasing is one of the most suitable methods to distinguish between balanced and structurally normal chromosome embryos from translocation carriers in PGT at present.
Similar content being viewed by others
Background
Balanced translocation is one of the most frequent indications for preimplantation genetic testing (PGT), which occurs at an incidence of 1/500 to 1/625 in the general population and even up to 1/20 in the patients who has a history of repeated IVF failure or recurrent miscarriages [1]. Although they often have normal phenotypes, but the risk of producing unbalanced gametes is high (typically approximately 70%) due to the abnormal segregation of rearranged chromosomes during meiosis [2, 3]. The unbalanced gametes will lead to apparent infertilities [4], recurrent miscarriages [5, 6] or other congenital abnormalities [4, 7]. Therefore, it is of great significance to prevent balanced translocations from being passed to the next generation by assisted reproductive technology (ART) and PGT.
The first PGT case for translocations reported in 1998 was used by fluorescence in situ hybridization (FISH) [8]. However, the application of FISH is limited by some technical problems, such as ambiguous signals and complex operation in detecting limited chromosomes [9,10,11]. With the development of technology, aCGH/SNP array [12, 13] and whole genome sequencing [14, 15] methods have been employed for balanced translocation detection. Although these traditional PGT methods can clearly identify embryos with chromosomally unbalanced translocation or aneuploidies, they can hardly further distinguish the balanced and structurally normal embryos. Over the past 5 years, researchers tried to use mate pair sequencing [16, 17], MicroSeq-PGD [18], MaReCs [19], and long read sequencing [20] to obtain the precise breakpoints of balanced translocation, followed by subsequent identification of normal embryos through PCR-Sanger sequencing or linkage analysis based on the breakpoints. However, it remains unstable and inaccurate to identify the precise breakpoints in the highly repetitive and variable translocation regions.
In our former study [21], we successfully utilized preimplantation genetic haplotyping (PGH) to distinguish balanced and structurally normal embryos prior to implantation for both reciprocal translocation and Robertsonian translocation carriers accurately, along with the genetic screening for all 23-pairs of chromosomes. However, the used SNP microarray contained relatively less SNP loci. In another study [22], the researchers chose Human CytoSNP-12 BeadChip, which also faced the same trouble. Therefore, it remains a challenge to obtain a highly efficient approach to distinguish the normal embryos from those with a balanced translocation karyotype in clinical. In this study, based on our previous PGH technology theory, here we established a new method named “Chromosomal Phasing on Base level” (BasePhasing). More SNP loci (700 K SNPs) bead chip (Illumina Infinium Asian Screening Array-24 v1.0, ASA) was used to perform BasePhasing and a new analysis pipeline was developed for ASA data. BasePhasing was validated by the whole genome amplification result of cell line GM16457, and two balanced translocation families were also analyzed. The accuracy of this method was validated by the conventional amniotic fluid karyotypes in the second trimester.
Methods
ASA assessment for BasePhasing
The basic Microarray technical data of ASA and Karyomap-12 was downloaded from Illumina official website (https://www.illumina.com). As we known, the number of informative SNPs is more valuable to linkage analysis, which allows for directly determining the accuracy of haplotype classification. For a SNP to be informative, one parent must have a heterozygous genotype and the other one should have a homozygous genotype. This is limited by specific families and may also be affected by the distribution of SNP frequency. For the same family, SNPs with high MAF are more likely to generate the informative SNP.
Usually, the region within ±2Mbp flanking region of the breakpoints was chosen to avoid misinterpretation from possible recombination events that might occur during meiosis. More importantly, sufficient number of informative SNPs could be obtained in the 2Mbp region to distinguish homologous recombination. Therefore, the whole human genome could be divided into large amounts of 2Mbp segments (called windows). By comparing the number of total SNPs and effective SNPs in each widow, we can get analytical performance of ASA bead chip.
