Abstract
Background
Borrelia burgdorferi sensu lato, the agent of Lyme borreliosis, is mainly maintained in natural foci through the transmission cycles of competent tick vectors (Ixodes sp.) and a vertebrate reservoir. Specific rodents have been identified as the principal reservoir of Borrelia burgdorferi sensu lato in Europe. Borrelia miyamotoi is the only relapsing fever spirochete transmitted by the same tick. The aim of the present study was to perform an epidemiological survey to determine the presence of B. burgdorferi sensu lato in rodents occurring in Slovenia and to explore the presence of Borrelia miyamotoi.
The study was performed in two parts, retrospective and prospective; a total of 297 rodents was analyzed. Detection and identification of borrelia was performed by molecular methods and additionally in the prospective study by isolation and genotyping (MluI-LRFP and MLST).
Results
During the prospective part of the study, borrelia was isolated from 2/46 (4.3 %) lung specimens and from 10/46 (21.7 %) heart specimens of rodents. All isolated strains were identified as B. afzelii subtype Mla1, and MLST analysis revealed 5 distinct sequence types. Borrelia DNA was successfully detected by one or other of the PCR methods in 18/46 (39.1 %) and 75/251 (29.9 %) samples in the prospective and retrospective studies, respectively. LightMix® was found to be more sensitive than the ‘’in-house” nested PCR (91/297 (30.6 %) vs 48/297 (16.1 %)).
Borrelia miyamotoi DNA was detected in 1/251 (0.4 %) and in 1/46 (2.2 %) heart specimens, in the retrospective and prospective parts of the study, respectively.
Conclusion
We determined the prevalence of B. afzelii in rodents and report for the first time the presence of B. miyamotoi in Slovenia.
Similar content being viewed by others
Background
Borrelia burgdorferi sensu lato is the causative agent of Lyme borreliosis, the most common tick transmitted infection in Slovenia: in 2012, the incidence of the disease was 239.8 cases/100,000 inhabitants [1]. At least four species are among the most important pathogenic species in Europe, Borrelia afzelii, Borrelia garinii, Borrelia burgdorferi sensu stricto and Borrelia spielmanii, forming part of the B. burgdorferi sensu lato complex [2]. All strains are mainly maintained in natural foci through the transmission cycles of competent tick vectors and a vertebrate reservoir. Specific rodents have been identified as the principal reservoir host of B. burgdorferi sensu lato in Europe, such as wood mouse (Apodemus sylvaticus), yellow-necked mouse (Apodemus flavicollis) and bank vole (Clethrionomys glareous) [3, 4]. Several other vertebrates have also been described as competent reservoir hosts, such as birds, dormice, hedgehogs, rats, squirrels, hares and lizards [5]. Borrelia is maintained for long time periods in a competent reservoir host, often lifelong [5].
Infected Ixodes ticks transmit borrelia to mammals through a bite. In Europe and Asia, Ixodes ricinus and Ixodes persulcatus ticks are the main vectors for B. garinii, B. afzelii, and B. burgdorferi sensu stricto transmission, while the deer tick Ixodes scapularis transmits B. burgdorferi sensu stricto in North America [6].
Borrelia miyamotoi is the only relapsing fever spirochete transmitted by hard ticks of the Ixodes species. Infection with B. miyamotoi has recently been reported to be associated with symptomatic febrile illnesses in humans in Eurasia and North America [7–9]. B. miyamotoi has been detected in urinary bladder and/or blood of wild rodents Apodemus argenteus, Apodemus speciosus, Myodes rufocanus and Myodes rutilus [10].
The aim of the present study was to perform an epidemiological survey to determine the presence of B. burgdorferi sensu lato in rodents occurring in Slovenia and to explore the presence of Borrelia miyamotoi. Additionally, we wanted to compare the results of two specific PCR, amplifying different parts of the ospA gene, in order to determine the presence of borrelia in two different rodent tissues: heart and lung.
Methods
The study was performed in two parts, retrospective and prospective.
