Abstract
Background
Klebsiella pneumoniae (K. pneumoniae) is a common cause of health-care associated infections (HAIs) and has high levels of antibiotic resistance. These bacteria are well-known for their ability to produce biofilm. The purpose of this study was to identify the antibiotic resistance pattern and biofilm-producing capacity of K. pneumoniae isolated from clinical samples in a tertiary care hospital in Klaten, Indonesia.
Methods
K. pneumoniae was isolated from inpatients in Soeradji Tirtonegoro Hospital Klaten from June 2017 to May 2018. Identification of K. pneumoniae isolate was done by analyzing colony morphology, microscopic examination, and by performing biochemical testing. Testing of antibiotics susceptibility and biofilm-producing capacity used the Kirby-Bauer disk diffusion method and adherence quantitative assays, respectively.
Results
A total of 167 (17.36%) K. pneumoniae isolates were isolated from 962 total clinical bacterial isolates during the study. Most of them were collected from patients aged more than 60 years old and were mainly obtained from respiratory specimens (51.50%). Most of K. pneumoniae isolates were extensively resistant to antibiotics. A more favorable profile was found only towards meropenem, amikacin, and piperacillin-tazobactam, showing 1.20%; 4.79% and 10.53% of resistance, respectively. The overall proportion of multidrug-resistant K. pneumoniae isolates was 54.49%. In addition, 148 (85.63%) isolates were biofilm producers, with 45 (26.95%) isolates as strong, 48 (28.74%) isolates as moderate, and 50 (29.94%) isolates as weak biofilm producers.
Conclusion
Most of the K. pneumoniae isolates demonstrated resistance to a wide range of antibiotics and are biofilm producers.
Similar content being viewed by others
Background
The emergence of antimicrobial resistance (AMR) is a rapidly increasing challenge in today’s healthcare institutions worldwide. Pathogenic bacteria, for example, Klebsiella pneumoniae are quickly developing multidrug resistant (MDR) strains and commonly pose a serious threat to the patients because of an increased fatality rate due to the reduced effectiveness of therapy options. K. pneumoniae is known to be responsible for community acquired infections although recently it is routinely observed as a major cause of hospital acquired pathogens. K. pneumoniae has been observed to develop resistance to antibiotics more easily than most bacteria through the production of enzymes such as Extended Spectrum β-Lactamase (ESBLs) and Carbapenemase [1,2,3]. The most important risk factor of AMR is antibiotic exposure. Some of the main contributors in the emergence and spread of highly resistant bacteria for health-care associated infections (HAIs) are the intensive and prolonged use of antibiotics in the hospital setting [4].
K. pneumoniae also plays an important role in spreading antimicrobial resistance genes from bacteria in the environment to clinically important bacteria [5]. There are multiple mechanisms of antimicrobial resistance which will negatively affect the therapeutic outcomes. The World Health Organization (WHO) declared antibiotic resistance as one of the three major problems in the world [3].
K. pneumoniae is a Gram-negative encapsulated bacteria that thrives in the mucosal surfaces of mammals and in the soil, vegetation, and water. In humans, K. pneumoniae typically will colonize the oropharynx and gastrointestinal tract, from where it can easily enter the circulation and other tissues causing infections such as bacteremia, septicemia, surgical site infection, urinary tract infection, hospital acquired pneumonia, and ventilator-associated pneumonia. It also contributes to the high frequency of opportunistic infections that occur among patients with immunocompromised situations, such as bladder neuropathy or diabetes mellitus [6, 7].
K. pneumoniae is also known for its capability to form biofilms, which are communities of bacteria embedded in an extracellular matrix. This matrix consists of proteins, exopolysaccharides, DNA, and lipopeptides [8]. In 1988, LeChevallier et al. first described K. pneumoniae and the biofilm-forming phenomenon [9]. K. pneumoniae has some virulence factors such as capsule polysaccharide, lipopolysaccharide, type 1 and type 3 fimbriae, outer membrane proteins and determinants for iron acquisition and nitrogen source usage. K. pneumoniae used these virulence factors for survival and for evade from immune system during infection as well as biofilm formation itself [10, 11]. K. pneumoniae can produce a thick layer of extracellular biofilm that supports the bacterial attachment to living or non-living surfaces, protecting antibiotics penetration and reducing its effects [12].
