Abstract
Background
The yellow jasmine flower (Jasminum humile L.) is a fragrant plant belonging to the Oleaceae family with promising phytoconstituents and interesting medicinal uses. The purpose of this study was to characterize the plant metabolome to identify the potential bioactive agents with cytotoxic effects and the underlying mechanism of cytotoxic activity.
Methods
First, HPLC–PDA-MS/MS was used to identify the potential bioactive compounds in the flowers. Furthermore, we assessed the cytotoxic activity of the flower extract against breast cancer (MCF-7) cell line using MTT assay followed by the cell cycle, DNA-flow cytometry, and Annexin V-FITC analyses alongside the effect on reactive oxygen species (ROS). Finally, Network pharmacology followed by a molecular docking study was performed to predict the pathways involved in anti-breast cancer activity.
Results
HPLC–PDA-MS/MS tentatively identified 33 compounds, mainly secoiridoids. J. humile extract showed a cytotoxic effect on MCF-7 breast cancer cell line with IC50 value of 9.3 ± 1.2 µg/mL. Studying the apoptotic effect of J. humile extract revealed that it disrupts G2/M phase in the cell cycle, increases the percentage of early and late apoptosis in Annexin V-FTIC, and affects the oxidative stress markers (CAT, SOD, and GSH-R). Network analysis revealed that out of 33 compounds, 24 displayed interaction with 52 human target genes. Relationship between compounds, target genes, and pathways revealed that J. humile exerts its effect on breast cancer by altering, Estrogen signaling pathway, HER2, and EGFR overexpression. To further verify the results of network pharmacology, molecular docking was performed with the five key compounds and the topmost target, EGFR. The results of molecular docking were consistent with those of network pharmacology.
Conclusion
Our findings suggest that J. humile suppresses breast cancer proliferation and induces cell cycle arrest and apoptosis partly by EGFR signaling pathway, highlighting J. humile as a potential therapeutic candidate against breast cancer.
Similar content being viewed by others
Background
Cancers are a group of diseases characterized by uncontrolled and unrestricted cell proliferation. Cancers may continue to progress, resulting in premature death, as they alter cell dynamics, cell growth, survival, and differentiation, causing them to become invasive. [1]. Despite the presence of numerous powerful anticancer agents, the majority of these agents have severe side effects. Some cancers respond well to surgery, while others benefit more from medications such as chemotherapy. Inducing apoptosis is considered a powerful cancer-fighting strategy [2]. Apoptosis can be induced by signals from inside the cell, such as genotoxic stress, or by extrinsic signals, such as the binding of ligands to cell surface death receptors [2]. Apoptosis, or programmed cell death, is mediated by an intracellular proteolytic cascade and is finely regulated at the gene level to produce the orderly addition to efficient elimination of impaired cells in a tremendously controlled manner without concomitant inflammatory reactions [3,4,5]. Although the accumulation of free radicals inside the body increases the susceptibility of cancer development, tumor cells are affected by the variations in reactive oxygen species (ROS) levels [6] as they are capable of eliciting oxidative stress. As a result of the release of pro-apoptotic proteins, macromolecular and organelle damage occurs, resulting in apoptosis and activation of antioxidant pathways in surviving cells.
Italian jasmine Jasminum humile L. (J. humile) is a fragrant plant belonging to the Oleaceae family, known as Italian jasmine or yellow jasmine. It is a small branched shrub with yellow fragrant flowers [7], its fragrant oil is used in the perfume industry and the yellow dye from the flowers and roots was used as a natural dye for solar cells [8]. Extracts of several species of the genus Jasminum revealed different pharmacological activities as anticancer [9,10,11], antioxidant [10, 12, 13], chemo-preventive, antibacterial [14, 15], analgesic [16], gastroprotective [17], and anti-inflammatory activities [10].
The present study explored the cytotoxic activity of a methanol extract of J. humile by MTT assay against breast cancer (MCF-7) cell line, combined with high-performance liquid chromatography/photodiode array coupled to mass spectroscopy-mass spectroscopy (HPLC–PDA-MS/MS) analysis to evaluate the role of the plant constituents in the plant cytotoxic activity. GO biological process analysis and KEGG pathway enrichment analysis were used to investigate the potential mechanisms underlying J. humile's anti-breast cancer activity. The top-hit compounds were molecular docked against the top-hit molecular target relevant to breast cancer identified in the constructed networks to learn more about how these compounds interact with the active regions of the target proteins. Such a strategy could help researchers better understand J. humile's molecular mechanism of action in breast cancer treatment.