In the retrospective part, we analyzed a total of 251 heart biopsies from rodents (105 females, 146 males) live trapped in spring and autumn of 2012 in the Central Slovenian region.
All rodents were identified to species level and heart biopsies were stored at −80 °C until tested.
The prospective part of the study took place in spring and autumn of 2013; 46 rodents (24 females, 23 males) were live trapped in the Central Slovenian region. After identification to species level, lungs and hearts were removed. Each specimen was dissected into two equal parts: one was immediately inoculated into modified Kelly-Pettenkofer medium and incubated at 33 °C for borrelia isolation, [11] while the other was frozen at −80 °C for PCR analysis.
Identification of rodents
Morphological species determination was confirmed by PCR and sequencing of the partial mitochondrial cytochrome b gene [12]. Rodent species were determined by sequence comparisons using the BLAST algorithm (www.ncbi.nlm.nih.gov).
Culture, isolation and characterization of borrelia strains
The lung and heart samples from the prospective study were incubated at 33 °C and examined weekly for the presence of spirochetes using dark-field microscopy. The samples were considered negative if no growth was detected after 9 weeks of incubation [13].
Genotypic characterization of isolated borrelia strains was performed using large restriction fragment pattern (LRFP) after MluI restriction and multilocus sequence typing (MLST) analysis.
MluI-LRFP was performed as previously described [13]. Briefly, borrelial DNA was isolated by the gel insert method, digested with restriction endonuclease MluI, and restricted fragments were separated by pulse-field gel electrophoresis (run time: 24 h, ramping time: 3–40 s). Borrelia species and subtypes were identified by their specific restriction profiles [13].
For MLST analyses, borrelial DNA was isolated using the InstaGene matrix (BioRad, USA) according to the manufacturer’s instructions. Eight chromosomal housekeeping genes were amplified by nested PCR and sequenced in both directions, as previously described by Margos et al. [14]. Sequences were analyzed using CLC Main Workbench 6.9.1, MLST Module (CLC bio, Denmark) and assigned allelic numbers and sequence types. New sequence types were added to the MLST database hosted by Imperial College London (London, UK) and are accessible at the MLST website (http://borrelia.mlst.net/).
Nucleic acid isolation from tissue specimens
DNA was isolated from heart and lung biopsies according to the manufacturer’s recommendations using a MagNA Pure Compact Total Nucleic Acid Isolation Kit, Roche, Germany and QIAamp DNA Mini Kit, Qiagen, in prospective and retrospective parts of the study, respectively.
Detection of Borrelia burgdorferi sensu lato DNA
The presence of B. burgdorferi sensu lato DNA was determined using two different PCRs, ‘’in-house” nested PCR and commercial real-time PCR (LightMix® Kit for detection of Borrelia spp., TIB MOLBIOL GMBh, Germany), both targeting the ospA gene.
LightMix® Kit for detection of Borrelia spp
A fragment from the Borrelia ospA gene was amplified with specific primers and detected with hybridization probes according to the manufacturer’s recommendations. In addition to amplification, the kit enables identification of Borrelia species by melting temperature (Tm) analysis. The PCR reaction is monitored by an additional PCR product to detect possible inhibition.
In-house nested PCR of ospA
Nested PCR was performed using primers for amplification of the ospA gene described by Guy and Stanek [15], under the following conditions. The reaction was carried out in 30 cycles of 95 °C for 45 s, 50 °C for 45 s and 72 °C for 60 s. Each sample was transferred to a second reaction and amplified under the same conditions for another 30 cycles [16]. One positive and three negative control samples were included in each experiment to control for amplification and contamination. To avoid PCR contamination and amplicon carry-over, samples were processed in separate rooms, and the use of plugged pipette tips was obligatory [17]. PCR amplification resulted in a 351 bp product. Amplification products were analyzed on SYBR Safe DNA gel stain (Invitrogen, USA) stained 1 % agarose gels. Identification of the species was done by sequencing the PCR product. Amplicons were purified and sequenced on ABI3500 (Applied Biosystem, California, USA). Sequences were analyzed with CLC Main Workbench 6.0 (CLC Bio, Denmark) and compared with the BLAST database (www.ncbi.nlm.nih.gov/blast/Blast.cgi).
