Abstract
Background
Mycoplasma bovis (M. bovis) is a highly contagious cattle pathogen spreading worldwide and especially in Ningxia Hui Autonomous Region in China.
Results
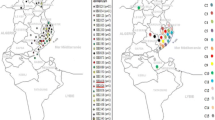
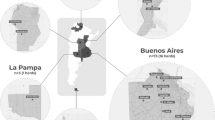
Two types of ST, ST10and ST134, were identified in Ningxia Hui Autonomous Region. Thirty-seven strains belonged to ST10 and 28 strains belonged to ST134. ST134 was a new ST and first found in 2009 and was only widely distributed in Ningxia Hui Autonomous Region at present. The M. bovis ST10 was widely spread in many provinces in China and was widespread in Ningxia Hui Autonomous Region since 2010. It is speculated that the prevalence of M. bovis ST10 in Ningxia Hui Autonomous Region began in 2010.
Conclusions
This study is the first report on the genetic diversity of M. bovis from 2009 to 2018 in Ningxia Hui Autonomous Region and provides the epidemiological information. These results may help further our understanding of the evolution of M. bovis and provide information that may be useful for the development of novel vaccines.
Similar content being viewed by others
Background
Mycoplasma bovis (M. bovis) is an important pathogen causing severe pneumonia, mastitis, and arthritis in the world. Especially, pneumonia caused by M. bovis has high morbidity and mortality. It is becoming one of the most widely recognized pathogens in the world[1, 2]. The pathogen is highly contagious and can spread rapidly throughout the herd. M. bovis can not only cause pneumonia, mastitis, arthritis, and otitis but also induce postpartum infection of the uterus with a mortality rate of 80%[3]. Since there are no effective vaccines and drugs to prevent and cure the disease caused by the pathogen, the incidence of the disease is on the rise[4, 5].
With the completion of whole-genome sequencing of M. bovis, a variety of highly repeatable molecular typing methods have been developed for molecular epidemiology and population structure research, including arbitrarily primed PCR (AP-PCR), random amplified polymorphic DNA (RAPD)[6], amplified fragment length polymorphism (AFLP)[7], pulsed-field gel electrophoresis (PFGE)[8], insertion sequence (IS)[9], variable number of tandem repeats (VNTR)[10, 11], multiple-locus variable-number tandem repeat (MLVA)[10] and multi-locus sequence typing (MLST)[5]. Although AP-PCR, RAPD, AFLP, and PFGE methods can obtain a large amount of genetic information, they are subjective in the analysis of DNA fragments and require special equipment[5], so it is difficult to establish a standardized method.
MLST is a rapidly developing molecular biology analysis method with high resolution in recent years. It is suitable for both molecular epidemiological studies and molecular advancement studies. The MLST method compares the nucleic acid sequences of the core fragments of several housekeeping genes and then compares the diversity of the alleles of the strains. Different strains correspond to different sequence types (ST)[4]. Through the STs of M. bovis pathogens can be used to understand the genetic diversity, population structure, and evolutionary trend, which will be beneficial to the control of M. bovis and the development of vaccines, as well as providing a theoretical basis for the prevention and control of M. bovis[4]. MLST is a typing technique based on seven housekeeping genes of M. bovis to study the genetic diversity, population structure, and evolutionary trend of M. bovis, including alcohol dehydrogenase-1 (adh-1), glutamate tRNA ligase (gltX), glycerol-3-phosphate dehydrogenase (gpsA), DNA gyrase subunit B (gyrB), phosphate acetyltransferase-2 (pta-2), thymidine kinase (tdk) and transketolase (tkt)[12]. The MLST data were used to populate a newly created and publicly available database (www.pubmlst.org/mbovis) intended to serve as a tool for epidemiologic studies and further investigating the population structure of M. bovis [5]. MLST is a powerful, scalable, and highly standardized method that makes it easy to clearly distinguish housekeeping genes among different strains[13].
Ningxia Hui Autonomous Region is one of the most important raising regions for cow and beef cattle in China[14]. The feeding level of bovine is currently ranked second in China. In 2009, our team isolated M. bovis for the first time from the lung of cows in Ningxia Hui Autonomous Region. From 2009 to 2018, a total of 65 strains of M. bovis were obtained from samples of lung, synovial fluid, nasal swab, and milk in Ningxia Hui Autonomous Region. However, there is no related study on the molecular epidemiology and population structure of M. bovis in Ningxia Hui Autonomous Region. In this study, The MLST method was used to classify 65 isolates from different cities of Ningxia Hui Autonomous Region from 2009 to 2018, aiming to investigate the population structure of M. bovis[2, 5] and to explore the evolutionary relationship of Ningxia Hui Autonomous Region isolates with Chinese isolates and global isolates, which will lay a foundation for further prevention and control of M. bovis in the world.