Materials and methods
Plant material and extraction
Jasminum humile L. in the flowering stage was collected on 20th May 2019 from EL-Keram farms, Moderayat al-Tahrir, El-Behaira government, Egypt. It was taxonomically identified by Dr. Mohammed El Gebaly (Consultant botanist-Orman Garden) and a voucher specimen (3.10.16.1) was kept in the herbarium of Faculty of Pharmacy, Cairo University. The plant flowers were collected, washed, and shadow dried. The flowers were then pulverized to a coarse powder (2–3 mm) with an electric blender before extraction. Pulverized flower (50 g) was extracted in a percolator with 80% methanol for 24 h at room temperature. Three washes with 100 mL of fresh solvent with a hold time of 5 h were carried out until the last extract was colorless. The combined extract was filtered Whatman No.1 filter paper and was further concentrated under vacuum using a rotary evaporator at 45 °C to yield 10.5 g dry extract. The crude solid extract was labeled and stored under cold conditions before analysis.
HPLC–PDA-MS/MS
A Thermo Finnigan LC system was used for the HPLC–PDA-MS/MS analysis of J. humile extract (Thermo Electron Corporation, Austin, TX, USA). The instrument utilized was a Zorbax Eclipse XDB-C18, Rapid resolution, 4.6 150 mm, with a 3.5 µm column (Agilent, Santa Clara, CA, USA). Acetonitrile concentration was raised from 5 to 30% in 60 min at a flow rate of 1 mL/min and a 1:1 split before the ESI source using gradient elution with water and acetonitrile (ACN), each containing 0.1% formic acid. Thermo Quest ESI source-equipped LCQ-Duo ion trap was utilized for MS analysis. The system was managed by Xcalibur software (XcaliburTM 2.0.7, Thermo Scientific, Waltham, MA, USA). The MS operating settings were employed in the negative mode [18].
Biological evaluation
MTT assay
MCF-7 cells were maintained in RPMI-1640 medium supplemented with 10% FBS, glutamine (2 raM), penicillin (100 units/mL), and streptomycin (100 µg/mL). The cells were cultured at 37 °C in a humidified 5% CO2 incubator. The extract of J. humile flower was tested for in vitro cytotoxicity, using MCF-7 cells by 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) assay. One hundred µL of (RMPI 1640) media was loaded into each of the 96-well plates (triplicate). The final volume for each well was 100 µL. The cultured MCF-7 cells were pooled in a 50 mL vial. Then, the cells were plated at a density of 1 × 106 cells/mL cells/well (100 µL) into 96-well microtiter plates. Each sample was replicated 3 times and the cells were incubated at 37 °C in a humidified 5% CO2 incubator for 24 h. After the incubation period, MTT (20 µL of 5 mg/mL) was added to each well and the cells were incubated for another 2–4 h until purple precipitates were clearly visible under a microscope. Flowingly, the medium together with MTT (190 µL) were aspirated off the wells, DMSO (100 µL) was added, and shake the plates for 5 min. Measure the absorbance spectrophotometry at 540 nm in a microtiter plate reader and the percentage cell viability was calculated manually using the formula:
A dose–response curve was plotted to enable the calculation of the concentrations that kill 50% of the MCF-7 cells (IC50) compared to the standard drug, etoposide, and the effect of the extract on normal keratinocyte cells (HaCaT).
DNA-flow cytometry analysis
The influence of J. humile extract (5, 10, and 20 µg/mL) on the cell cycle distribution of MCF-7 cell line was evaluated using the CycleTEST™ PLUS DNA Reagent Kit (Becton Dickinson Immunocytometry Systems, San Jose, CA) according to the manufacture instructions. PBMC cells served as control cells with known DNA content for determining the DNA Index (DI) of the inspected samples. Propodium iodide (PI) was used as a DNA-binding dye before running on the DNA cytometer [19]. CELLQUEST software was used for analyzing Cell-cycle distribution.
Annexin V-FITC apoptosis assay
Annexin V-FITC/DAPI assay (Cayman Chemical, Ann Arbor, MI) was used to analyze apoptotic cells. After culturing MCF-7 cells into a monolayer, they were treated with J. humile extract at 5, 10, and 20 µg/mL. Cells were subsequently collected through trypsinization, double washed in phosphate buffer saline (PBS) followed by the binding buffer. Afterward, cells were re-suspended in 100 µL of binding buffer with the addition of 5 µL of FITC-Annexin V (Becton Dickinson BD PharmingenTM, Heidelberg, Germany) followed by a 30 min. incubation period at 4 °C. Cells were then washed in binding buffer and re-suspended in 150 µL of binding buffer with the addition of 1 µL of DAPI (1 µg/µL in PBS) (Invitrogen, Life Technologies, Darmstadt, Germany). The flow cytometer BD FACS Canto II (BD Biosciences, San Jose, CA) was used to analyze the cells and the results were deduced with FlowJo7.6.4 software (Tree Star, FlowJo LLC, Ashland, OR) [20].