Detection of Borrelia miyamotoi
The presence of B. miyamotoi DNA was detected using a real-time protocol and specific primers targeting the 16S rRNA gene, as described by Platonov et al. [8]. To confirm the specificity of the real-time protocol, an amplified flagellin gene was direct sequenced into all positive samples [8]. Nucleotide sequences were aligned, compared and analyzed by CLC Main Workbench 6.0 (CLC Bio, Denmark) and BLAST (www.ncbi.nlm.nih.gov/blast/Blast.cgi).
Statistical analysis
Statistical analyses were performed using SPSS Statistics 18.0 (Chicago: SPSS Inc). A 95 % confidence interval (CI) was calculated for the prevalence of Borrelia sp. in rodents.
Ethical statement
Rodents used in this study were collected from nature in collaboration with the Slovenian Museum of Natural History, with the approval of the Ministry of Agriculture, Forestry and Food, and the Administration of the Republic of Slovenia for Food Safety, Veterinary Sector and Plant Protection (323-02-251/2004/7).
Results
Identification of rodents
The study included 297 rodent species, 46 in the prospective and 251 in the retrospective part.
During the prospective study, eighteen out of 46 (39.1 %) rodents were identified as yellow necked mice (Apodemus flavicollis), 22/46 (47.8 %) as bank voles (Myodes glareolus), 5/46 (10.9 %) as Mediterranean water shrews (Neomys anomalus) and 1/46 (2.2 %) as a lesser white-toothed shrew (Crocidura suaveolens).
The retrospective study included 251 rodents, of which 155 (61.8 %) were identified as yellow necked mice (Apodemus flavicollis), 94 (37.5 %) as bank voles (Myodes glareolus), and 2 (0.8 %) as wood mice (Apodemus sylvaticus).
Cultivation, isolation and genotypic characterization of borrelia strains
Cultivation was performed on 46 rodents collected in the prospective part of the study. Borrelia sp. was isolated from 2/46 (4.3 %) lung specimens and from 10/46 (21.7 %) heart specimens of 7 Myodes glareolus and 4 Apodemus flavicollis. Borrelia sp. was simultaneously isolated from heart and lung specimens of 1 Myodes glareolus (Table 1).
MluI-LRFP analysis revealed all isolates as B. afzelii Mla1. Results are shown in Table 1.
MLST analysis of 12 isolated B. afzelii strains revealed 5 distinct sequence types (ST); 3 have been previously reported, 2 were assigned a new ST number (Table 1).
Detection of borrelial DNA
Prospective study
In lung specimens, borrelial DNA was detected in 10/46 (21.7 %) and 8/46 (17.4 %) rodents, using LightMix® and ‘’in-house” nested PCR, respectively; the concordance of the two PCR methods was 42/46 (91.3 %) (Table 2). In heart specimens, borrelial DNA was detected in 17/46 (37.0 %) and 18/46 (39.1 %) rodents with LightMix® and ‘’in-house” nested PCR, respectively; the concordance of the two tests was 45/46 (97.8 %). Borrelia sp. DNA was detected by one or other of the applied methods in 18/46 (39.1 %, 95 % CI: 25.0–53.2 %) rodent samples.
Identification according to Tm using LightMix® revealed the presence of B. afzelii/B. valaisiana; LightMix® kit does not allow a distinction between these two species based on differences in melting temperatures [18]. Sequencing of ‘’in-house” OspA PCR products revealed the presence of B. afzelii, only.
Retrospective study
Borrelial DNA was detected in 74/251 (29.5 %) and 30/251 (12.0 %) heart biopsies with LightMix® and ‘’in-house” nested PCR, respectively; the concordance of the two tests was 209/251 (83.3 %) (Table 2). Borrelia sp. DNA was detected by one or other of the applied methods in 75/251 (29.9 %, 95%CI: 24.2–35.6 %) rodent samples.