Ethics statement
Written informed consent was obtained from each family and the study protocol was approved by the Ethics Committee for Human Subject research of the Obstetrics and Gynecology Hospital, Fudan University.
Samples preparation and DNA isolation
Cell line GM16457 was obtained from the NIGMS Human Genetic Cell Repository at the Coriell Institute for Medical Research, with karyotype 46,XX,del(18)(q22.3) and a known 5.3Mbp deletion in chr18. Two translocation carrier families that would undergo assisted reproductive were enrolled in Shanghai Ji Ai Genetics & IVF Institute in May 2018. Both families had a history of recurrent spontaneous abortion, infertility or pregnancies with chromosome anomalies. The translocation karyotypes were 46,XY,t(1:5)(q21;q35) and 46,XY,t(6;7)(q23;q34) respectively. Five milliliter peripheral blood from each couple and family members was collected at recruitment.
For cell line GM16457 and peripheral blood samples, the high molecular weight DNA was isolated as described in the manufacturers’ protocol (DNeasy Blood & Tissue Kit, QIAGEN, Germany).
Single cell preparation and WGA
Three to ten cells of the GM16457 cell line were isolated by micromanipulating under a dissection microscope (Olympus CKX41, Japan) using a finely pulled glass Pasteur pipette. For embryos at the blastocyst stage, three to ten cells were removed from the trophectoderm on day five of embryonic development. The biopsied cells were placed into 0 .2mL PCR tubes in a total volume of less than 4 .0μL. Whole genome amplification (WGA) was conducted by means of multiple displacement amplification (MDA) according to the manufacturer’s instructions (Repli-g single cell kit, QIAGEN, Germany). The isothermal amplification was carried out at 30 °C for 8 h and followed by enzyme inactivation at 65 °C for 3 min.
SNP-array and analysis
The isolated DNA and WGA products were treated according to the manufacturer’s instructions (Illumina, San Diego, CA, USA), which were then scanned using an Illumina iScan Bead Array Reader. The microarray scanning results were processed using the B allele frequency and Log R Ratio of Genome Studio software (Illumina) and Karyo Studio software (Illumina) to analyze the copy number of the chromosomes. Genome wide preimplantation genetic haplotyping (PGH) analysis based on Illumina Human Karyomap-12 V1.0 microarray was performed as our previous description [21].
BasePhasing
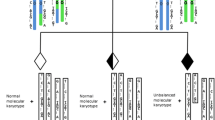
Based on informative SNPs and chromosomal phasing principles we developed BasePhasing pipeline, which was programmed in Practical Extraction and Reporting Language (Perl), and was capable of obtaining the clear haplotypes of each family member in linkage analysis (Fig. 1). The raw scanning data would be imported into the BasePhasing to produce the accurate chromosomal aneuploidy and haplotype results, once all the family samples were detected by ASA bead chip in a single test. To avoid misinterpretation precious statement, the region within ±2Mbp flanking region of the breakpoints was chosen to analyze the balanced translocation carriers.
Overview of the BasePhasing pipeline. Summary of the most important features of the BasePhasing pipeline. Black arrows show the flow of information from the samples to the final identification results. ASA bead chip scanning data and quality control (yellow background), BasePhasing analysis (green background), and candidate embryos transfer (blue background). CNVP, CNV partition analysis algorithm
Additionally, BasePhasing was a universal pipeline with compatibility for other bead chips such as Human Karyomap-12, Infinium Global Screening Array-24 (GSA), Human CytoSNP-12, and Infinium Omini series microarrays.
Results
ASA basic performance for BasePhasing
ASA bead chip contained approximately 700 K SNPs, more than double what those for Karyomap-12 bead chips. Although half amount of ASA SNPs MAF was lower than 0.1, the total number of high frequency SNPs was more than Karyomap-12 (Additional file 1: Table S1, Fig. 2a). Such as in MAF 0.25–0.5, ASA had total SNPs number 149,462 and Karyomap-12 was 136,690. In sum, ASA entirely had more SNPs than Karyomap-12 at every MAF segment.