Oxidative stress parameters
Spectrophotometric assessment of Superoxide dismutase (SOD) activity at 560 nm following the method of [21] was done based on inhibition of nitro blue tetrazolium- NADH and phenazinemethosulphate (PMS)-mediated formazan formation. Spectrophotometric assessment of Catalase (CAT) was done using the method of [21]. The assay measures the decomposition of H2O2 by CAT at 240 nm. Glutathione reductase (GSH-R) was assessed spectrophotometrically [21] based on the reduction of oxidized glutathione by GSH-R with the help of NADPH. Afterward, the thiol group of reduced glutathione reacts with the chromogen to yield a colored complex measured at 405 nm.
Network Pharmacology
Target Genes Associated with Breast cancer and Selected Compounds
Binding DB (https://www.bindingdb.org/bind/index.jsp) was used using the "homo sapiens" setting to predict target genes for selected compounds based on SMILES. The "minimum needed interaction score" was set to "high confidence (0.700)" during Binding DB prediction. The public database DisGeNET (http://www.disgenet.org/) was used to identify disease-related target genes [22].
Interactions between Compounds and Overlapping Genes: Network Construction
Cytoscape ver. 3.9.1 (https://cytoscape.org/) was used to construct, display, and analyze the network of interactions based on the Binding DB prediction results for constituents and overlapping genes. Nodes in the network indicate bioactive components and genes, while edges show interactions between compounds and genes. Anti-breast cancer components and hub genes were identified by analyzing the network's topological structure and setting the "Degree value" of compounds or genes. A compound's or a gene's degree value represents how many phytoconstituents or genes are present in a network. The therapeutic effects of compounds are enhanced if the compounds target more disease-inducing genes [22].
Building a Protein–Protein Interaction Network
An online database called STRING (https://string-db.org/) was used to gather information on protein–protein interactions between the target proteins of selected J.humile components (PPI). The website calculated a score for each protein's mutual information. The stronger the contact between the two target proteins, the higher the score. Since high-confidence data > 0.7 were used to ensure accuracy and reliability, the study was considered reliable. The obtained protein interaction data was imported into the Cytoscape 3.9.1 application to generate a PPI protein interaction network. The CytoHubba plug-in was employed for the identification of Hub genes [23]. The cyto hubba plg-in has 12 topological features among which we used the “Degree” parameters to screen out the top-ranked proteins according to their degree value of interaction.
Target Protein Gene Ontology and KEGG Enrichment Analysis
It was found that proteins that interact with the active components of J.humile play a role in gene function and signaling pathways by using the database for annotation, visualization, and integrated discovery (David) v 6.8 to evaluate gene function and KEGG pathway enrichment, respectively [24]. The functional enrichment database DAVID, which is accessible online, aids researchers in understanding the bioactivity of a large number of genes. In the current study, p-value ≤ 0.01 was chosen, and the top 10 KEGG pathways and GO enrichments were picked for further investigation. Cellular components (CC), molecular functions, biological processes, and pathways were all included in the list of target proteins. KEGG pathway enrichment results were used to decipher the possible molecular mechanisms of J.humile against breast cancer. KEGG pathway bubble charts made with SRPLOT (http://bioinformatics.com.cn/) were of particular interest.
Molecular docking
The current study used the MOE-Dock from Chemical Computing Group Inc. to carry out computational experiments. The crystal structures of EGFR were obtained using the Protein Data Bank (PDB id 4HJ0). After editing the crystal structures to remove the water molecules and add hydrogen atoms to the protein, MOPAC 7.0 was used to minimize the energy. Then, the alpha spheres produced were used to create dummy atoms by identifying the active site. In docked regions, RMSD values of less than 1.3 were clustered together. The docked complex in the lowest energy-minimized stance was chosen for further investigation. Ten different conformations were carefully chosen. The MMFF94x force field energy computation was then used for the resulting docked complex model to determine the energy parameters and estimate the docked interactions at the active site [25].
Statistical analysis
All data are displayed as the mean ± S.D of three separate determinations. The arithmetic mean and the standard deviation (SD) of all assessments were calculated by the software program Microsoft Excel 2007. Statistical analysis was performed using GraphPad Prism version 5.01. To establish the statistical significance in relation to the reference standard, a one-way ANOVA was employed, followed by a Tukey-posthoc test.