In 2017, there were 213 (19.28%) K. pneumoniae isolates from 1105 bacteria isolated in Soeradji Tirtonegoro hospital in Klaten, Indonesia. In 2005, a study conducted in a hospital in Surabaya, Indonesia reported the prevalence of ESBLs was 28% among clinical isolates of K. pneumoniae [13]. These antimicrobial-resistant bacteria have become a worldwide problem and there is still very limited data regarding biofilm producing capacity and antimicrobial resistence of K. pneumoniae in Indonesia. Biofilm forming capacity is demonstrated to have a greater resistance to antibiotics. Further studies are needed to describe the pattern of antibiotic resistance and the capacity to produce biofilm of K. pneumoniae clinical isolates in Indonesia. Thus, the purpose of this study was to identify the antibiotic resistance patterns, and the biofilm-producing capacity of K. pneumoniae clinical isolates in the Soeradji Tirtonegoro hospital in Klaten, Indonesia.
Methods
Subjects
Using a cross-sectional design, this observational research studied K. pneumoniae clinical isolates from inpatients in Soeradji Tirtonegoro Hospital, a tertiary care hospital Klaten, Indonesia, during June 2017 – May 2018.
Isolation, identification and antibiotic susceptibility tests of K. pneumoniae
Identification of K. pneumoniae isolate was conducted by culturing on McConkey agar, Gram staining and biochemical testing using Microbact™ GNB 24E (Oxoid, UK). Kirby Bauer method was used to perform antibiotic susceptibility tests. The Clinical and Laboratory Standards Institute 2015 was used to classify as sensitive, intermediate or resistant bacteria [14]. K. pneumoniae isolates that showed résistance to three or more different classes of antimicrobials were classified as multidrug-resistant (MDR) K. pneumoniae [15].
Biofilm formation assay
A quantitative adherence assay was employed to perform biofilm formation assay [16]. An overnight culture at 37 °C in trypticase soy broth (TSB) was conducted for each isolate. Subsequently, 2 μL of cell suspension was inoculated in sterile 96 well-flat bottom polystyrene microtitre plates contained 198 μl of TSB. Negative control wells that contained 200 ul of un-inoculated TSB also included in each test. Incubation was done at 37 °C for 24 h. With 200 μL phosphate-buffered saline (PBS), the wells were gently washed 3 times. The wells were dried in an inverted position. Biofilm mass was stained with 50 μL of 0.1% crystal violet. With 200 μL of distilled water, the wells were gently washed 3 times and dried in an inverted position. Finally, the wells were dissolved in 200 μL of 5% isopropanol acid to solubilize the stained biofilm mass. Optical density (OD) measurement was done by using a microplate reader at 570 nm. Each isolate or negative control was tested for 8–12 wells, and the mean OD was determined. Optical density cut-off (ODc) was assigned as an average OD of negative controls + (3× standard deviation (SD) of negative controls). Isolate with OD ≤ ODc categorized as non-biofilm producer. Meanwhile, the isolate was categorized as biofilm producer consisting of weak biofilm producer if 2 × ODc < OD ≤4 × ODc; moderate biofilm 2 × ODc < OD ≤4 × ODc; and strong biofilm producer if OD > 4 × ODc [16].
Statistical analysis
Frequencies and percentages were used to describe the variables in this study. Associations between the antibiotic sensitivity pattern and the biofilm producing capacity of K. pneumoniae were tested by Chi-square tests using STATA 13 software (STATA, College Station, Texas). The results were presented as prevalence ratios with a 95% confidence interval. Statistical significance was set if p-value < 0.05.
Ethical approval
The Medical and Health Research Ethics Committee, Faculty of Medicine, Public Health and Nursing, Universitas Gadjah Mada, Yogyakarta, Indonesia permitted this study with number: KE/FK/0862/EC/2017.
