Abstract
Background
Extracellular matrix secretion and odontoblastic differentiation in human dental pulp stem cells (hDPSCs) are the cellular bases for reparative dentinogenesis. Osteomodulin (OMD) is a member of the small leucine-rich proteoglycan family distributed in the extracellular matrix but little is known about its role in osteo/odontogenic differentiation. The objective of this study was to investigate the role of OMD during osteo/odontoblastic differentiation of hDPSCs.
Methods
hDPSCs were selected using immune-magnetic beads and their capability of multi-differentiation was identified. OMD knockdown was achieved using short hairpin RNA (shRNA) lentivirus and was confirmed by western blot. Gene expression was measured by real-time qPCR and osteo/odontoblastic differentiation of hDPSCs was determined by alizarin red S staining.
Results
Compared with uninduced cells, the transcription of OMD was up-regulated by 35-fold at the late stage of osteo/odontogenic differentiation. shRNA-mediated gene silencing of OMD decreased the expression of odontoblastic genes, such as alkaline phosphatase (ALP), dentin matrix acidic phosphoprotein 1 (DMP1) and dentin sialophosphoprotein (DSPP). Besides, knockdown of OMD attenuated the mineralized nodules formation induced by osteo/odontogenic medium.
Conclusions
These results implied that OMD may play a pivotal role in modulating the osteo/odontoblastic differentiation of hDPSCs.
Similar content being viewed by others
Background
The dental pulp contains a unique precursor population of mesenchymal stem cells (MSCs) [1]. MSCs are multipotent, highly proliferative and have the ability to differentiate into odontoblast/odontoblast-like cells in response to the stimuli such as caries or dental trauma [2, 3]. The odontoblasts can secret reactionary/reparative dentine matrix which underlies the formation of dentinal bridge. Cultured dental pulp stem cells can also differentiate into odontoblast-like cells and form calcium nodules under certain circumstances in vitro [4, 5]. Identification of the factors that regulate these processes is of instructive significance.
Dentinogenesis is highly regulated by the expression of the extracellular matrix (ECM) proteins. The dentin contains structural macromolecules and other proteins as extracellular matrix components, including type I collagen, osteonectin, osteopontin and dentin sialoprotein [6, 7]. An important family of molecules with regulatory functions is the small leucine-rich proteoglycans (SLRPs), which are extensively involved in the dentinal biomineralization [8]. In particular, it has been confirmed that biglycan and decorin are identified in the matrices of dentin and implicated in dentinogenesis [9].
Osteomodulin (OMD), a heterologous protein of Osteoadherin, belongs to SLRPs and was originally isolated as a keratan sulfate proteoglycan from bovine long bone [10]. SLRPs normally distribute in extracellular matrices, but OMD is the only member specifically restricted to mineralized tissues [11, 12]. Not only does OMD have a high affinity for hydroxyapatite crystals via its large and acidic C-terminal domain [10, 13], but also it can directly regulate diameter and alter shape of type I collagen fibrils [14]. However, the functions of OMD on osteo/odontoblastic differentiation and mineralization have yet to be fully determined, nevertheless it has been shown that OMD expression starts in the polarized odontoblasts and increases in the odontoblast cell layer and alveolar bone during early crown formation [15, 16]. Therefore, it was hypothesized that OMD may be positively correlated with the osteo/odontogenic differentiation and accordingly, the purpose of the present study was to investigate the influence of OMD deficiency on the biomineralization of hDPSCs.
Methods
Isolation and culture of hDPSCs
Healthy human third molars extracted for orthodontic treatment purpose were obtained from 17- to 20-year-old individuals at the oral surgery clinic of the Ninth People’s Hospital affiliated to Shanghai Jiao Tong University School of Medicine. The primary cultured human dental pulp cells were isolated from ten molars using explant method. The cells were pooled together to select hDPSCs by STRO-1-labelled magnetic beads as described previously [17, 18]. Cells were cultured in growth medium (GM): high-glucose Dulbecco’s modified Eagle’s medium (DMEM; Gibco-BRL, Grand Island, NY, USA) supplemented with 10% fetal bovine serum (Gibco-BRL, Life Technologies, Paisley, UK), 100 U/mL penicillin, and 100 mg/mL streptomycin. The medium was renewed every 2 or 3 days. Cell cultures between the second and fifth passages were used.