Results
HPLC–PDA-MS/MS metabolites profiling
The aqueous methanolic extract of J. humile flower was analyzed by HPLC–PDA-MS/MS to characterize its metabolome and identify potential bioactive agents. The analysis tentatively identified 33 compounds (Fig. 1 & Table 1) belonging to various classes [26,27,28,29,30,31,32,33,34,35,36,37,38,39,40,41]. The major phytochemical classes are secoiridoids, phenylethanoids, Phenolic acids, flavonoids, and lignans. The major compounds were identified as oleoside derivatives (Fig. 2) and kaempferol glucosides.
Biological evaluation
MTT assay
J. humile extract showed a cytotoxic effect on MCF-7 breast cancer cell line with IC50 = 9.3 ± 1.2 µg/mL, the extract induced no cytotoxicity toward the normal cell line (HaCaT) with IC50 496.2 ± 4.88 µg/mL.
Cell-cycle analysis
The outcome of treating MCF-7 cells with J. humile extract at three different concentration levels: 5, 10 and 20 µg/mL (Figs. 3 & 4) revealed that J. humile extract significantly reduced the number of cells in the G0-G1 and S phases compared to untreated control cells in a concentration-dependent manner, with 57.42%, 52.81%, and 48.34% compared to 61.32%, and 28.02%, 15.97% and 16.05 compared to 31.42% for 5, 10, and 20 µg/mL, respectively. Whereas data showed a significant increase in G2/M phase cell population as compared to untreated control cells in a concentration-dependent manner recording, 14.56%, 31.22%, and 35.61% as compared to 7.26% for 5, 10, and 20 µg/mL respectively. In addition, results represented a significant increase in cell cycle population in Pre-G1 phase when compared with untreated control cells in a concentration-dependent manner recording, 3.64%, 5.88%, and 9.92% compared to 1.72% for 5, 10, and 20 µg/mL, respectively.
Effects of different concentrations of J. humile extract on Cell Cycle Analysis of MCF-7 cells. a Untreated MCF-7 cells, b MCF-7 cells treated with 5 µg/ml J. humile extract, c MCF-7 cells treated with 10 µg/ml J. humile extract, d MCF-7 cells treated with 20 µg/ml J. humile extract. Light blue indicates % apoptosis, orange indicates G0-G1 phase, shaded part indicates S phase and red indicates G2-M phase
Annexin V-FITC apoptosis
In the current study J. humile extract displayed cytotoxic activity against MCF-7 cancer cell line with the use of different concentrations (5, 10, and 20 µg/mL). Analyzing Fig. 3 revealed that J. humile extract induced a significant increase in the rate of both total apoptosis and necrosis in 3.64%, 5.88%, and 9.92% of cells at 5, 10, and 20 µg/mL, respectively compared to 1.72% in untreated control MCF-7 cells. Mutually early and late apoptosis results displayed a significant increase from control in a concentration-dependent manner (P < 0.05). Whereas no significant change in the rate of necrosis at 5 µg/mL (P < 0.05) was detected, although the 10 and 20 µg/mL showed a significant increase from control in the rate of necrosis (P < 0.05) as represented in (Figs. 5 & 6).
Effects of different concentrations of J. humile extract on Annexin V-FITC analysis of MCF-7 cells. a Untreated MCF-7 cells, b MCF-7 cells treated with 5 µg/ml J. humile extract, c MCF-7 cells treated with 10 µg/ml J. humile extract, d MCF-7 cells treated with 20 µg/ml J. humile extract. Early apoptosis (lower right quadrant), late apoptosis (upper right quadrant), viable cells (lower left quadrant), Necrosis (upper left quadrant). Values are presented as means ± SD with significance level at (p < 0.05)
Oxidative stress parameters
The results in this study (Fig. 7) revealed that treatment of the MCF-7 cells with 10 µg/mL of the J. humile extract significantly elevated the level of SOD with concomitant depression of CAT and GSH-R as compared to the control cells causing the accumulation of ROS, thus triggering apoptosis (Table 2).
Network Pharmacology
Scrutinizing anti-breast cancer target
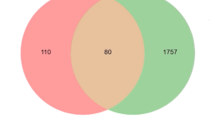
A total of 231 genes were related to 24 compounds from the above-mentioned 33 compounds using Binding DB prediction (Table S1), while 9 compounds exhibited no human target interactions. A total of 6776 disease-related genes were found after retrieving the DisGeNET database. 52 overlapped genes were discovered after comparing 231 compound-related genes with 6776 disease-related genes. The names of overlapping genes targeted by J.humile drugs and breast cancer are shown in Fig. 8. A network was constructed for active compounds and overlapping genes which includes 76 nodes and 225 edges (Fig. 9).