Results
Characteristics of clinical samples
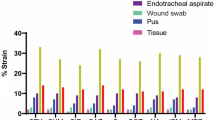
From June 2017 to May 2018, 167 (17.36%) K. pneumoniae isolates were examined from 962 total clinical bacterial isolates at Soeradji Tirtonegoro Hospital, Klaten, Indonesia. K. pneumoniae isolates were isolated from 107 male (64.07%) and 60 female (35.93%) patients. Most of K. pneumoniae were isolated from patients aged more than 60 years old (38.10%) (Table 1). K. pneumoniae samples were mostly isolated from respiratory specimens (51.50%) and pus (16.17%) (Table 1).
Antibiotic susceptibility pattern
Most of K. pneumoniae were resistant to a wide range of antibiotics. Among biofilm producer isolates, K. pneumoniae had only a good sensitivity to meropenem (98.60%), amikacin (95.80%), and piperacillin-tazobactam (90.00%). In contrast, among non-biofilm producer isolates, K. pneumoniae showed good sensitivity to meropenem, levofloxacin, amikacin, piperacillin-tazobactam and ciprofloxacin with 100.00%; 95.83%; 91.67%; 87.50%, and 86.67% of sensitivity respectively (Table 2).
Biofilm formation detection
In this study, among the 167 K. pneumoniae isolates tested, there were 143 (85.63%) isolates as biofilm producer and 24 (14.37%) isolates that were not biofilm producers. Among biofilm producers, there were 45 (26.95%) isolates as strong, 48 (28.74%) isolates as moderate, and 50 (29.94%) isolates identified as weak biofilm producers (Table 3). K. pneumoniae MDR isolates were found in 91 (54.5%) isolates and 76 (45.5%) isolates were non-MDR (Table 4). There was no significant association between K. pneumoniae MDR and biofilm production capacity based on the statistical analysis using chi-square tests (Table 4).
Discussion
In this study, the proportion of K. pneumoniae isolates was 17.36% of the total clinical bacterial isolates during June 2017–May 2018. This proportion should raise serious concerns because K. pneumoniae is one of the most important causes of MDR infections in the world. These bacteria are often associated with HAIs and highly contagious outbreaks in hospitals with increased mortality rate and longer stays in the hospitals, all of which results in inflated healthcare costs [17].
Most of the K. pneumoniae isolates in this study were collected from male patients. This result was in line with Osagie et al. [18] who collected the samples from 5 primary health care centers in Nigeria and reported that K. pneumoniae infection was higher in males than females. Akter et al. [19] also reported that male patients had a higher risk to get Klebsiella infection than females. The association between sex and the incident of K. pneumoniae was associated with poor lifestyle choices in the form of smoking and alcoholism [18]. However, no statistically significant differences between female and male subjects were reported in those studies.
Most of K. pneumoniae in this study were obtained from patients aged 18 to 65 years of age. This finding differs from a previous study that revealed most K. pneumoniae isolates were isolated from patients over 70 years old [20]. Meanwhile, another recent study suggested that a greater number of K. pneumoniae isolates were obtained from patients aged between 40 to 65 years old [21]. The differences in terms of patient’s age distribution could be related to the strength of the immune system response, which is expected to decline in senescence. Patients aged under 40 years tend to have stronger immune systems, thus giving more pressure to K. pneumoniae to fight the immunity of the host [22]. On the contrary, an increased age leads to a higher risk of K. pneumoniae infection because of increased incident of comorbid illness [23].
K. pneumoniae isolates were mainly isolated from respiratory specimens. Ashurst and Dawson [7] emphasized that K. pneumoniae typically colonizes human mucosal surfaces of the oropharynx and gastrointestinal tract. For this reason, K. pneumoniae is considered to be the most common cause of hospital-acquired pneumonia in the United States. This finding is similar to a study by Wang et al. [24] which reported that the respiratory tract was the main infection site of K. pneumoniae in the Republic of China. In comparison, Seifi et al. [6] who collected samples from 2 hospitals in Tehran reported that K. pneumoniae samples were isolated from urine, surgical wounds, sputum and blood with the percentage of 61.7, 18.1, 11.7, and 8.5% respectively.