As previously reported, identification using Tm analysis revealed the presence of B. afzelii/B. valaisiana; sequencing of ‘’in-house” OspA PCR products revealed the presence of B. afzelii, only.
Detection of Borrelia miyamotoi
Borrelia miyamotoi DNA was detected in 1/251 (0.4 %) heart specimens of Apodemus flavicollis in the retrospective part of the study and in 1/46 (2.2 %) heart specimens of Apodemus flavicollis in the prospective part of the study.
Direct sequencing of PCR product of the flagellin gene of both positive samples revealed 99.58 % and 99.33 % identity with B. miyamotoi strain KT12F-IR and B. miyamotoi strain OS179m-07, respectively.
Discussion
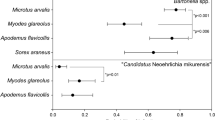
Borrelia burgdorferi sensu lato utilizes an extremely wide range of hosts, including rodents, birds and lagomorphs [19]. Birds are mainly host for B. garinii, whereas B. afzelii, B. burgdorferi sensu stricto and B. bavariensis utilize rodents as their main reservoir host [20]. In Europe, Apodemus spp. and M. glareolus have been reported to transmit B. miyamotoi to 23.8 % of xenodiagnostic ticks [21].
Since the study by Zore et al. [22], no epidemiological data concerning the presence B. burgdorferi sensu lato in rodents and ticks have been reported from Slovenia. In addition, the presence of Borrelia miyamotoi in Slovenia was neither screened nor previously reported.
In the prospective part of the study, detection of borrelia DNA was successful in 18/46 (39.1 %) samples, while the corresponding result for the retrospective part of the study was 75/251 (29.9 %), detected by one or other of the applied methods. Comparing the two PCR methods, LightMix® was found to be more sensitive than the ‘’in-house” nested PCR (91/297 (30.6 %) vs 48/297 (16.1 %)). Benefits of LightMix® also include ease of performance, rapidity, ability of species discrimination and lower chance of contamination.
The prevalence of borrrelia infection in host animals in our study is comparable with that reported in the study by Zore et al. [22], in which borrelial DNA was detected in 17/34 rodents (50 % prevalence). The prevalence reported in our study is higher than in the study by Schmidt et al. [23], in which borrelia DNA was detected in 14.8 % (16/110) of rodents trapped in the northern part of Austria. Generally, the reasons for differences in prevalence among different studies can be explained by different methods of sample collection, sample sizes, detection methods, geographic locations and time periods [24]. A reliable and standardized approach for such studies is not defined.
Identification according to Tm analysis of the ospA gene using LightMix® revealed the presence of B. afzelii or B. valaisiana; such an analysis does not allow a distinction between these two species, which is a great disadvantage of the kit [18]. Since LightMix is a commercial real-time PCR and sequences of the primers and hybridization probes are not available, sequencing of the product is not possible, which is the main limitation of the assay. On the other hand, comparison of the sequence of our in-house PCR, targeting the ospA gene, with the NCBI Blast database revealed the presence of B. afzelii, only. These results are in accordance with the study by Schmidt et al., in which B. afzelii was identified in the majority of the rodents skin samples [23]. In the study by Perez et al., [25] B. afzelii was also the dominant species and the authors concluded that rodents transmitted only B. afzelii to ticks. In the previously performed Slovenian study, the most commonly found species was B. afzelii, but B. burgdorferi sensu stricto and B. garinii were also detected [22].
We were able to isolate borrelia from heart and/or lung of 4/18 (22.2 %) yellow-necked mice (Apodemus flavicollis), 6/22 (27.3 %) bank voles (Myodes glareolus) and from none of the Mediterranean water shrews (Neomys anomalus) or lesser white-toothed shrew (Crocidura suaveolens). All isolated strains were identified using MluI-LRFP as B. afzelii subtype Mla1. Although we expected B. garini and B. burgdorferi sensu stricto to be isolated in our study, already reported in Slovenian rodents [22], we did not find them.