Results
Strains identification
Sixty-seven isolates were identified as M. bovis by PCR amplification using 16S rRNA and uvrC primers and sequencing of amplified products.
MLST analysis of M. bovis isolates
.
A total of 3 STs were identified among the 67 strains of M. bovis. Among the 65 isolates from Ningxia Hui Autonomous Region, 37 isolates belonged to ST10 and 28 isolates belonged to ST134. The HB0801 and PG45 isolates belonged to ST10 and ST17, respectively (Table 1).
According to Table 1, ST10 and ST134 strains could be isolated from samples of clinical mastitis (n = 9), arthritis (n = 22), and pneumonia (n = 34). The main clinical symptoms of calves were arthritis and pneumonia. The clinical symptoms of dairy cattle were mainly mastitis and pneumonia, while beef cattle were mainly pneumonia and arthritis.
The NX001 strain isolated from Ningxia Hui Autonomous Region for the first time in 2009 was ST134, and the NX002 strain isolated in 2010 was ST10. Since then, the ST10 and ST134 have been isolated from different lesions of bovine in different cities in Ningxia Hui Autonomous Region.
Phylogenetic analysis
The phylogenetic tree constructed from the concatenated sequences of the seven target genes revealed two distinct lineages (Fig. 1). The ST173 and ST17 were in the same lineage. The other STs were in the other lineage including ST10, ST26, ST32, ST43, ST172, and the Ningxia Hui Autonomous Region isolates of ST134.
Discussion
Molecular epidemiological studies are of great significance to reveal the population structure, genetic diversity, and prevalence of Mycoplasma spp [4, 15], which facilitates the formulation of effective prevention and control measures, including the development of vaccines and diagnostic methods[4, 16]. MLST studies performed on 44 strains from nine Chinese provinces from 2008 to 2014 showed that ST10, ST32, and ST43 were found in Hubei province (n= 25) and that ST10 was also found in Anhui (n = 1), Fujian (n = 2), Hunan (n = 1), Henan (n = 8), Inner Mongolia (n = 1), Jiangxi (n = 3), Guangzhou (n = 2), and Shandong (n = 1) province [4]. However, there are no reports about the MLST study of M. bovis in Ningxia Hui Autonomous Region. Therefore, this study is the first report on the molecular epidemiology of M. bovis from 2009 to 2018 in Ningxia Hui Autonomous Region.
At present, there are seven STs in M. bovis in China including ST10, ST26, ST32, ST43, ST134, ST172, and ST173[4]. ST10 is widely prevalent in all provinces reported in China, including Ningxia Hui Autonomous Region (n = 37, 56.9%) where are no MLST reports. After the ST10 strain was first isolated in 2010 in Ningxia Hui Autonomous Region, the ST10 of M. bovis could be isolated and identified every year. And it has been widely diffused in different cities in Ningxia Hui Autonomous Region. Interestingly, the ST10 strains have been reported that were widely distributed in American, Australia, and Israel[4]. Previously, the international spread of contagious bovine pleuropneumonia was shown to be linked to the movement of cattle[4, 17, 18]. So, it is a similar transfer probably that through international movement of cattle and domestic movement of cattle caused the widespread distribution of the M. bovis ST10 in China and even in Ningxia Hui Autonomous Region. The reason for the widespread prevalence of ST10 strain in Ningxia Hui Autonomous Region may be that the Ningxia Hui Autonomous Region government strongly supports the construction of large-scale cattle breeding parks. For their expansion, intensive cattle farms had to purchase cattle from different provinces of China and different countries in the world, but M. bovis was ignored, which led to the widespread presence of exogenous M. bovis in Ningxia Hui Autonomous Region. It is speculated that the prevalence of M. bovis ST10 in Ningxia Hui Autonomous Region began in 2010.
In 2009, our lab isolated M. bovis from the lung tissue of cows for the first time, which was ST134. From 2009 to 2018, ST134 (n = 28, 43.1%) was isolated and identified from different cities of Ningxia Hui Autonomous Region. However, it has not been identified in other provinces in China. Therefore, it confirmed that ST134 strains were closely related strains with the same origin in Ningxia Hui Autonomous Region, China, and has been widely distributed in Ningxia Hui Autonomous Region for many years.