Most of K. pneumoniae was resistant to various antibiotics, with ampicillin, cefazolin, and cefuroxime being the least effective for K. pneumoniae while amikacin, piperacillin-tazobactam, and meropenem had the most favorable profile. This report is supported by the study conducted by Madahiah [25] that found K. pneumoniae isolates were 100% resistant to ampicillin and 100% sensitive to amikacin. Ciprofloxacin and amoxicillin-clavulanic acid showed 38.75% and 36.69% resistance, respectively. This finding is similar to that of Cepas et al. [26] that reported 40% of K. pneumoniae strains were resistant to ciprofloxacin and amoxicillin-clavulanic acid.
Antibiotic exposure is the most crucial factor of antimicrobial resistance. The growth of antibiotic resistance are involving many factors such as the use of antibiotics in the hospital, in community, even in animal production, agriculture, and environment. As a consequence of the capability in purchasing antibiotics freely without prescription, therefore, antibiotics are used excessively. In the health service setting, intensive and prolonged use of antibiotics are very likely the main underlying factor in the widespread transmission of difficult-to-cure antibiotic-resistant nosocomial infections [4].
The overall proportion of MDR K. pneumoniae isolates in this study was 54.49%. This high percentage of MDR K. pneumoniae is supported by some previous studies. Cepas et al. [26] informed that 38% of K. pneumoniae strains were MDR. Manjula et al. [27] showed that 37 (90.2%) of 41 isolates were MDR. It was also found that the majority of the MDR K. pneumoniae isolates showed high resistance towards penicillin, cephalosporin, fluoroquinolone, aminoglycoside, and sulfonamide. This result is in line with a previous study by Moini et al. [28] which reported that 46.6% of the K. pneumoniae isolates were MDR and the high resistance was shown towards ampicillin, third-generation cephalosporins, and aminoglycosides. This MDR pattern exhibited by microbes is causing a great challenge in managing infections, and consequently, it is very important to monitor and optimize antibiotic use through antibiotic stewardship programs. Several studies have shown that treatment with a combination of antibiotics can help to prevent the emergence of new resistant strains, as therapy failures are commonly found in individuals who only receive single antibiotic therapy. It is also important for clinicians and microbiologists to collaborate to warrant effective management of infections as promoted in the Rational Use of Medicine (RUM) Program [29].
In this study, 148 (85.63%) of 167 isolates were biofilm producers. A similar study reported by Hassan et al. [16] stated that out of 110 K. pneumoniae tested, 70 isolates (64.7%) were identified as high or moderate biofilm producers and 40 isolates (35.3%) were identified as weak biofilm producers. In another study, Cepas et al. [26] reported that 37.6% of K. pneumoniae strains were biofilm producers. Yang and Zhang [20] reported that 62.5% of K. pneumoniae isolated from urine, sputum, wound swabs and blood were biofilm producers. Seifi et al. [6] reported that the majority of K. pneumoniae (93.6%) were biofilm producer and only 6.4% were not biofilm producers. Among biofilm producer strains, 33% were categorized as strong biofilm producer, 52.1% as moderately biofilm producers and 8.5% as weak biofilm producers. The capacity to form biofilms was different from each isolate because in general several factors are influencing the capacity such as the physicochemical characteristic of K. pneumoniae, physical interaction between the constituents, the type of surface where the biofilm attaches, temperature, pH, etc. [30]. Vuotto et al. [31] highlighted extensively drug resistant (XDR) K. pneumoniae biofilm-producing ability correlate to antibiotic resistance profile.
Cepas et al. [26] looked for possible relation of antimicrobial resistance and the ability to form biofilm between the collected samples of E. coli, K. pneumoniae, and P. aeruginosa. There was no statistically significant relationship between general MDR and biofilm formation in three Gram-negative species because the MDR isolates did not show any greater disposition to becoming a stronger biofilm producer compared to non-MDR isolates. However, they reported some correlations between biofilm formation and resistance to specific antibiotics. Resistance to gentamicin and ceftazidime was correlated with biofilm formation in E. coli, as well as ciprofloxacin in P. aeruginosa and piperacillin-tazobactam and colistin in K. pneumoniae. This pattern of resistance should raise grave concerns because colistin is now considered to be the last line treatment choice for K pneumoniae [17].