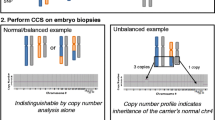
The performance of ASA bead chip in terms of SNP and CNV analysis. a SNPs distribution of ASA and Karyomap-12 bead chips. MAF: minor allele frequency. b A 5.3Mbp deletion in cell line gDNA and its single cell MDA products. Black arrows indicate the 5.3Mbp deletion in chr18. B allele frequency (BAF, genotyping information) represented by blue dots. Smoothed log R ratio represented by a red line. In addition, there is an ideogram of the chromosome, found and known regions, information from the DGV (Database of Genomic Variants), and gene information. c 136 ASA and Karyomap-12 bead chip data on the CNV performance for CCS. % Defect: Score given to each sample based on the number of detected regions. This value is the sum of the length of all detected regions per sample divided by the length of the genome. LogRDev: Standard deviation of the log R ratios of the sample. Sample type: 52 genome DNA and 84 MDA products
The total number of windows in the whole genome was 1561, but some regions like centromeres and satellites had no SNP probes distribution, so the effective number of windows was 1476. The number of uniquely mapped SNPs in each window was determined (Table 1). To get enough number of informative SNPs for haplotype analysis, knowing that in typical cases approximately only 10–20% of SNPs were informative SNPs, we considered there should be at least 50 SNPs in each window. In the whole genome, ASA with 98.85% region met the requirements, and the average number of SNPs in each window was up to 473.2.
Evaluation of BasePhasing feasibility
To assess ASA performance with single cell MDA products, we used both gDNA and single cell MDA products of cell line GM16457. Cell line GM16457 gDNA gave call rate of 98.7% and heterozygous call rate of 16.1%. SNP call rates were a little lower in single cells, about 96.3% and heterozygous call rates 14.8% (Table 2). We also compared the SNPs accordance between cell line gDNA and its single cell MDA products. It showed that the accordance rate was up to ~ 97%, not only between gDNA and its MDA products but also between the MDA products themselves. In addition, the known 5.3Mbp deletion in cell line GM16457 was clearly detected in MDA products (Fig. 2b).
Except the SNPs amount and distribution, ASA could also perform well for comprehensive chromosome screening (CCS). The CCS analysis mainly relied on the SNP allele frequency analysis in the whole genome. We gathered and compared 136 previous samples data of bead chip in our laboratory, including 43 samples by ASA and 93 samples by Karyomap-12. We found that there were no differences between ASA and Karyomap-12 on the LogRdev versus %Defects (Fig. 2c), which signified the copy number variation (CNV) performance for CCS.
BasePhasing clinical test
In this study, two balanced translocation families were collected. In family 1, the carrier’s parents were normal, therefore their unbalanced embryos were used as reference. In family 2, the translocation was inherited from his mother, therefore paternal grandparent as reference. With our method BasePhasing, we obtained molecular karyotypes from all the 18 biopsied blastocysts. Of the 18 diagnosed blastocysts, 8 were unbalanced, 10 were balanced or normal (such rate might be a little higher than the published data due to the small sample size). BasePhasing analysis was performed in the 10 blastocysts, which verified that 5 were balanced carriers and 5 were normal embryos. Sample information and BasePhasing results of the two families are listed in Table 3. Specifically, the results of BasePhasing and PGH were exactly identical (Fig. 3). Such as in family 2, both of the two methods indicated embryo-2, embryo-3 and embryo-5 were translocation carrier embryos and embryo-1 and embryo-4 were structurally normal embryos.