Flow cytometric analysis
The cell surface markers present on hDPSCs were detected by flow cytometric analysis [19, 20]. Briefly, hDPSCs were incubated with fluorescence-conjugated antibodies for CD73-phycoerythrin (PE), CD105-PE, CD166-PE, CD34-PE, CD90-fluorescein iso thioocyanide (FITC) and CD45-FITC (BD Biosciences, San Jose, CA, USA, and Biolegend, San Diego, CA, USA). Cell suspensions in phosphate buffered saline (PBS) without antibodies served as controls. The cells were then washed three times with PBS to remove unbound antibodies and finally resuspended with 300 μL PBS. Cells were sorted using a flow cytometer (FACSCalibur; BD Biosciences, Mountain View, CA, USA) and analysed with FlowJo software (Tree Star, San Carlos, CA, USA).
Alizarin red S staining and oil red O staining assay
For osteo/odontogenic differentiation, hDPSCs were subcultured in human mesenchymal stem cell osteogenic differentiation medium (OM) (Cyagen Biosciences, Santa Clara, CA, USA) which contained dexamethasone, L-ascorbic acid and beta-glycerophosphate for up to 21 days. Cells cultured in GM were kept as a control group. After 3 weeks of differentiation, cells were fixed in 4% paraformaldehyde for 30 min. After being washed with PBS for three times, calcium deposition was visualized by staining with alizarin red S solution for 3–5 min. Excess stain was removed by washing with distilled water. To study adipogenesis, hDPSCs were cultured in GM until they reached 100% confluence, following which the medium was changed to adipogenic differentiation medium (AM) (Cyagen Biosciences) which contained Insulin, IBMX, Rosiglitazone and Dexamethasone according to the manufacturer’s instructions. After three to five cycles of induction/maintenance, cells were fixed in 4% paraformaldehyde for 30 min and stained with fresh oil red O solution.
Lentivirus production and transduction
Sequences for constructing shRNA targeting human OMD were obtained from the RNAi Consortium (Broad institute) [21]. shRNAs against the OMD gene and a non-target gene were generated with PLKO.1 vector (sequences were shown in Table 1). 9 μg of the ViralPower™ Packaging Mix (Invitrogen, Carlsbad, CA, USA) and 3 μg of the constructed PLKO.1-shOMD or PLKO.1-Ctrl vector were used to co-transfect 6 × 106 293FT cells in the presence of 36 μL Lipofectamine™ 2000 (Invitrogen). Lentivirus was harvested from the culture supernatant at 48 h and 72 h after transfection and filtered through a 0.45 μm filter. For infection, hDPSCs cultured to 30–40% confluence were exposed to recombinant lentivirus in the presence of 10 μg/mL polybrene for 24 h. After incubated in GM for another 24 h, cells were treated with 1 μg/mL puromycin for 48 h to generate stable cell lines.
Western blot analysis
Western blot was performed as described before [22]. Briefly, cells were washed with PBS and harvested with EBC lysis buffer (50 mM Tris HCl, pH 8.0, 120 mM NaCl, 0.5% Nonidet P-40) supplemented with protease inhibitors (Selleck Chemicals, Houston, TX, USA). Protein fractions were collected by centrifugation at 10,000 g at 4 °C for 10 min. The supernatants which contained 50 μg of protein samples were subjected to 10% sodium dodecyl sulfate polyacrylamide gel electrophoresis, following protein quantitation using BCA protein assay. Protein was transferred onto the polyvinylidene difluoride membrane. After blocking in 1 × TBS-0.05%Tween 20 (TBST) containing 5% dry milk powder, membranes were incubated with OMD antibodies (1: 1000; Abgent, San Diego, CA, USA) and α-tubulin antibodies (1: 1000; Santa Cruz Biotechnology, Santa Cruz, CA, USA) overnight at 4 °C. After the membranes were incubated with HRP-conjugated secondary antibody (DAKO, Glostrup, Denmark) for 1 h and extensive washing with 1 × TBST, specific bands were reacted with an ECL chemiluminescence detection system (Thermo Fisher Scientific, Waltham, MA, USA) and visualized with X-ray films (Carestream, Xiamen, Fujian, China).