To evaluate the evolutionary relationship of M. bovis between isolates from Ningxia Hui Autonomous Region and isolates from other provinces in China, a phylogenetic tree was constructed based on concatenated sequences. The evolutionary analysis showed that all ST10 strains were in the same lineage as ST26, ST32, ST43, ST172, including Ningxia Hui Autonomous Region ST134 strains. However, the ST173 strains were in the same lineage as the ST17 strain. The study results indicate that the M. bovis strains with STs different from ST173 and ST17 were closely related strains with the same origin.
Conclusions
This study revealed the genetic diversity of M. bovis from 2009 to 2018 in Ningxia Hui Autonomous Region and provides epidemiological information. ST10 strains were widely prevalent in Ningxia Hui Autonomous Region as well as all provinces of China that have been reported, and ST134 strains were also widely distributed in Ningxia Hui Autonomous Region.
These results may help further our understanding of the evolution of M. bovis and provide information that may be useful for the development of novel vaccines.
Methods
Strains
The detailed information of sixty-seven strains of M. bovis was listed in Table 1, including the host, isolation site, geographical location, and the number of strains. Sixty-five strains came from different farms and were isolated from nasal swab, or milk, or joint fluid, or lung of M. bovis-infected cattle in Ningxia Hui Autonomous Region from 2009 to 2018. HB0801 strain was isolated from the lungs of beef cattle in Hubei Province in 2008 when M. bovis was first reported in China[19]. PG45 strain (ATCC 25,523) was donated by Professor Aizhen Guo of Huazhong Agricultural University of China.
Cultivation and identification of M. bovis.
The M. bovis strains were cultured in PPLO broth(BD DifcoTM, US California)[4, 20]. M. bovis genomic DNA was extracted using a bacterial DNA extraction kit (Tiangen, Beijing, China)[21]. The16S rRNA gene[22] and uvrC gene[23] were amplificated using two pairs of primers (Table 2). The PCR reaction mixture was 50 µL (5 µL 10 × PCR buffer, 4 µL dNTP mixture, 0.25 µL rTaq, 2 µL primers, 1 µL DNA, 37.75µL ddH2O). The reaction mixture was incubated at 95 ℃ for 2 min, 35 cycles of 95 ℃ for 30 s, 50 ℃ for 20 s and 72 ℃ for 2 min, then a final incubation at 72 ℃ for 8 min[22, 23].
Multi-locus sequence typing
The genes of 67 isolates of M. bovis were amplified by PCR using MLST scheme (adh-1, gltX, gpsA, gyrB, pta-2, tdk and tkt)[4, 5] (Table 2). The PCR reaction mixture was 50 µL (5 µL 10 × PCR buffer, 4 µL dNTP mixture, 0.25 µL rTaq, 2 µL primers, 1 µL DNA, 37.75µL ddH2O). The reaction mixture was incubated at 95 ℃ for 2 min, 35 cycles of 95 ℃ for 30 s, 55 ℃ for 30 s and 72 ℃ for 1 min, then a final incubation at 72 ℃ for 5 min[5]. The PCR amplification products were sent to Shanghai Bioengineering Co., Ltd for sequencing. The assembly sequences were aligned by the http://pubmlst.org/mbovis/ database to obtain the allele number and STs.
Phylogenetic analysis
Seven gene sequences of 65 strains of M. bovis in Ningxia Hui Autonomous Region and the strains of different STs in China and PG45 reference strains were concatenated. A phylogenetic tree was constructed from concatenated sequences. The evolutionary history was inferred by using the Maximum Likelihood method based on the Kimura 2-parameter model[3] of MEGA 10.0. Initial tree(s) for the heuristic search was obtained automatically by applying Neighbor-Join and BioNJ algorithms to a matrix of pairwise distances estimated using the Maximum Composite Likelihood (MCL) approach and then selecting the topology with superior log likelihood value[3].
Availability of data and materials
The datasets generated and/or analysed during the current study are available in the National Center for Biotechnology Information repository (MW19432 - MW194385, MW194386 - MW194450, MW194451 - MW194515, MW194516 - MW194580, MW194581 - MW194645, MW194646 - MW194710, MW194711 - MW194775).