This study showed that antibiotic resistance was greater among K. pneumoniae which is biofilm producer than non-biofilm producer. This finding has been reported in many studies. A study by Saha et al. [32] demonstrated that all the biofilm-producing isolates presented more resistant patterns in comparison to non-biofilm producers, however, despite that finding, the protective mechanisms in biofilms are different from those responsible for conventional antibiotics resistance. In biofilms, the protective covering of the adhesive biomaterial that leads to poor antibiotics penetration, the adaptive responses to stress, and the formation of persister cells are hypothesized to constitute a multilayered defense, thus increasing the difficulty in eradication especially when combined with the resistant nature of the bacteria itself [33, 34]. Another study by Alcantar-Curiel et al. [35] proposed that while it appears that antibiotics resistance and bacterial capability to form biofilm play an important role in the global spread of K. pneumoniae, the clear relationship between these factors has not been fully identified and elaborated. This finding is confirmed by de Campos et al. [36] explaining that the capacity to form biofilms was not clearly correlated with clonal types of MDR bacteria.
It has been confirmed by several studies that in the majority of cases, a single antibiotic therapy is inadequate to annihilate biofilm-forming infections. In consequence, managing infections with currently accessible antibiotics and evaluating the outcomes have become important and urgent protocols for the successful treatment of biofilm-associated infections [29]. Several studies recommend a combination of antibiotic therapy with macrolides such as erythromycin, clarithromycin, and azithromycin as the main antibiotics for biofilm associated infection due to Gram-negative bacteria because of their high antibiofilm activity inside and outside of living organisms [37]. Taking into account the currently known environmental and bioecological aspects, Wu et al. [29] suggested that apart from the administration of combined antibiotics, removal of infected foreign bodies and source of infection as well as the administration of bacterial quorum sensing inhibitors or biofilm dispersal agents would result in a more effective management for biofilm infections.
Our study showed the antibiotic-resistant bacteria problems in hospital setting which are also reported by some studies [38, 39]. This situation is alarming when considering the quality and quantity of antibiotics prescriptions in most hospitals. Hospital surveillance in Surabaya conducted in 2012 demonstrated that 30.6% of antibiotics were administered without indications supported by susceptibility tests [40]. Thus, the prescription of antibiotics continues to become a formidable challenge in the world, including Indonesia. In the Netherlands, where antibiotic usage and antibiotic resistant bacteria are low, the antibiotics prescription protocols are not optimal as reported by van der Meer [41]. They reported that 15% of antibiotic therapy in surgical and internal medicine wards was not adequate [41].
Resistance mediated by ESBLs consists of penicillins, cephalosporins (including third-generation cephalosporins) and aztreonam. Dumaru et al. [42] performed a study to identify biofilm formation by Gram-negative bacteria and figured out their antibiograms along with the detection of ESBLs and Metallo-beta-lactamases (MBLs) production. There was a statistically significant association between MBL and biofilm production. However, the association between ESBL and biofilm production was not significant [42].
The major cause of inappropriate antibiotic prescribing is due to a lack of education about infection and antibiotic usage. One of the most relevant steps in antibiotic prescribing is an adjustment of initial antibiotic therapy based on the clinical microbiology result [43]. Therefore, it is essential to perform antibiotic susceptibility testing. Collecting clinical samples before antibiotic administration is also a critical point. Many physicians who prescribe antibiotics do not completely understand if their inappropriate prescriptions can have an impact on bacterial resistance development. Adjusting the initial antimicrobial therapy based on the clinical microbiology result will diminish the selection pressure to the microorganism in hospital-based infections. Thus, it is of paramount importance for each hospital to have an antibiotic guidance or stewardship program for all pharmacists and the physicians based on the most accurate microbiological data. In conjunction with this guidance, a continuous effort in hospital surveillance, infection control, and clinical audits must be conducted to fight against the rapid development of antibiotic-resistant pathogens [43].
Conclusions
Most of the K. pneumoniae isolates showed resistance to a wide range of antibiotics. Most of these were also shown to be biofilm producers of various capacities. Global efforts should be intensified to prevent the spread of multi-drug resistant bacteria and eliminate the hospital-born microbes that are causing a dramatic rise in mortality.