RNA isolation and determination
Total RNA was extracted at designated time points using TRIzol reagent (Invitrogen) according to the manufacturer’s instructions. Then, 500 ng extracted RNA was reverse transcribed using Ommiscript Reverse Transcription kit (QIAGEN, Valencia, CA, USA) and the resulting cDNA was diluted to 5 ng/μL. 1 μL of this diluted product was used as a template for real-time quantitative PCR (real-time qPCR) and 0.5 μL for reverse transcription PCR (RT-PCR). Real-time qPCR was performed using the DNA Engine OPTICON™ 2 system (MJ Research, Waltham, MA, USA) with SYBR Green I fluorogenic dye (Molecular Probes, Eugene, OR, USA) under the following conditions: 40 cycles each involving 5 s of denaturation at 95°C and 30 s of amplication at 60°C. RT-PCR was performed using Go Taq1 Flexi DNA polymerase (Promega, Madison, WI, USA) under the following conditions: 25 cycles each involving 30 s of denaturation at 94°C, 30 s of annealing at 55°C and 20 s of extension at 72°C. The primers were listed in Table 1. The mRNA levels of target genes were analysed according to the comparative Cq method [23] and normalized to β-actin.
Statistical analysis
Data were expressed as means ± standard deviation (SD) from at least three independent experiments. The statistical significance of difference was assessed using Student’s two-tailed t-test. P < 0.05 indicated a significant difference between groups.
Results
Characterization of the hDPSCs
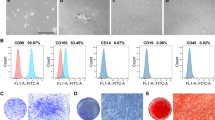
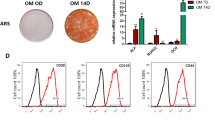
The hDPSCs possess many in vitro phenotypic characteristics of bone marrow-derived MSCs [24, 25]. Flow cytometric analysis was one of the methods based on cell surface molecules. hDPSCs showed the characteristic pattern of MSC-associated surface markers, including CD73, CD90, CD105 and CD166 and were negative for hematopoietic stem cell surface markers CD34 and CD45. Isolated cells that highly expressed CD73, CD90, CD105 and CD166 were used for subsequent experiments (Fig. 1a). The hDPSCs retained multilineage differentiation capacity. Calcium deposition was confirmed by alizarin red S staining (Fig. 1b) and lipid formation was revealed by oil red O staining (Fig. 1c), which indicated hDPSCs’ differentiation into cells like osteoblasts and adipocytes.
Up-regulation of OMD during osteo/odontogenic differentiation
To investigate the expression pattern of OMD mRNA during osteo/odontogenic differentiation, hDPSCs were cultured in OM for 3 weeks, after which samples were analysed by real-time qPCR and RT-PCR. Compared with the control group in GM, hDPSCs which were incubated in induction medium showed a significant 35-fold up-regulation in OMD gene expression (Fig. 2).
Knockdown of OMD in hDPSCs
Puromycin treatment for 2 days almost completely eliminated uninfected hDPSCs without affecting the growth rate and morphology of successfully infected hDPSCs. hDPSCs which were infected with lentiviral constructs harbouring OMD shRNA and non-target shRNA could survive puromycin treatment and were indicated as shOMD-hDPSCs and Ctrl-hDPSCs, respectively. The efficiency of the shRNA-mediated knockdown was confirmed by real-time qPCR and Western blot without induction. The results showed that OMD mRNA in shOMD-hDPSCs was reduced with a concomitant decrease in OMD protein levels in GM (Fig. 3a and b).