Abbreviations
- adh-1:
-
Alcohol dehydrogenase-1
- AFLP:
-
Amplified fragment length polymorphism
- AP-PCR:
-
Arbitrarily primed PCR
- ddH2O:
-
Doble distilled water
- DNA:
-
Deoxyribonucleic acid
- gltX:
-
Glutamate tRNA ligase
- gpsA:
-
Glycerol-3-phosphate dehydrogenase
- gyrB:
-
DNA gyrase subunit B
- IS:
-
Insertion sequence
- M. bovis:
-
Mycoplasma bovis
- MLST:
-
Multi-locus sequence typing
- PCR:
-
Polymerase chain reaction
- PFGE:
-
Pulsed-field gel electrophoresis
- PPLO:
-
Pleuropneumonia-like organisms
- pta-2:
-
Phosphate acetyltransferase-2
- RAPD:
-
Random amplified polymorphic DNA
- RNA:
-
Ribosomal ribonucleic acid
- ST:
-
Sequence types
- tdk:
-
Thymidine kinase
- tkt:
-
Transketolase
- VNTR:
-
Variable number of tandem repeats
References
Gille L, Pilo P, Valgaeren BR, Van Driessche L, Van Loo H, Bodmer M, Burki S, Boyen F, Haesebrouck F, Deprez P, Pardon B. A new predilection site of Mycoplasma bovis: Postsurgical seromas in beef cattle. Vet Microbiol. 2016;186:67–70.
Register KB, Woodbury MR, Davies JL, Trujillo JD, Perez-Casal J, Burrage PH, Clark EG, Windeyer MC. Systemic mycoplasmosis with dystocia and abortion in a North American bison (Bison bison) herd. J Vet Diagn Invest. 2013;25(4):541–5.
Burki S, Spergser J, Bodmer M, Pilo P. A dominant lineage of Mycoplasma bovis is associated with an increased number of severe mastitis cases in cattle. Vet Microbiol. 2016;196:63–6.
Menghwar H, He C, Zhang H, Zhao G, Zhu X, Khan FA, Faisal M, Rasheed MA, Zubair M, Memon AM, Ridley A, Robertson ID, Chen Y, Guo A. Genotype distribution of Chinese Mycoplasma bovis isolates and their evolutionary relationship to strains from other countries. Microb Pathog. 2017;111:108–17.
Register KB, Thole L, Rosenbush RF, Minion FC. Multilocus sequence typing of Mycoplasma bovis reveals host-specific genotypes in cattle versus bison. Vet Microbiol. 2015;175(1):92–8.
Butler JA, Pinnow CC, Thomson JU, Levisohn S, Rosenbusch RF. Use of arbitrarily primed polymerase chain reaction to investigate Mycoplasma bovis outbreaks. Vet Microbiol. 2001;78(2):175–81.
Soehnlen MK, Aydin A, Murthy KS, Lengerich EJ, Hattel AL, Houser BA, Fenton GD, Lysczek HR, Burns CM, Townsend AM, Brooks JW, Wolfgang DR, Jayarao BM. Epidemiology of Mycoplasma bovis in Pennsylvania veal calves. J Dairy Sci. 2012;95(1):247–54.
Arcangioli MA, Aslan H, Tardy F, Poumarat F, Le Grand D. The use of pulsed-field gel electrophoresis to investigate the epidemiology of Mycoplasma bovis in French calf feedlots. Vet J. 2012;192(1):96–100.
Miles K, McAuliffe L, Persson A, Ayling RD, Nicholas RA. Insertion sequence profiling of UK Mycoplasma bovis field isolates. Vet Microbiol. 2005;107(3–4):301–6.
Pinho L, Thompson G, Rosenbusch R, Carvalheira J. Genotyping of Mycoplasma bovis isolates using multiple-locus variable-number tandem-repeat analysis. J Microbiol Methods. 2012;88(3):377–85.
Spergser J, Macher K, Kargl M, Lysnyansky I, Rosengarten R. Emergence, re-emergence, spread and host species crossing of Mycoplasma bovis in the Austrian Alps caused by a single endemic strain. Vet Microbiol. 2013;164(3–4):299–306.
Manso-Silvan L, Dupuy V, Lysnyansky I, Ozdemir U, Thiaucourt F. Phylogeny and molecular typing of Mycoplasma agalactiae and Mycoplasma bovis by multilocus sequencing. Vet Microbiol. 2012;161(1–2):104–12.
Yeo M, Mauricio IL, Messenger LA, Lewis MD, Llewellyn MS, Acosta N, Bhattacharyya T, Diosque P, Carrasco HJ, Miles MA. Multilocus sequence typing (MLST) for lineage assignment and high resolution diversity studies in Trypanosoma cruzi. PLoS Negl Trop Dis. 2011;5(6):e1049.
Tan C, Yang J, Li M. Temporal-Spatial Variation of Drought Indicated by SPI and SPEI in Ningxia Hui Autonomous Region, China. Atmosphere. 2015;6(10):1399–421.