GO and KEGG analysis
To show the interaction of target proteins with their relevant components, DAVID analysis of GO enrichment and KEGG analysis were used. The Benjamini–Hochberg procedure was used to correct P-values, and the top 10 significantly enriched items in the BP, MF, and CC categories (Fig. 10 & Table S2) were picked based on p ≤ 0.05. According to GO functional analysis, J. humile's principal targets were inflammatory response regulation, protein kinase B regulation, steroid hormone response, intracellular receptor signaling pathway, and so on. Relevant signaling pathways related with J. humile's anti-breast cancer action were identified using KEGG pathway analysis. Pathways in cancer (16), chemical carcinogenesis-receptor activation (10), estrogen signaling system (8), proteoglycans in cancer (7), lipid and atherosclerosis (6), and P13-Akt signaling pathway (6) had the most genes (Table S3). The substantially enriched genes from the 22 KEGG signaling pathways results (Fig. 11) were EGFR, ESR1, ESR2, HSP90AA1, HSP90AB1, MMP9, and MMP1 [42]. Breast cancer pathways had been targeted by target genes thereby KEGG pathway for breast cancer is depicted in Fig. 12, where target genes were represented with red color. On the basis of this pathway, it can be concluded that J. humile exerts the anti-breast cancer effect by targeting the estrogen signaling pathway, HER2 proteins, and triple-negative breast cancer process.
KEGG pathway for breast cancer (hsa05224), figure obtained under permission and guidelines [42]
Molecular docking
For molecular docking, the top most hub gene EGFR was selected based on PPI interactions analysis. The top most active compounds Quercetin 3,7-diglucoside, Quercetin xylosyl glucoside, Kaempferol 3-xyloside-7-glucoside, kaempferol-3-O-D-glucoside and Isorhamnetin-3-O-glucoside were docked with EGFR. The two crucial factors were used while analyzing docking results: (i) the best docked pose binding energy prediction using MOE scoring system and (ii) Hydrogen bond information of top-ranked posture. Each compound was docked in 10 distinct positions throughout the docking run. The recovered molecules were first sorted using the pre-validated methodology described above, and then the visualization approach was used to determine the inhibitor binding mode that is best based on the inhibitor's critical interactions with the active site residues. Table 3 summarizes the docking information for the top-ranked poses.
All the selected compounds displayed significant interaction with the EGFR. All of them bind in the same binding pocket where the attached inhibitor was present thus showing that these compounds also possess inhibitory attributes on EGFR. Compounds interacted with the active site of EGFR by forming bonds with the following amino acid residues: Thr766, Thr830, Lys 721, Glu738, Lys 692, Asp 831 and Thr 830. Among all compounds, Isorhamnetin-3-O-glucoside displayed maximum binding interaction with EGFR as shown in Fig. 13.
Discussion
Breast cancer is considered one of the most common causes of cancer related death among women [43]. Extracts of several species of the genus Jasminum revealed different pharmacological activities as anticancer [9,10,11]. In the present study, we evaluated the effect of Jasminum humile on breast cancer cells. Our results demonstrated that J. humile significantly inhibited breast cancer growth and was inactive on healthy cells suggesting a potential selective anti-cancer activity of the extract. J. humile extract caused a dose-dependent inhibition of cell growth with S phase arrest, a reduction in G0/G1 phase, and an increase in pre-G1 cells. An increase of cells in the pre-G1 phase was recognized as an indicator of DNA fragmentation and hence apoptosis. The results could prove that J. humile extract may exert its cytotoxic effect in MCF-7 cells by arresting its proliferation in G2/M phase. In order to determine if cell death occurred through apoptosis or necrosis Annexin V-based flow cytometry analysis was performed. Annexin-V FTIC assay showed that J. humile extract displayed a noticeable rise in apoptotic cell percentage including both early and late apoptotic phases in a concentration-dependent manner. Additionally, humile extract had a cytotoxic effect by interfering with the redox homeostasis of the cell between ROS production and the antioxidant system. All these results indicate that J. humile may serve as a potential therapeutic agent against breast cancer, however the identities of the effective compounds remain unclear. HPLC–PDA-MS/MS metabolites profiling identified 33 compounds. Utilizing network pharmacology, out of 33 compounds, 24 displayed interaction with 52 human target genes