The identification of shOMD-hDPSCs cultured in GM. a The transcription of OMD gene in Ctrl-hDPSCs and shOMD-hDPSCs by RT-PCR. b The expression of OMD protein in Ctrl-hDPSCs and shOMD-hDPSCs by western blot. Ctrl: the control group; shOMD: the OMD knockdown group. *Significant difference (P < 0.05) versus Ctrl. **Significant difference (P < 0.01) versus Ctrl
Inhibition of OMD impairs osteo/odontoblastic differentiation of hDPSCs
The stable gene knockdown was achieved throughout the differentiation period (Fig. 4a). The transcriptional level of OMD in Ctrl-hDPSCs was progressively increased with induction. The results of real-time qPCR showed that the expression of the odontoblast markers DMP1 and DSPP in Ctrl-hDPSCs were substantially up-regulated during 7 days of induction and ALP mRNA in Ctrl-hDPSCs reached its peak on day 14. As is shown in Fig. 4, the mRNA levels of DMP1, DSPP and ALP in shOMD-hDPSCs were significantly lower than those in the control group at each given time point. Specifically speaking, DMP1, DSPP and ALP mRNA levels in Ctrl-hDPSCs were at least twice as high as those in shOMD-hDPSCs at early induction period. After osteo/odontogenic induction for 14 days, DMP1, DSPP and ALP mRNA levels in Ctrl-hDPSCs were at least three times higher than those in shOMD-hDPSCs. In addition, shOMD-hDPSCs could not form calcified nodules or differentiate into osteo/odontoblast-like cells after 21 days of culture in osteo/odontogenic medium compared with the control groups (Fig. 4e and f).
The mRNA levels of a OMD, b DMP1, c DSPP and d ALP in Ctrl-hDPSCs and shOMD-hDPSCs during osteo/odontogenic induction by real-time qPCR. e Alizarin red S staining in Ctrl-hDPSCs and shOMD-hDPSCs after induction. f The relative quantification of the mineralization nodules in (e). **Significant difference (P < 0.01) versus Ctrl
Discussion
hDPSCs play an essential role in dentinogenesis and dental repair [26]. The capacity of self-renewal and multilineage differentiation into cell types such as odontoblasts/osteoblasts, adipocytes, and neuron-like cells attracts researchers a lot. hDPSCs have been considered as an alternative therapeutic cell source for dental tissue and whole-tooth regeneration [27]. Therefore, the identification of genes regulating the hDPSCs into osteo/odontogenic fate will help clarify the mechanisms of regenerative strategies.
Heretofore multiple signaling molecules and transcription factors, including bone morphogenetic proteins, fibroblast growth factors, Wnt proteins, Hedgehog families and Cbfa1/Runx2 protein, have been shown to be implicated in mediating differentiation and organization of osteogenic tissues when responding to inductive signals [28]. However, there still existed mechanisms that have not been disclosed. To the best of our knowledge, OMD was one of the genes which had seldomly been investigated with regard to its expression patterns and functions in hDPSCs’ cytodifferentiation.
In this study, the hDPSC subpopulation expressing STRO-1 surface marker was used. In accordance with previous reports [29, 30], the isolated hDPSCs expressed MSC-specific cell surface antigens such as CD73, CD90, CD105 and CD166, and were negative for the hematopoietic surface markers CD34 and CD45. In our previous study, it was found that the expression of STRO-1 declined gradually with the continuing passage of cells (data not shown). Thus cell cultures between the second and fifth passages were used. In this study, mineral nodules (considered as a late marker of osteo/odontogenic differentiation [31]) were found to increase with the induction period of hDPSCs. ALP, DMP1 and DSPP (also referred as osteo/odontogenic differentiation markers [32, 33]) were up-regulated in the cells of the control group during the early induction period.
DSPP is a pre-proprotein secreted by odontoblasts and its cleaved products named dentin sialoprotein and dentin phosphoprotein are found in significant quantity in the extracellular matrix of dentin [34]. DMP1 has been reported to be expressed during the initial stages of mineralized matrix formation in bone and dentin [35]. The expression levels of DSPP and DMP1 suggest the dentinogenesis ability of dental pulp cells (DPCs) [36]. In the studies of Lin et al. and Qi et al., the transcription levels of DSPP and DMP1 increased continuously during odontoblastic differentiation of DPCs and achieved the highest level after 14 days of induction [37, 38], while in this study they reached the highest point at day 7. It is supposed that the difference in induction medium and cellular condition of hDPSCs may explain the discrepancies. Meanwhile, the expression of OMD was up-regulated in mRNA level after 21 days of induction. So it was proposed a hypothesis that OMD may regulate the osteo/odontoblastic differentiation of hDPSCs.