BasePhasing results of family 2 in the two balanced translocation breakpoints related chromosomes. a BasePhasing results of family 2 in the balanced translocation breakpoint related chromosome 6. The left smooth bar charts were performed with Blue-fuse-Multi software from Karyomap-12 bead chip data, the right scatter bar charts were performed with BasePhasing from ASA bead chip data. b BasePhasing results of family 2 in the balanced translocation breakpoint related chromosome 7. The left smooth bar charts were performed with Blue-fuse-Multi software from Karyomap-12 bead chip data, the right scatter bar charts were performed with BasePhasing from ASA bead chip data. SR region: the breakpoint of balanced translocation (gray box labeled). The different colorful histograms represented different haplotypes. The blue and red histograms represented the father’s haplotypes. The orange and green histograms represented the mother’s haplotypes. In the embryos, the gray column represented the haplotype that was inherited from the normal parent. And in the carrier’s family number, the gray column represented the haplotype that wasn’t passed on to the carrier
In the meantime, the number of informative SNP loci within ±2Mbp regions flanking the breakpoints and the whole related chromosome was calculated (Table 4). It was clearly shown that there were more informative SNPs in the ASA bead chip than Karyomap-12 both in the related whole chromosomes and breakpoints flanking regions. For example, in chromosome 5 (0–180,915,260), there were 3774 SNPs available on the ASA chip, and the average number of informative SNPs per window was 83.4 (ranging from 13 to 151). While there were only 2732 SNPs available on the Karyomap-12 chip, and the average number of key SNPs per window was 60.4 (ranging from 6 to 118).
We also calculated the number of informative SNP loci of family 2 in the whole human genome. The average number of informative SNPs was 85.9 in ASA bead chip and 59.4 in Karyomap-12 bead chip within ±2Mbp region along. The number of informative SNPs on ASA bead chip was a little bit more (Fig. 4). Based on this property of ASA bead chip, we asserted BasePhasing can perform better in balanced translocation carriers’ diagnosis.
After completing the BasePhasing analysis, embryo E1 in family 1 and embryo E4 in family 2 were transferred back to patients. For the two women that were pregnant after embryo transfer, cytogenetic analysis of amniotic fluid was required to be performed in the second trimesters. It was confirmed that the results predicted by BasePhasing and cytogenetic analysis of amniotic fluid cells were totally consistent.
Discussion
Theoretically, only two kinds of gametes from alternate segregation, one with a normal karyotype and another with a balanced karyotype, can produce a viable conceptus in balanced translocation carriers, the remaining unbalanced gametes from other segregation patterns may lead to repeated miscarriage, infertility or newborns with congenital malformations [23,24,25]. Therefore, balanced translocation carriers are generally suggested to get a successful pregnancy with PGT. In 2016, Treff et al. [26] reported a new method using unbalanced embryos as a reference to distinguish between balanced translocation and normal blastocysts based upon SNP genotype. Recently Zhang et al. [21] and Wang et al. [22] utilized PGH to successfully distinguish between balanced and normal embryos prior to implantation from balanced translocation carriers accurately. Any SNP marker-based data can be applied for PGH analysis only if there are sufficient informative SNPs flanking the breakpoint to establish haplotype and program family linkage analysis. Meanwhile, the comprehensive chromosome screening (CCS) can also be completed with SNP allele frequency analysis in the single test.
In this study, we applied the ASA bead chip in BasePhasing pipeline and got accurate PGT results. By comparing the number of SNP loci in different MAFs and in 2Mbp continuous windows of ASA chip and karyomap-12 chip, it was verified that ASA could be used to perform haplotype linkage analysis of the whole genome. The average number of SNP loci in each window of ASA was 473.2, and up to 98.85% regions had more than 50 SNPs. Informative SNPs was 85.9 in ASA and 59.4 in Karyomap-12 bead chip within ±2Mbp region along of family 2. Through the analysis of whole gene amplification products of 3–10 cells of GM16457 cell line, we verified that ASA chip could be used for CNV testing of PGT, even a 5.3Mbp deletion was positive.