Availability of data and materials
All data analyzed or generated during this study are included in the submission. The raw data are available from the corresponding author on reasonable request.
Abbreviations
- AMR:
-
Antimicrobial resistance
- ESBLs:
-
Extended Spectrum β-Lactamases
- MBLs:
-
Metallo-beta-lactamases
- MDR:
-
Multi-drug resistant
- OD:
-
Optical density
- ODc:
-
Optical density cut-off
References
Podschun R, Ullmann U. Klebsiella spp. as nosocomial pathogens: epidemiology, taxonomy, typing methods, and pathogenicity factors. Clin Microbiol Rev. 1998;11(4):589–603.
Bengoechea JA, Sa Pessoa J. Klebsiella pneumoniae infection biology: living to counteract host defences. FEMS Microbiol Rev. 2019;43(2):123–44.
Munita JM1, Arias CA. Mechanisms of Antibiotic Resistance. Microbiol Spectr. 2016;4(2). https://doi.org/10.1128/microbiolspec.VMBF-0016-2015.
Prestinaci F, Pezzotti P, Pantosti A. Antimicrobial resistance: a global multifaceted phenomenon. Pathog Glob Health. 2015;109(7):309–18.
Wyres KL, Holt KE. Klebsiella pneumoniae as a key trafficker of drug resistance genes from environmental to clinically important bacteria. Curr Opin Microbiol. 2018;45:131–9.
Seifi K, Kazemian H, Heidari H, Rezagholizadeh F, Saee Y, Shirvani F, Houri H. Evaluation of biofilm formation among Klebsiella pneumoniae isolates and molecular characterization by ERIC-PCR. Jundishapur J Microbiol. 2016;9(1):e30682.
Ashurst JV, Dawson A. Klebsiella Pneumonia. In: StatPearls. Treasure Island (FL); 2019.
Donlan RM. Biofilms: microbial life on surfaces. Emerg Infect Dis. 2002;8(9):881–90.
LeChevallier MW, Cawthon CD, Lee RG. Factors promoting survival of bacteria in chlorinated water supplies. Appl Environ Microbiol. 1988;54(3):649–54.
Li B, Zhao Y, Liu C, Chen Z, Zhou D. Molecular pathogenesis of Klebsiella pneumoniae. Future Microbiol. 2014;9(9):1071–81.
Murphy CN, Clegg S. Klebsiella pneumoniae and type 3 fimbriae: nosocomial infection, regulation and biofilm formation. Future Microbiol. 2012;7(8):991–1002.
Vuotto C, Longo F, Balice MP, Donelli G, Varaldo PE. Antibiotic resistance related to biofilm formation in Klebsiella pneumoniae. Pathogens. 2014;3(3):743–58.
Parathon H, Kuntaman K, Widiastoety TH, Muliawan BT, Karuniawati A, Qibtiyah M, Djanun Z, Tawilah JF, Aditama T, Thamlikitkul V, et al. Progress towards antimicrobial resistance containment and control in Indonesia. BMJ. 2017;358:j3808.
Clinical and Laboratory Standards Institute (CLSI) Performance Standards for Antimicrobial Susceptibility Testing: Twenty-Fifth Informational Supplement. CLSI document M100-S25 2015.
Magiorakos AP, Srinivasan A, Carey RB, Carmeli Y, Falagas ME, Giske CG, Harbarth S, Hindler JF, Kahlmeter G, Olsson-Liljequist B, et al. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: an international expert proposal for interim standard definitions for acquired resistance. Clin Microbiol Infect. 2012;18(3):268–81.
Hassan A, Usman J, Kaleem F, Omair M, Khalid A, Iqbal M. Evaluation of different detection methods of biofilm formation in the clinical isolates. Braz J Infect Dis. 2011;15(4):305–11.
Kidd TJ, Mills G, Sa-Pessoa J, Dumigan A, Frank CG, Insua JL, Ingram R, Hobley L, Bengoechea JA. A Klebsiella pneumoniae antibiotic resistance mechanism that subdues host defences and promotes virulence. EMBO Mol Med. 2017;9(4):430–47.