Our results are consistent with the results of previous studies, in which it was shown that rodent sera is borreliacidal for B. garinii [26]. It was additionally found that the wood mouse is far more competent for B. afzelii than for B. burgdorferi sensu stricto [27] and that B. afzelii is specifically maintained by European rodents [28].
Borrelia sp. can be isolated and/or PCR detected in various organs of small mammals [29]. In the study by Khanakah et al., it was demonstrated that Borrelia strains were more frequently cultured from specimens of the bladder wall than from heart muscle. On the other hand, PCR of heart specimens was more often positive than culture of bladder wall and heart muscle [30]. To our knowledge, no study comparing PCR detection in heart, lung and urinary bladder has been published. The main limitation of our study is the absence of urinary bladder samples.
MLST analysis of 12 isolates from rodent specimens revealed 5 distinct sequence types (Table 1); 3 ST have been previously identified, while 2 ST were assigned new ST numbers (Table 1). B. afzelii ST 335, which was found in 6 Slovenian isolates, was also previously identified in two Italian isolates from I. ricinus ticks, B. afzelii ST 215 (found in one Slovenian isolate) in three isolates originating from Latvia and one from Italy, B. afzelii ST 342 (found in one Slovenian isolate) in one isolate from I. ricinus from Austria (http://borrelia.mlst.net/sql/burstspadvanced.asp).
Both B. afzelii isolates with new sequence types (ST549 and 550) originated from Myodes glareous and were added to the MLST database.
Borrelia miyamotoi DNA was detected in only two heart samples of A. flavicollis mice, resulting in a low overall prevalence of 2/297 (0.7 %); more precisely in 0.4 % and 2.2 % in the retrospective and prospective parts of the study, respectively. The results of the study provide the first evidence of B. miyamotoi presence in Slovenia.
In the study by Cosson et al. [31], the prevalence of B. miyamotoi in ticks and bank voles was 3.0 % (8/267 ticks) and 5.55 % (4/72 bank voles), respectively. A prevalence of 1.5 % was determined in the study by Taylor et al. [10], in which urinary bladders of rodents were tested. In terms of the prevalence of Lyme borreliosis borrelia, B. miyamotoi is rarely found in reservoir hosts in Slovenia.
Conclusion
We determined the prevalence of B. burgdorferi sensu lato in various rodents in Slovenia. Using molecular methods, detection of borrelia DNA was successful in 18/46 (39.1 %) and 75/251 (29.9 %) samples, in the prospective and retrospective parts of the study, respectively. LightMix® was found to be more sensitive than ‘’in-house” nested PCR (91/297 (30.6 %) vs 48/297 (16.1 %)), easy to perform, fast and not prone to contamination.
Identification of Borrelia species revealed the presence of B. afzelii. We report the presence of B. miyamotoi for the first time in Slovenia.
Abbreviations
- PCR:
-
Polymerase chain reaction
- DNA:
-
Deoxyribonucleic acid
- BLAST:
-
Basic local alignment search tool
- ospA :
-
gen for the major outer surface protein A
- LRFP:
-
Large restriction fragment pattern
- MLST:
-
Multilocus sequence typing
- Tm:
-
Melting temperature
- ST:
-
Sequence type
References
Sočan M, Klavs I, Frelih T, Grilc E, Grgič Vitek M, Učakar V, et. al. Epidemiološko spremljanje nalezljivih bolezni v Sloveniji v letu 2012. Edited by Kraigher A. SM, Klavs I., Frelih T., Grilc E., Grgič Vitek M., Učakar V., Kolman J. Ljubljana; 2013.
Stanek G, Wormser GP, Gray J, Strle F. Lyme borreliosis. Lancet. 2012;379(9814):461–73.
Kurtenbach K, Dizij A, Seitz HM, Margos G, Moter SE, Kramer MD, et al. Differential immune responses to Borrelia burgdorferi in European wild rodent species influence spirochete transmission to Ixodes ricinus L. (Acari: Ixodidae). Infect Immun. 1994;62(12):5344–52.