Compared with previous studies, several advantages of this research could be concluded. First, large numbers of SNPs could be used to perform the chromosome aneuploidies and haplotype linkage analysis, guaranteeing the accuracy of results. Second, without the need of precise translocation breakpoint location and personalized design, our method was universal for any kind of translocation. Third, the bead chip experiment was relatively simple and data analysis was convenient, which was suitable for clinical work. In addition, ASA based BasePhasing had over twice loci while only took about one quarter (22.5%) cost of Kayomap-12 bead chip (Additional file 2: Table S2). So BasePhasing would be one of the most suitable methods for PGT of balanced translocation carriers in clinical application at present. Nonetheless, for performing the analysis, one carrier’s family member or an unbalanced embryo should be used as a reference. Therefore, one limitation of our research was that the method didn’t apply to these patients both with de novo translocation and without an unbalanced embryo. To our knowledge, no current methods could effectively overcome this difficulty.
Conclusions
Infinium ASA bead chip based BasePhasing pipeline shows good performance in PGT of balanced translocation. With the characteristics of simple operation procedure and accurate results, we demonstrate that BasePhasing is one of the most suitable methods to distinguish between balanced and structurally normal chromosome embryos from translocation carriers at present. In the meanwhile, we show a referable strategy to effectively expand the newly developed bead chips to clinical application. However, the sensitivity and specificity of BasePhasing should be further validated in a larger sample size. Furthermore, whether BasePhasing could be used to detect other single-gene diseases or other genetic diseases is worth further verifying.
Abbreviations
- ART:
-
assisted reproductive technology
- ASA:
-
Illumina Infinium Asian Screening Array-24 v1.0
- CCS:
-
comprehensive chromosome screening
- CNV:
-
Copy number variation
- DGV:
-
Database of Genomic Variants
- FISH:
-
Fluorescent in-situ hybridization
- GSA:
-
Infinium Global Screening Array-24
- IVF:
-
In vitro fertilization
- MAF:
-
minor allele frequency
- MDA:
-
Multiple displacement amplification
- PGD:
-
Preimplantation genetic diagnosis
- PGH:
-
Preimplantation genetic haplotyping
- PGT:
-
preimplantation genetic testing
- SNP:
-
Single nucleotide polymorphism
- WGA:
-
Whole genome amplification
References
Alfarawati S, Fragouli E, Colls P, Wells D. First births after preimplantation genetic diagnosis of structural chromosome abnormalities using comparative genomic hybridization and microarray analysis. Hum Reprod. 2011;26(6):1560–74.
Lim CK, Cho JW, Song IO, Kang IS, Yoon YD, Jun JH. Estimation of chromosomal imbalances in preimplantation embryos from preimplantation genetic diagnosis cycles of reciprocal translocations with or without acrocentric chromosomes. Fertil Steril. 2008;90(6):2144–51.
Ottolini CS, Newnham L, Capalbo A, Natesan SA, Joshi HA, Cimadomo D, Griffin DK, Sage K, Summers MC, Thornhill AR, et al. Genome-wide maps of recombination and chromosome segregation in human oocytes and embryos show selection for maternal recombination rates. Nat Genet. 2015;47(7):727–35.
Ethics Committee of American Society for Reproductive M. Use of preimplantation genetic diagnosis for serious adult onset conditions: a committee opinion. Fertil Steril. 2013;100(1):54–7.
Fragouli E, Wells D, Delhanty JD. Chromosome abnormalities in the human oocyte. Cytogenet Genome Res. 2011;133(2–4):107–18.
Arumugam B, Samuel CR, Thyagarajan SS. Balanced autosomal translocations in two women reporting recurrent miscarriage. J Clin Diagn Res. 2016;10(12):GD01–3.
Zhang Y, Zhu S, Wu J, Liu S, Sun X. Quadrivalent asymmetry in reciprocal translocation carriers predicts meiotic segregation patterns in cleavage stage embryos. Reprod BioMed Online. 2014;29(4):490.