Osagie RN, Eyaufe AA, Iserhienrhien O, Okodua M, Onuabonah F, Daibo OO. Antibiotic suscepbility profile of Klebsiella pneumoniae isolated from sputum samples amongst hospitalized adults in parts of Edo state, south-south. Niger Merit Res J. 2017;5(8):378–83.
Akter J, Chowdhury AMMA, Al FM. Study on prevalence and antibiotic resistance pattern of Klebsiella isolated from clinical samples in south east region of Bangladesh. American J Drug Discov Dev. 2014;4:73–9.
Yang D, Zhang Z. Biofilm-forming Klebsiella pneumoniae strains have greater likelihood of producing extended-spectrum beta-lactamases. J Hosp Infect. 2008;68(4):369–71.
Zheng JX, Lin ZW, Chen C, Chen Z, Lin FJ, Wu Y, Yang SY, Sun X, Yao WM, Li DY, et al. Biofilm formation in Klebsiella pneumoniae bacteremia strains was found to be associated with CC23 and the presence of wcaG. Front Cell Infect Microbiol. 2018;8:21.
Gunn JS, Bakaletz LO, Wozniak DJ. What's on the outside matters: the role of the extracellular polymeric substance of gram-negative biofilms in evading host immunity and as a target for therapeutic intervention. J Biol Chem. 2016;291(24):12538–46.
Meatherall BL, Gregson D, Ross T, Pitout JD, Laupland KB. Incidence, risk factors, and outcomes of Klebsiella pneumoniae bacteremia. Am J Med. 2009;122(9):866–73.
Wang C, Yuan Z, Huang W, Yan L, Tang J, Liu CW. Epidemiologic analysis and control strategy of Klebsiella pneumoniae infection in intensive care units in a teaching hospital of People's Republic of China. Infect Drug Resist. 2019;12:391–8.
Madahiah BM, Noor US, Abdul S, Dan Ali AQ. Klebsiella pneumoniae Urinary Tract Infections Associated with Long-term Catheterization and Spinal Cord Injuries. J Med Sci. 2002;2:227–9.
Cepas V, Lopez Y, Munoz E, Rolo D, Ardanuy C, Marti S, Xercavins M, Horcajada JP, Bosch J, Soto SM. Relationship between biofilm formation and antimicrobial resistance in gram-negative Bacteria. Microb Drug Resist. 2019;25(1):72–9.
GM N, CM G. Nagshetty K, Patil SA, Gaddad SM, Shivannavar CT: antibiotic susceptibility pattern of ESbetaL producing Klebsiella pneumoniae isolated from urine samples of pregnant women in Karnataka. J Clin Diagn Res. 2014;8(10):DC08–11.
Moini AS, Soltani B, Taghavi Ardakani A, Moravveji A, Erami M, Haji Rezaei M, Namazi M. Multidrug-resistant Escherichia coli and Klebsiella pneumoniae isolated from patients in Kashan. Iran Jundishapur J Microbiol. 2015;8(10):e27517.
Wu H, Moser C, Wang HZ, Hoiby N, Song ZJ. Strategies for combating bacterial biofilm infections. Int J Oral Sci. 2015;7(1):1–7.
Cherif-Antar A, Moussa-Boudjemaa B, Didouh N, Medjahdi K, Mayo B, Florez AB. Diversity and biofilm-forming capability of bacteria recovered from stainless steel pipes of a milk-processing dairy plant. Dairy Sci Technol. 2016;96:27–38.
Vuotto C, Longo F, Pascolini C, Donelli G, Balice MP, Libori MF, Tiracchia V, Salvia A, Varaldo PE. Biofilm formation and antibiotic resistance in Klebsiella pneumoniae urinary strains. J Appl Microbiol. 2017;123(4):1003–18.
Saha A, Devi KM, Damrolien S, Devi KS. Krossnunpuii, Sharma KT. Biofilm production and its correlation with antibiotic resistance pattern among clinical isolates of Pseudomonas aeruginosa in a a tertiary care hospital in north-East India. Int J Adv Med. 2018;5(4):964–8.