To test our hypothesis, OMD knockdown hDPSCs were established by infecting them with a lentiviral construct harboring shRNA targeting OMD. It is worth noting that the OMD transcriptional level in Ctrl-hDPSCs after induction for 21 days was three times higher than that in Ctrl-hDPSCs cultured on day 0 (Fig. 4a), while the OMD mRNA of the induced and uninfected hDPSCs was around 35 times higher than that in uninduced and uninfected hDPSCs (Fig. 2a). Reasons bringing about this phenomenon may be the lentivirus process and its potential influence on gene expression. Nonetheless, it could be noticed that OMD knockdown dramatically suppressed the differentiation of hDPSCs into osteo/odontoblasts according to the low expression of ALP, DMP1 and DSPP and reduction of calcified nodules formation. Studies have explored the potential mechanisms of OMD and it has been reported that the expression of OMD is regulated by the cytokines TGFβ1 and BMP2: TGFβ1 down-regulates OMD, while BMP-2 up-regulates OMD [11]. Therefore, it is proposed that OMD may plays a certain role in TGFβ and BMP signaling during osteo/odontogenic differentiation. However, the exact function of OMD during mineralization remains to be fully elucidated.
Conclusions
This study demonstrates that OMD knockdown can inhibit the osteo/odontoblastic differentiation of hDPSCs by suppressing mineralization and the expression of osteo/odontoblast-related genes. OMD may promote the osteo/odontogenic differentiation of hDPSCs. Further investigation is required to elucidate the mechanisms by how OMD regulates the biological characteristics of hDPSCs.
Abbreviations
- ALP:
-
alkaline phosphatase
- AM:
-
adipogenic differentiation medium
- DMP1:
-
dentin matrix acidic phosphoprotein 1
- DSPP:
-
dentin sialophosphoprotein
- FITC:
-
fluorescein iso thioocyanide
- GM:
-
growth medium
- hDPSCs:
-
human dental pulp stem cells
- mRNA:
-
messenger ribonucleic acid
- OM:
-
osteo/odontogenic differentiation medium
- OMD:
-
Osteomodulin
- PBS:
-
phosphate buffered saline
- PE:
-
phycoerythrin
- qPCR:
-
quantitative PCR
- RT-PCR:
-
reverse transcription PCR
- shRNA:
-
short hairpin RNA
- SLRP:
-
small leucine-rich proteoglycan
References
Gronthos S, Mankani M, Brahim J, Robey PG, Shi S. Postnatal human dental pulp stem cells (DPSCs) in vitro and in vivo. Proc Natl Acad Sci U S A. 2000;97(25):13625–30.
Shi S, Gronthos S. Perivascular niche of postnatal mesenchymal stem cells in human bone marrow and dental pulp. J Bone Miner Res. 2003;18(4):696–704.
Liu H, Gronthos S, Shi S. Dental pulp stem cells. Methods Enzymol. 2006;419:99–113.
Tonomura A, Sumita Y, Ando Y, Iejima D, Kagami H, Honda MJ, et al. Differential inducibility of human and porcine dental pulp-derived cells into odontoblasts. Connect Tissue Res. 2007;48(5):229–38.
Ajlan SA, Ashri NY, Aldahmash AM, Alnbaheen MS. Osteogenic differentiation of dental pulp stem cells under the influence of three different materials. BMC Oral Health. 2015;15:132.
Papagerakis P, Berdal A, Mesbah M, Peuchmaur M, Malaval L, Nydegger J, et al. Investigation of osteocalcin, osteonectin, and dentin sialophosphoprotein in developing human teeth. Bone. 2002;30(2):377–85.
Butler WT, Brunn JC, Qin C. Dentin extracellular matrix (ECM) proteins: comparison to bone ECM and contribution to dynamics of dentinogenesis. Connect Tissue Res. 2003;44(Suppl 1):171–8.
Waddington RJ, Hall RC, Embery G, Lloyd DM. Changing profiles of proteoglycans in the transition of predentine to dentine. Matrix Biol. 2003;22(2):153–61.