Talleklint L, Jaenson TG, Mather TN. Seasonal variation in the capacity of the bank vole to infect larval ticks (Acari: Ixodidae) with the Lyme disease spirochete, Borrelia burgdorferi. J Med Entomol. 1993;30(4):812–5.
Piesman J, Gern L. Lyme borreliosis in Europe and North America. Parasitology. 2004;129(Suppl):S191–220.
Mason LM, Veerman CC, Geijtenbeek TB, Hovius JW. Menage a trois: Borrelia, dendritic cells, and tick saliva interactions. Trends Parasitol. 2014;30(2):95–103.
Krause PJ, Narasimhan S, Wormser GP, Rollend L, Fikrig E, Lepore T, et al. Human Borrelia miyamotoi infection in the United States. N Engl J Med. 2013;368(3):291–3.
Platonov AE, Karan LS, Kolyasnikova NM, Makhneva NA, Toporkova MG, Maleev VV, et al. Humans infected with relapsing fever spirochete Borrelia miyamotoi, Russia. Emerg Infect Dis. 2011;17(10):1816–23.
Gugliotta JL, Goethert HK, Berardi VP, Telford 3rd SR. Meningoencephalitis from Borrelia miyamotoi in an immunocompromised patient. N Engl J Med. 2013;368(3):240–5.
Taylor KR, Takano A, Konnai S, Shimozuru M, Kawabata H, Tsubota T. Borrelia miyamotoi infections among wild rodents show age and month independence and correlation with Ixodes persulcatus larval attachment in Hokkaido, Japan. Vector Borne Zoonotic Dis. 2013;13(2):92–7.
Ruzic-Sabljic E, Strle F. Comparison of growth of Borrelia afzelii, B. garinii, and B. burgdorferi sensu stricto in MKP and BSK-II medium. Int J Med Microbiol. 2004;294(6):407–12.
Korva M, Knap N, Rus KR, Fajs L, Grubelnik G, Bremec M, et al. Phylogeographic diversity of pathogenic and non-pathogenic hantaviruses in slovenia. Viruses. 2013;5(12):3071–87.
Ruzic-Sabljic E, Maraspin V, Lotric-Furlan S, Jurca T, Logar M, Pikelj-Pecnik A, et al. Characterization of Borrelia burgdorferi sensu lato strains isolated from human material in Slovenia. Wien Klin Wochenschr. 2002;114(13–14):544–50.
Margos G, Gatewood AG, Aanensen DM, Hanincova K, Terekhova D, Vollmer SA, et al. MLST of housekeeping genes captures geographic population structure and suggests a European origin of Borrelia burgdorferi. Proc Natl Acad Sci U S A. 2008;105(25):8730–5.
Guy EC, Stanek G. Detection of Borrelia burgdorferi in patients with Lyme disease by the polymerase chain reaction. J Clin Pathol. 1991;44(7):610–1.
Cerar T, Ogrinc K, Cimperman J, Lotric-Furlan S, Strle F, Ruzic-Sabljic E. Validation of cultivation and PCR methods for diagnosis of Lyme neuroborreliosis. J Clin Microbiol. 2008;46(10):3375–9.
Schmidt BL. PCR in laboratory diagnosis of human Borrelia burgdorferi infections. Clin Microbiol Rev. 1997;10(1):185–201.
Wolff D, Gerritzen A. Evaluation of a real-time PCR assay for the detection, genotyping, and quantification of Borrelia burgdorferi sensu lato in Ixodes ticks in a routine laboratory setting. Clin Lab. 2011;57(1–2):67–73.
Piesman J, Schwan TG. Ecology of borreliae and their arthropod vectors. In: Samuels DS RJ, editor. Borrelia: molecular biology, host interaction and pathogenesis. Norfolk: Caister Academic Press; 2010. p. 251–78.
Franke J, Hildebrandt A, Dorn W. Exploring gaps in our knowledge on Lyme borreliosis spirochaetes--updates on complex heterogeneity, ecology, and pathogenicity. Ticks Tick Borne Dis. 2013;4(1–2):11–25.