Stewart PS. Mechanisms of antibiotic resistance in bacterial biofilms. Int J Med Microbiol. 2002;292(2):107–13.
Agarwal A, Jain A. Association between drug resistance & production of biofilm in staphylococci. Indian J Med Res. 2012;135(4):562–4.
Alcántar-Curiel MD, Ledezma-Escalante CA, Jarillo-Quijada MD, et al. Association of Antibiotic Resistance, cell adherence, and biofilm production with the Endemicity of nosocomial Klebsiella pneumoniae. Biomed Res Int. 2018;7012958.
de Campos PA, Royer S, Batistao DW, Araujo BF, Queiroz LL, de Brito CS, Gontijo-Filho PP, Ribas RM. Multidrug resistance related to biofilm formation in Acinetobacter baumannii and Klebsiella pneumoniae clinical strains from different Pulsotypes. Curr Microbiol. 2016;72(5):617–27.
Ichimiya T, Takeoka K, Hiramatsu K, Hirai K, Yamasaki T, Nasu M. The influence of azithromycin on the biofilm formation of Pseudomonas aeruginosa in vitro. Chemotherapy. 1996;42(3):186–91.
Radji M, Fauziah S, Aribinuko N. Antibiotic sensitivity pattern of bacterial pathogens in the intensive care unit of Fatmawati hospital. Indonesia Asian Pac J Trop Biomed. 2011;1(1):39–42.
Saharman YR, Lestari DC. Phenotype characterization of Beta-lactamase producing enterobacteriaceae in the intensive care unit (ICU) of Cipto Mangunkusumo hospital in 2011. Acta Med Indones. 2013;45(1):11–6.
Hadi U. Kuntaman, Qibtiyah M, Paraton H. problem of antibiotic use and antimicrobial resistance in Indonesia: are we really making progress? Ind J Trop Infect Dis. 2013;4(4):5–8.
van der Meer JW, Gyssens IC. Quality of antimicrobial drug prescription in hospital. Clin Microbiol Infect. 2001;7(Suppl 6):12–5.
Dumaru R, Baral R, Shrestha LB. Study of biofilm formation and antibiotic resistance pattern of gram-negative bacilli among the clinical isolates at BPKIHS. Dharan BMC Res Notes. 2019;12(1):38.
Sudarmono P. Bacteria fight back, also in Indonesia! Acta Med Indones. 2013;45(1):1–2.
Acknowledgments
We thank dr. Hesty Lusinta Sp.MK, Margono, Erna YS, Retno W, Mulyani for their technical assistance. We also thank dr. Haniffah Pradipta Siwi for critical editing of the manuscript and dr. Yuli Mawarti, MPH for statistical analysis.
About this supplement
This article has been published as part of BMC Proceedings Volume 13 Supplement 11, 2019: Selected articles from the 3rd International Symposium on Congenital Anomaly and Developmental Biology 2019 (ISCADB 2019). The full contents of the supplement are available online at https://bmcproc.biomedcentral.com/articles/supplements/volume-13-supplement-11.
Funding
This study and its publication cost were supported by Faculty of Medicine, Public Health, and Nursing, Universitas Gadjah Mada
Author information
Authors and Affiliations
Contributions
HN, KS, FF, FW, SN, VPH, MSH, AM, ATA, TN conceived the study. HN, KS, FF, FW, SN, VPH, MSH, AM collected the data. HN,KS analyzed the data. HN, MSH, ATA, TN outlined the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Ethics approval for this study was obtained from Faculty of Medicine, Public Health and Nursing, Universitas Gadjah Mada, Yogyakarta, Indonesia with number KE/FK/0862/EC/2017. Written informed consent was obtained from all patients participating in this study.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Nirwati, H., Sinanjung, K., Fahrunissa, F. et al. Biofilm formation and antibiotic resistance of Klebsiella pneumoniae isolated from clinical samples in a tertiary care hospital, Klaten, Indonesia. BMC Proc 13 (Suppl 11), 20 (2019). https://doi.org/10.1186/s12919-019-0176-7
Published:
DOI: https://doi.org/10.1186/s12919-019-0176-7