Conn CM, Harper JC, Winston RML, Delhanty JDA. Infertile couples with Robertsonian translocations: preimplantation genetic analysis of embryos reveals chaotic cleavage divisions. Hum Genet. 1998;102(1):117–23.
Willadsen S, Levron J, Munne S, Schimmel T, Marquez C, Scott R, Cohen J. Rapid visualization of metaphase chromosomes in single human blastomeres after fusion with in-vitro matured bovine eggs. Hum Reprod. 1999;14(2):470–5.
Pehlivan T, Rubio C, Rodrigo L, Remohi J, Pellicer A, Simon C. Preimplantation genetic diagnosis by fluorescence in situ hybridization: clinical possibilities and pitfalls. J Soc Gynecol Investig. 2003;10(6):315–22.
Liss J, Kiewisz J, Zabielska J, Kulwikowska P, Lukaszuk K. Application of FISH method for preimplantation genetic diagnostics of reciprocal and Robertsonian translocations. Folia Histochem Cytobiol. 2015;53(2):162–8.
Yang Z, Lin J, Zhang J, Fong WI, Li P, Zhao R, Liu X, Podevin W, Kuang Y, Liu J. Randomized comparison of next-generation sequencing and array comparative genomic hybridization for preimplantation genetic screening: a pilot study. BMC Med Genet. 2015;8:30.
Huang CC, Chang LJ, Tsai YY, Hung CC, Fang MY, Su YN, Chen HF, Chen SU. A feasible strategy of preimplantation genetic diagnosis for carriers with chromosomal translocation: using blastocyst biopsy and array comparative genomic hybridization. J Formos Med Assoc. 2013;112(9):537–44.
Talkowski ME, Ernst C, Heilbut A, Chiang C, Hanscom C, Lindgren A, Kirby A, Liu S, Muddukrishna B, Ohsumi TK, et al. Next-generation sequencing strategies enable routine detection of balanced chromosome rearrangements for clinical diagnostics and genetic research. Am J Hum Genet. 2011;88(4):469–81.
Dong Z, Jiang L, Yang C, Hu H, Wang X, Chen H, Choy KW, Hu H, Dong Y, Hu B, et al. A robust approach for blind detection of balanced chromosomal rearrangements with whole-genome low-coverage sequencing. Hum Mutat. 2014;35(5):625–36.
Wang L, Shen J, Cram DS, Ma M, Wang H, Zhang W, Fan J, Gao Z, Zhang L, Li Z. Preferential selection and transfer of euploid noncarrier embryos in preimplantation genetic diagnosis cycles for reciprocal translocations. Fertil Steril. 2017;108(4):620–27.
Aristidou C, Koufaris C, Theodosiou A, Bak M, Mehrjouy MM, Behjati F, Tanteles G, Christophidouanastasiadou V, Tommerup N, Sismani C. Accurate breakpoint mapping in apparently balanced translocation families with discordant phenotypes using whole genome mate-pair sequencing. PLoS One. 2017;12(1):e0169935.
Hu L, Cheng D, Gong F, Lu C, Tan Y, Luo K, Wu X, He W, Xie P, Feng T, et al. Reciprocal translocation carrier diagnosis in preimplantation human embryos. EBioMedicine. 2016;14:139–47.
Xu J, Zhang Z, Niu W, Yang Q, Yao G, Shi S, Jin H, Song W, Chen L, Zhang X, et al. Mapping allele with resolved carrier status of Robertsonian and reciprocal translocation in human preimplantation embryos. Proc Natl Acad Sci U S A. 2017;114(41):E8695–702.