Burri C, Schumann O, Schumann C, Gern L. Are Apodemus spp. mice and Myodes glareolus reservoirs for Borrelia miyamotoi, Candidatus Neoehrlichia mikurensis, Rickettsia helvetica, R. monacensis and Anaplasma phagocytophilum? Ticks Tick Borne Dis. 2014;5(3):245–51.
Zore A, Petrovec M, Prosenc K, Trilar T, Ruzic-Sabljic E, Avsic-Zupanc T. Infection of small mammals with Borrelia burgdorferi sensu lato in Slovenia as determined by polymerase chain reaction (PCR). Wien Klin Wochenschr. 1999;111(22–23):997–9.
Schmidt S, Essbauer SS, Mayer-Scholl A, Poppert S, Schmidt-Chanasit J, Klempa B, et al.: Multiple Infections of Rodents with Zoonotic Pathogens in Austria. Vector Borne Zoonotic Dis 2014. Vector Borne Zoonotic Dis. 2014 Jul;14(7):467–75.
Mannelli A, Bertolotti L, Gern L, Gray J. Ecology of Borrelia burgdorferi sensu lato in Europe: transmission dynamics in multi-host systems, influence of molecular processes and effects of climate change. FEMS Microbiol Rev. 2012;36(4):837–61.
Perez D, Kneubuhler Y, Rais O, Gern L. Seasonality of Ixodes ricinus ticks on vegetation and on rodents and Borrelia burgdorferi sensu lato genospecies diversity in two lyme borreliosis-endemic areas in Switzerland. Vector Borne Zoonotic Dis. 2012;12(8):633–44.
Huegli D, Hu CM, Humair PF, Wilske B, Gern L. Apodemus species mice are reservoir hosts of Borrelia garinii OspA serotype 4 in Switzerland. J Clin Microbiol. 2002;40(12):4735–7.
Richter D, Schlee DB, Allgower R, Matuschka FR. Relationships of a novel Lyme disease spirochete, Borrelia spielmani sp. nov., with its hosts in Central Europe. Appl Environ Microbiol. 2004;70(11):6414–9.
Hanincova K, Schafer SM, Etti S, Sewell HS, Taragelova V, Ziak D, et al. Association of Borrelia afzelii with rodents in Europe. Parasitology. 2003;126(Pt 1):11–20.
Gern L, Humair F. Ecology of Borrelia burgdorferi sensu lato in Europe. In: Gray J, Kahl O, Lane RS, Stanek G, editors. Lyme Borreliosis: Biology, Epidemiology and Control. Oxon, United Kingdom: CAB International; 2002. p. 149–74.
Khanakah G, Kocianova E, Vyrostekova V, Rehacek J, Kundi M, Stanek G. Seasonal variations in detecting Borrelia burgdorferi sensu lato in rodents from north eastern Austria. Wien Klin Wochenschr. 2006;118(23–24):754–8.
Cosson JF, Michelet L, Chotte J, Le Naour E, Cote M, Devillers E, et al. Genetic characterization of the human relapsing fever spirochete Borrelia miyamotoi in vectors and animal reservoirs of Lyme disease spirochetes in France. Parasit Vectors. 2014;7(1):233.
Acknowledgements
This study was supported by the Ministry of Higher Education, Science and Sport of Slovenia (grant no. P3-0083).
Author information
Authors and Affiliations
Corresponding author
Additional information
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
TC, MK, TAZ and ERS conducted the research. TC performed the experimental part and drafted the manuscript. All co-authors evaluated the results, reviewed the analyses and contributed to writing the manuscript. All authors read and approved the final manuscript.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Cerar, T., Korva, M., Avšič-Županc, T. et al. Detection, identification and genotyping of Borrellia spp. in rodents in Slovenia by PCR and culture. BMC Vet Res 11, 188 (2015). https://doi.org/10.1186/s12917-015-0501-y
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12917-015-0501-y