as potential target of J. humile responsible for its inhibitory effect against breast cancer. Then, DAVID analysis of GO enrichment and KEGG analysis were used to show the interaction of target proteins with their relevant components. According to GO functional analysis, J. humile's principal targets were inflammatory response regulation, protein kinase B regulation, steroid hormone response, intracellular receptor signaling pathway, and so on. Relevant signaling pathways related with J. humile's anti-breast cancer action were identified using KEGG pathway analysis. Pathways in cancer (16), chemical carcinogenesis-receptor activation (10), estrogen signaling system (8), proteoglycans in cancer (7), lipid and atherosclerosis (6), and P13-Akt signaling pathway (6) had the most genes. The substantially enriched genes from the 22 KEGG signaling pathways results were EGFR, ESR1, ESR2, HSP90AA1, HSP90AB1, MMP9, and MMP1. On the basis of this pathway, it can be concluded that J. humile exerts the anti-breast cancer effect by targeting the estrogen signaling pathway, HER2 proteins, and triple-negative breast cancer process. Notably, the epidermal growth factor receptor (EGFR) signaling pathway is one of the most important pathways that regulate growth, survival, proliferation, and differentiation in mammalian cells [44]. EGFR is a receptor tyrosine kinase that is commonly upregulated in cancers such as in non-small-cell lung cancer, metastatic colorectal cancer, glioblastoma, head and neck cancer, pancreatic cancer, and breast cancer [45]. To further verify the results of network pharmacology, molecular docking was performed with the five key compounds (Quercetin 3,7-diglucoside, Quercetin xylosyl glucoside, Kaempferol 3-xyloside-7-glucoside, kaempferol-3-O-D-glucoside and Isorhamnetin-3-O-glucoside) and the top most target, EGFR. All the selected compounds displayed significant interaction with the EGFR. Among all compounds, Isorhamnetin-3-O-glucoside displayed maximum binding interaction with EGFR. Interestingly, isorhamnetin-3-O-rhamnoside showed strong inhibitory effects on human breast adenocarcinoma cell line MCF-7 proliferation [46]. The results of molecular docking were consistent with those of network pharmacology.
Conclusions
In conclusion, our study provides the first clear evidence that Jasminum humile L. flowers have significant antitumor activity against breast cancer. Furthermore, Jasminum humile L. significantly inhibited cell growth, prompted cell apoptosis, and caused cell cycle arrest partly via EGFR signaling pathway. Additionally, we identified many active compounds from Jasminum humile that can be used to develop new therapeutic agents to fight breast cancer.
Availability of data and materials
All data generated or analysed during this study are included in this published article [and its supplementary information files].
Change history
31 May 2023
A Correction to this paper has been published: https://doi.org/10.1186/s12906-023-04011-x
References
Tor YS, Yazan LS, Foo JB, et al. Induction of apoptosis in MCF-7 cells via oxidative stress generation, mitochondria-dependent and caspase-independent pathway by ethyl acetate extract of Dillenia suffruticosa and its chemical profile. PLoS One. 2015;10(6):e0127441.
Maltecca F, Casari G. In vivo detection of oxidized proteins: a practical approach to tissue-derived mitochondria. Protein Misfolding and Cellular Stress in Disease and Aging: Concepts and Protocols. 2010;257-267.
Reed JC. Mechanisms of apoptosis. Am J Pathol. 2000;157(5):1415–30.
Elmore S. Apoptosis: a review of programmed cell death. Toxicol Pathol. 2007;35(4):495–516.
Sunilkumar D, Drishya G, Chandrasekharan A, et al. Oxyresveratrol drives caspase-independent apoptosis-like cell death in MDA-MB-231 breast cancer cells through the induction of ROS. Biochem Pharmacol. 2020;173:113724.
Khan A, Farooq U, Ullah F, et al. Determination of biological activities and total phenolic contents of flowers of Jasminum humile and roots of Dorema aucheri. J Chem Soc Pak. 2014;36(2):291–5.
Krishnaswamy N, Raman VJT, sambac cnofvoJ, Soc osJIB: Cytogenetical studies of the Indian Jasmines. I. 1948, 27:77-83.
Atli A, Atilgan A, Altinkaya C, et al. yellow jasmine, and madder berries as novel natural sensitizers for dye-sensitized solar cells. Int J Energy Res. 2019;43(8):3914–22.
El-Hawary SS, El-Hefnawy HM, El-Raey MA, et al. Jasminum azoricum L. leaves: HPLC-PDA/MS/MS profiling and in-vitro cytotoxicity supported by molecular docking. Nat Prod Res. 2021;35(23):5518–20.
El-Hawary SS, El-Hefnawy HM, Osman SM, et al. Phenolic profiling of different Jasminum species cultivated in Egypt and their antioxidant activity. Nat Prod Res. 2021;35(22):4663–8.