Orsini G, Ruggeri A Jr, Mazzoni A, Papa V, Mazzotti G, Di Lenarda R, et al. Immunohistochemical identification of decorin and biglycan in human dentin: a correlative field emission scanning electron microscopy/transmission electron microscopy study. Calcif Tissue Int. 2007;81(1):39–45.
Wendel M, Sommarin Y, Heinegard D. Bone matrix proteins: isolation and characterization of a novel cell-binding keratan sulfate proteoglycan (osteoadherin) from bovine bone. J Cell Biol. 1998;141(3):839–47.
Rehn AP, Chalk AM, Wendel M. Differential regulation of osteoadherin (OSAD) by TGF-beta1 and BMP-2. Biochem Biophys Res Commun. 2006;349(3):1057–64.
Sommarin Y, Wendel M, Shen Z, Hellman U, Heinegard D. Osteoadherin, a cell-binding keratan sulfate proteoglycan in bone, belongs to the family of leucine-rich repeat proteins of the extracellular matrix. J Biol Chem. 1998;273(27):16723–9.
Ramstad VE, Franzen A, Heinegard D, Wendel M, Reinholt FP. Ultrastructural distribution of osteoadherin in rat bone shows a pattern similar to that of bone sialoprotein. Calcif Tissue Int. 2003;72(1):57–64.
Tashima T, Nagatoishi S, Sagara H, Ohnuma S, Tsumoto K. Osteomodulin regulates diameter and alters shape of collagen fibrils. Biochem Biophys Res Commun. 2015;463(3):292–6.
Buchaille R, Couble ML, Magloire H, Bleicher F. Expression of the small leucine-rich proteoglycan osteoadherin/osteomodulin in human dental pulp and developing rat teeth. Bone. 2000;27(2):265–70.
Petersson U, Hultenby K, Wendel M. Identification, distribution and expression of osteoadherin during tooth formation. Eur J Oral Sci. 2003;111(2):128–36.
Calarco A, Di Salle A, Tammaro L, De Luca I, Mucerino S, Petillo O, et al. Long-term fluoride release from dental resins affects STRO-1+ cell behavior. J Dent Res. 2015;94(8):1099–105.
Miura M, Gronthos S, Zhao M, Lu B, Fisher LW, Robey PG, et al. SHED: stem cells from human exfoliated deciduous teeth. Proc Natl Acad Sci U S A. 2003;100(10):5807–12.
T LR, Sanchez-Abarca LI, Muntion S, Preciado S, Puig N, Lopez-Ruano G, et al. MSC surface markers (CD44, CD73, and CD90) can identify human MSC-derived extracellular vesicles by conventional flow cytometry. Cell Commun Signal. 2016;14:2.
Niu C, Yuan K, Ma R, Gao L, Jiang W, Hu X, et al. Gold nanoparticles promote osteogenic differentiation of human periodontal ligament stem cells via the p38 MAPK signaling pathway. Mol Med Rep. 2017;16(4):4879–86.
Hu H, Zhu W, Qin J, Chen M, Gong L, Li L, et al. Acetylation of PGK1 promotes liver cancer cell proliferation and tumorigenesis. Hepatology. 2017;65(2):515–28.
Wei Y, Ye Q, Tang Z, Tian G, Zhu Q, Gao H, et al. Calcitonin induces collagen synthesis and osteoblastic differentiation in human periodontal ligament fibroblasts. Arch Oral Biol. 2017;74:114–22.
Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25(4):402–8.
Liu J, Yu F, Sun Y, Jiang B, Zhang W, Yang J, et al. Concise reviews: characteristics and potential applications of human dental tissue-derived mesenchymal stem cells. Stem Cells. 2015;33(3):627–38.
Sharpe PT. Dental mesenchymal stem cells. Development. 2016;143(13):2273–80.
Sloan AJ, Smith AJ. Stem cells and the dental pulp: potential roles in dentine regeneration and repair. Oral Dis. 2007;13(2):151–7.
Yen AH, Yelick PC. Dental tissue regeneration - a mini-review. Gerontology. 2011;57(1):85–94.
Zhang YD, Chen Z, Song YQ, Liu C, Chen YP. Making a tooth: growth factors, transcription factors, and stem cells. Cell Res. 2005;15(5):301–16.