Dutta UR, Rao SN, Pidugu VK, V SV, Bhattacherjee A, Bhowmik AD, Ramaswamy SK, Singh KG, Dalal A. Breakpoint mapping of a novel de novo translocation t(X;20)(q11.1;p13) by positional cloning and long read sequencing. Genomics. 2018. https://doi.org/10.1016/j.ygeno.2018.07.005. [Epub ahead of print]
Zhang S, Lei C, Wu J, Zhou J, Sun H, Fu J, Sun Y, Sun X, Lu D, Zhang Y. The establishment and application of preimplantation genetic haplotyping in embryo diagnosis for reciprocal and Robertsonian translocation carriers. BMC Med Genet. 2017;10(1):60.
Wang J, Zeng Y, Ding C, Cai B, Lu B, Li R, Xu Y, Xu Y, Zhou C. Preimplantation genetic testing of Robertsonian translocation by SNP array-based preimplantation genetic haplotyping. Prenat Diagn. 2018;38(8):547–54.
Morin SJ, Eccles J, Iturriaga A, Zimmerman RS. Translocations, inversions and other chromosome rearrangements. Fertil Steril. 2017;107(1):19–26.
Wang YZ, Ding CH, Wang J, Zeng YH, Zhou W, Li R, Zhou CQ, Deng MF, Xu YW. Number of blastocysts biopsied as a predictive indicator to obtain at least one normal/balanced embryo following preimplantation genetic diagnosis with single nucleotide polymorphism microarray in translocation cases. J Assist Reprod Genet. 2017;34(1):51–9.
Ye Y, Qian Y, Xu C, Jin F. Meiotic segregation analysis of embryos from reciprocal translocation carriers in PGD cycles. Reprod BioMed Online. 2012;24(1):83–90.
Treff NR, Thompson K, Rafizadeh M, Chow M, Morrison L, Xin T, Garnsey H, Reda CV, Metzgar TL, Neal S. SNP array-based analyses of unbalanced embryos as a reference to distinguish between balanced translocation carrier and normal blastocysts. J Assist Reprod Genet. 2016;33(8):1–5.
Acknowledgements
We express deepest gratitude to all the families enrolled in our research. We also thank the staff of the Basecare Medical Device Co., Ltd. and Genetics Laboratory and IVF Laboratory for their invaluable efforts and contribution relating to the experiment.
Funding
The study design and collection, analysis, and interpretation of data and preparation of the manuscript was supported by National Key R&D Program of China (2016YFC1303100) and the Shanghai Science and Technology Innovation Action Plan Program (18411953800). The publication costs for this article were funded by the Shanghai Science and Technology Innovation Action Plan Program (18411953800).
Availability of data and materials
The datasets generated and analyzed during the current study are not publicly available due to a concern to protect individual patient confidentiality but are available from the corresponding author on reasonable request.
Author information
Authors and Affiliations
Contributions
BL, XS, and CX designed the research, SZ and DZ wrote the manuscript; DZ and JZ performed the bioinformatics analysis; YM and LK provided the analysis pipeline; SZ and DZ performed the microarray experiments; SZ performed the intra-cytoplasmic sperm injection and blastocyst biopsy experiments; SZ and DZ collected the cases; YZ and SZ made prenatal cytogenetic analysis; XS and CX substantively revised the manuscript. All authors have read and approved the final manuscript.
Corresponding authors
Ethics declarations
Ethics approval and consent to participate
Written informed consent was obtained from each family and this study was approved by the Ethics Committee for Human Subject research of the Obstetrics and Gynecology Hospital (2018–22), Fudan University.
Consent for publication
We obtained the consent to publish their clinical data from the patients in this study.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Additional files
Additional file 1:
Table S1. Comparison of ASA and Karyomap-12 SNPs numbers at different minor allele frequency. (DOCX 17 kb)
Additional file 2:
Table S2. The low cost of BasePhasing in PGT. (DOCX 16 kb)
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Zhang, S., Zhao, D., Zhang, J. et al. BasePhasing: a highly efficient approach for preimplantation genetic haplotyping in clinical application of balanced translocation carriers. BMC Med Genomics 12, 52 (2019). https://doi.org/10.1186/s12920-019-0495-6
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12920-019-0495-6