El-Hawary SS, El-Hefnawy HM, Osman SM, Mostafa ES, Mokhtar F, El-Raey M. Chemical profile of two jasminum sambac L.(AIT) cultivars cultivated in egypt-their mediated silver nanoparticles synthesis and selective cytotoxicity. 2019.
Kumaresan M, Kannan M, Sankari A, et al. Phytochemical screening and antioxidant activity of Jasminum multiflorum (pink Kakada) leaves and flowers. J Pharmacogn Phytochem. 2019;8(3):1168–73.
Khidzir KM, Cheng SF, Chuah CH. Interspecies variation of chemical constituents and antioxidant capacity of extracts from Jasminum sambac and Jasminum multiflorum grown in Malaysia. Ind Crops Prod. 2015;74:635–41.
Nain P, Kumar A, Sharma S, et al. In vitro evaluation of antimicrobial and antioxidant activities of methanolic extract of Jasminum humile leaves. Asian Pac J Trop Med. 2011;4(10):804–7.
El-Sayed MA, Al-Gendy AA, Hamdan DI, et al. Phytoconstituents, LC-ESI-MS profile, antioxidant and antimicrobial activities of Citrus x limon L. Burm. f. Cultivar Variegated Pink Lemon. J Pharm Sci Res. 2017;9(4):375.
Bhowmik D, Chatterjee DP, Mallik A, et al. Study of the analgesic activity of methanolic extract of jasmine root (Jasminum sambac). Indian J Res Pharm Biotechnol. 2013;1(1):14.
AlRashdi AS, Salama SM, Alkiyumi SS, et al. Mechanisms of gastroprotective effects of ethanolic leaf extract of Jasminum sambac against HCl/ethanol-induced gastric mucosal injury in rats. Evid Based Complement Alternat Med. 2012;2012:786426.
Anaya‐Esparza LM, Ramírez‐Marez MV, Montalvo‐González E, Sánchez‐Burgos JA: Cherimoya (Annona cherimola Mill.). Fruit and Vegetable Phytochemicals: Chemistry and Human Health, 2nd Edition 2017;93–1002.
Biselli R, Ferlini C, Fattorossi A, et al. Inflammatory myofibroblastic tumor (inflammatory pseudotumor): DNA flow cytometric analysis of nine pediatric cases. Cancer. 1996;77(4):778–84.
Schutte B, Nuydens R, Geerts H, Ramaekers F. Annexin V binding assay as a tool to measure apoptosis in differentiated neuronal cells. J Neurosci Methods. 1998;86(1):63–9.
Machado-Vieira R, Andreazza AC, Viale CI, et al. Oxidative stress parameters in unmedicated and treated bipolar subjects during initial manic episode: a possible role for lithium antioxidant effects. Neurosci Lett. 2007;421(1):33–6.
Alzarea SI, Qasim S, Uttra AM, et al. Network Pharmacology and Molecular Docking Based Prediction of Mechanism of Pharmacological Attributes of Glutinol. Processes. 2022;10(8):1492.
Ahmed SR, Al-Sanea MM, Mostafa EM, et al. A Network Pharmacology Analysis of Cytotoxic Triterpenes Isolated from Euphorbia abyssinica Latex Supported by Drug-likeness and ADMET Studies. ACS Omega. 2022;7(21):17713–22.
Kanehisa M, Furumichi M, Sato Y, Kawashima M, Ishiguro-Watanabe M. KEGG for taxonomy-based analysis of pathways and genomes. Nucleic Acids Res. 2023;51(D1):D587–92.
Qasim S, Kalsoom S, Shahzad M, Irfan HM, Zafar MS, Bukhari IA, Vohra F, Afzal S. Appraisal of disease-modifying potential of amlodipine as an anti-arthritic agent: new indication for an old drug. Inflammopharmacology. 2020;28(4):1121-36.18.
Omar SH. Oleuropein in olive and its pharmacological effects. Sci Pharm. 2010;78(2):133–54.
Lommen A, Godejohann M, Venema DP, et al. Application of directly coupled HPLC− NMR− MS to the identification and confirmation of quercetin glycosides and phloretin glycosides in apple peel. Anal Chem. 2000;72(8):1793–7.
Alam MA, Subhan N, Hossain H, et al. Hydroxycinnamic acid derivatives: a potential class of natural compounds for the management of lipid metabolism and obesity. Nutr Metab. 2016;13(1):1–3.
Tomassini L, Ventrone A, Frezza C, et al. Lignans and secoiridoid glycosides from the stem barks of Jasminum tortuosum. Nat Prod Res. 2018;32(15):1853–7.
Harborne JB, Mabry TJ. The flavonoids: advances in research. New York: Springer; 2013.