Yalvac ME, Ramazanoglu M, Rizvanov AA, Sahin F, Bayrak OF, Salli U, et al. Isolation and characterization of stem cells derived from human third molar tooth germs of young adults: implications in neo-vascularization, osteo-, adipo- and neurogenesis. Pharmacogenomics J. 2010;10(2):105–13.
Tasli PN, Tapsin S, Demirel S, Yalvac ME, Akyuz S, Yarat A, et al. Isolation and characterization of dental pulp stem cells from a patient with Papillon-Lefevre syndrome. J Endod. 2013;39(1):31–8.
Yu J, Wang Y, Deng Z, Tang L, Li Y, Shi J, et al. Odontogenic capability: bone marrow stromal stem cells versus dental pulp stem cells. Biol Cell. 2007;99(8):465–74.
Yokose S, Kadokura H, Tajima Y, Fujieda K, Katayama I, Matsuoka T, et al. Establishment and characterization of a culture system for enzymatically released rat dental pulp cells. Calcif Tissue Int. 2000;66(2):139–44.
Narayanan K, Srinivas R, Ramachandran A, Hao J, Quinn B, George A. Differentiation of embryonic mesenchymal cells to odontoblast-like cells by overexpression of dentin matrix protein 1. Proc Natl Acad Sci U S A. 2001;98(8):4516–21.
Prasad M, Butler WT, Qin C. Dentin sialophosphoprotein in biomineralization. Connect Tissue Res. 2010;51(5):404–17.
He G, George A. Dentin matrix protein 1 immobilized on type I collagen fibrils facilitates apatite deposition in vitro. J Biol Chem. 2004;279(12):11649–56.
Ching HS, Luddin N, Rahman IA, Ponnuraj KT. Expression of odontogenic and osteogenic markers in DPSCs and SHED: a review. Curr Stem Cell Res Ther. 2017;12(1):71–9.
Qi SC, Cui C, Yan YH, Sun GH, Zhu SR. Effects of high-mobility group box 1 on the proliferation and odontoblastic differentiation of human dental pulp cells. Int Endod J. 2013;46(12):1153–63.
Lin H, Xu L, Liu H, Sun Q, Chen Z, Yuan G, et al. KLF4 promotes the odontoblastic differentiation of human dental pulp cells. J Endod. 2011;37(7):948–54.
Acknowledgements
The authors would like to thank the affected individuals for participating in the study. The authors would like to show sincere appreciation to the staff at the Dr. Daming Gao’s lab, Shanghai Institute of Biochemistry and Cell Biology for providing technical support in this study.
Funding
This work was supported by grants from the National Natural Science Foundation of China (No. 81570964/81371143/81190133/81401844), Chinese Academy of Sciences (No. XDA01030502), Science and Technology Commission of Shanghai Municipality (No.14431900900/16430723500), and was partly supported by Science and Technology Commission of Shanghai Municipality (No.17140903500) and Shanghai Summit & Plateau Disciplines. The authors declare that the funding bodies played no role in: the design of the study; the collection, analysis, and interpretation of data; and the writing of the manuscript.
Availability of data and materials
All data generated or analysed during this study are included in this published article.
Author information
Authors and Affiliations
Contributions
WL prepared the basic study design and contributed in writing and revising the manuscript. LG participated in different aspects of laboratory studies and prepared of the primary draft for this paper. WJ and CN provided general technical support especially in lentivirus production. KY and XH helped get all basic laboratory materials. RM and ZH helped in the development of the main research idea and supervised the experimental process. All authors read and approved the final version of manuscript.
Corresponding authors
Ethics declarations
Ethics approval and consent to participate
Procedures involving human subjects were carried out in accordance with The Code of Ethics of the World Medical Association (Declaration of Helsinki). This study was approved by the Ethics Committee of Shanghai Ninth People’s Hospital affiliated with Shanghai Jiao Tong University, School of Medicine, China (Document No. 201769). Written informed consent was obtained from each patient.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Lin, W., Gao, L., Jiang, W. et al. The role of osteomodulin on osteo/odontogenic differentiation in human dental pulp stem cells. BMC Oral Health 19, 22 (2019). https://doi.org/10.1186/s12903-018-0680-6
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12903-018-0680-6