Olaogun OM, Kanci A, Barber SR, Tivendale KA, Markham PF, Marenda MS, Browning GF. Genetic diversity of Mycoplasma arginini isolates based on multilocus sequence typing. Vet Microbiol. 2015;180(1–2):123–8.
Dos Santos LF, Sreevatsan S, Torremorell M, Moreira MA, Sibila M, Pieters M. Genotype distribution of Mycoplasma hyopneumoniae in swine herds from different geographical regions. Vet Microbiol. 2015;175(2–4):374–81.
Fisher J. The origins, spread and disappearance of contagious bovine pleuro-pneumonia in New Zealand. Aust Vet J. 2006;84(12):439–44.
Xin J, Li Y, Nicholas RA, Chen C, Liu Y, Zhang MJ, Dong H. A history of the prevalence and control of contagious bovine pleuropneumonia in China. Vet J. 2012;191(2):166–70.
Qi J, Guo A, Cui P, Chen Y, Mustafa R, Ba X, Hu C, Bai Z, Chen X, Shi L, Chen H. Comparative geno-plasticity analysis of Mycoplasma bovis HB0801 (Chinese isolate). PLoS One. 2012;7(5):e38239.
Rasheed MA, Qi J, Zhu X, Chenfei H, Menghwar H, Khan FA, Zhao G, Zubair M, Hu C, Chen Y, Chen H, Guo A. Comparative Genomics of Mycoplasma bovis Strains Reveals That Decreased Virulence with Increasing Passages Might Correlate with Potential Virulence-Related Factors. Front Cell Infect Microbiol. 2017;7:177.
Li J, Zuo X, Lv X, Kong F, Xu W, Yang S. Association of DNA repair gene polymorphisms with response to chemotherapy and prognosis of gastric cancer in a Chinese population. Tumour Biol. 2014;35(8):7569–74.
Konigsson MH, Bolske G, Johansson KE. Intraspecific variation in the 16S rRNA gene sequences of Mycoplasma agalactiae and Mycoplasma bovis strains. Vet Microbiol. 2002;85(3):209–20.
Subramaniam S, Bergonier D, Poumarat F, Capaul S, Schlatter Y, Nicolet J, Frey J. Species identification of Mycoplasma bovis and Mycoplasma agalactiae based on the uvrC genes by PCR. Mol Cell Probes. 1998;12(3):161–9.
Acknowledgements
The authors thank the staff of the Clinical Veterinary Laboratory, School of Agricultural of Ningxia University for their unremitting efforts in the isolation and identification of M. bovis since 2009. Thanks to Professor Aizhen Guo from Huazhong Agricultural University for donating PG45 and HB0801 strains. Thanks to the Ningxia Hui Autonomous Region Science and Technology Department for providing funds for this work.
Funding
This work was funded by Ningxia Hui Autonomous Region Department of Science and Technology for supporting the project funding (2019BBF02005). This funding body plays no role in the design of the study and collection, analysis, and interpretation of data and in writing the manuscript.
Author information
Authors and Affiliations
Contributions
YNG, SHH, and YL conceived and designed the experiments; YNG and HFL analyzed the data and drafted the manuscript; YNG, YYL, and SQG performed experiments and acquired data. YNG, SHH, and HFL revised the manuscript. All authors read and approved the final manuscript.
Authors’ information
YNG, HFL, and SHH: Department of Veterinary Clinical Sciences, School of Agriculture, Ningxia University, Yinchuan 750,021, Ningxia Hui Autonomous Region, China. SQG: Technology Center Department, Yinchuan Customs, Yinchuan 750,000, Ningxia Hui Autonomous Region, China. YYL: Animal Disease Prevention and Control Center of Yan’an, Yan’an 716,000, Shaanxi, China YL: Department of Life Science and Technology, Ningxia Polytechnic College, Yinchuan 750,021, Ningxia Hui Autonomous Region, China.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
This study was submitted to and approved by the Laboratory Animal Ethical and Welfare Committee of Ningxia University (Approval No. NXU-ACAU-2018-124). All owners of farms who participated in this study provided their verbal consent based on the long and tight cooperative fellowship between the owners and our department, which was approved by Laboratory Animal Ethical and Welfare Committee of Ningxia University.
Consent for publication
Not applicable.
Competing interests
All authors declare that they have no conflict of interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Guo, Y., Luo, H., Guo, S. et al. Multi-locus sequence typing of Mycoplasma bovis to assess its genetic diversity from 2009 to 2018 in Ningxia Hui Autonomous Region, China. BMC Vet Res 16, 454 (2020). https://doi.org/10.1186/s12917-020-02668-x
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12917-020-02668-x