Koike A, Barreira JC, Barros L, et al. Edible flowers of Viola tricolor L. as a new functional food: Antioxidant activity, individual phenolics and effects of gamma and electron-beam irradiation. Food Chemistry. 2015;179:6–14.
Yang JH, Kondratyuk TP, Marler LE, et al. Isolation and evaluation of kaempferol glycosides from the fern Neocheiropteris palmatopedata. Phytochemistry. 2010;71(5–6):641–7.
Shen YC, Chen CH. Multiflorin, A New Secoiridold Lactone from Jasminum Multiflorum. J Chin Chem Soc. 1994;41(4):473–6.
Damtoft S, Franzyk H, Jensen SR. Excelsioside, a secoiridoid glucoside from Fraxinus excelsior. Phytochemistry. 1992;31(12):4197–201.
Tanahashi T, Takenaka Y, Nagakura N, et al. Three secoiridoid glucosides from Jasminum nudiflorum. J Nat Prod. 1999;62(9):1311–5.
Shen YC, Lin SL. New secoiridoid glucosides from Jasminum lanceolarium. Planta Med. 1996;62(06):515–8.
Takenaka Y, Tanahashi T, Taguchi H, Nagakura N, Nishi T. Nine new secoiridoid glucosides from Jasminum nudiflorum. Chem Pharm Bull. 2002;50(3):384–9.
Cecchi L, Migliorini M, Cherubini C, et al. Phenolic profiles, oil amount and sugar content during olive ripening of three typical Tuscan cultivars to detect the best harvesting time for oil production. Food Res Int. 2013;54(2):1876–84.
Tanahashi T, Takenaka Y, Akimoto M, et al. Six secoiridoid glucosides from Jasminum polyanthum. Chem Pharm Bull. 1997;45(2):367–72.
Zhang YJ, Liu YQ, Pu XY, et al. Iridoidal glycosides from Jasminum sambac. Phytochemistry. 1995;38(4):899–903.
Pérez-Bonilla M, Salido S, van Beek TA, et al. Isolation of antioxidative secoiridoids from olive wood (Olea europaea L.) guided by on-line HPLC–DAD–radical scavenging detection. Food Chemistry. 2011;124(1):36–41.
Kanehisa M, Goto S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28(1):27–30.
Watkins EJ. Overview of breast cancer. J Am Acad PAs. 2019;32(10):13–7.
Oda K, Matsuoka Y, Funahashi A, Kitano H. A comprehensive pathway map of epidermal growth factor receptor signaling. Mol Syst Biol. 2005;1(1):2005–10.
Wee P, Wang Z. Epidermal growth factor receptor cell proliferation signaling pathways. Cancers. 2017;9(5):52.
Chen A, Gu N, Pei J, Su E, Duan X, Cao F, Zhao L. Synthesis of isorhamnetin-3-O-rhamnoside by a three-enzyme (rhamnosyltransferase, glycine max sucrose synthase, UDP-rhamnose synthase) cascade using a UDP-rhamnose regeneration system. Molecules. 2019;24(17):3042.
Acknowledgements
The authors extend their appreciation to the Deanship of Scientific Research at King Khalid University for funding this work through large Groups (Project under grant number R.G.P. 2/ 59/44).
Funding
This research received no external fund.
Author information
Authors and Affiliations
Contributions
Conceptualization: F.A.M., M.A.A.; formal analysis: A.O.M, S.R.A.; Software: S.Q. & F.A.M.; Investigation F.A.M. & S.E.E.; Methodology: A.A.S., M.Y.A., O.F.H.; Project administration: F.A.M. & A.A.S.; Supervision: S.S.E. & A.A.S.; Writing – original draft: F.A.M., M.A.A., A.O.M., S.R.A., S.Q., S.E.E.; Writing – review & editing: S.S.E., A.A.S., M.Y.A. All the authors revised and approved the final version of the manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
The original online version of this article was revised: Remove the affiliation ‘Cell Culture Lab, Egyptian Organization for Biological Products and Vaccines (VACSERA Holding Company), Giza, Egypt’ to author Serag Eldin I. Elbehairi.
Supplementary Information
Additional file 1:
Table S1. Genes related to 24compounds. Table S2. GO Analysis. Table S3. KEGG Pathwaysanalysis.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
El-Hawary, S.S., Albalawi, M.A., Montasser, A.O.S. et al. Network pharmacology and molecular docking study for biological pathway detection of cytotoxicity of the yellow jasmine flowers. BMC Complement Med Ther 23, 164 (2023). https://doi.org/10.1186/s12906-023-03987-w
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12906-023-03987-w