Abstract
Background
Mutations in the GLI-similar 3 (GLIS3) gene encoding the transcription factor GLIS3 are a rare cause of neonatal diabetes and congenital hypothyroidism with 12 reported patients to date. Additional features, previously described, include congenital glaucoma, hepatic fibrosis, polycystic kidneys, developmental delay, facial dysmorphism, osteopenia, sensorineural deafness, choanal atresia, craniosynostosis and pancreatic exocrine insufficiency.
Case presentation
We report a new case for consanguineous parents with homozygous novel mutation in GLIS3 gene who presented with neonatal diabetes mellitus, severe resistant congenital hypothyroidism, cholestatic liver disease, bilateral congenital glaucoma and facial dysmorphism. There were associated abnormalities in the external genitalia in form of bifid scrotum, bilateral undescended testicles, microphallus and scrotal hypospadias which might be a coincidental finding.
Conclusions
We suggest that infants with neonatal diabetes associated with dysmorphism should be screened for GLIS3 gene mutations.
Similar content being viewed by others
Background
The Gli-similar family of Kruppel-like zinc finger proteins is comprised of three proteins, Glis1-3. Glis1 was first identified by a yeast-two-hybrid screening using the ligand-binding domain of the retinoic acid-related orphan receptor γ (RORγ) as bait [1]. Subsequently, two additional members of the family were identified that possessed high levels of homology with the zinc fingers of Glis1 and were termed Glis2 and Glis3 [2, 3]. The human GLIS3 gene is located on chromosome 9p24.2 and encodes a protein that is approximately 90 kD in size [4]. Mutations in GLIS3 (9p24.2, OMIM#610192) have been described in the literature as a rare cause of neonatal diabetes. GLI-similar 3 (GLIS3) is identified transcription factor containing five Kruppel-like zinc finger motifs [5]. GLIS3 expression occurs early in embryogenesis and is thought to play a critical role in the cellular regulation of development by functioning as a repressor or activator of transcription [5, 6]. In 2003, Taha et al described two siblings for consanguineous Saudi Arabian family had intrauterine growth retardation, neonatal diabetes and hypothyroidism, progressive hepatic fibrosis, renal cystic dysplasia, facial dysmorphism and congenital glaucoma [7]. In 2006, Senee et al performed Genome wide linkage analysis and sequencing of candidate genes on this family and they identified a homozygous frame shift mutation (c.1873dupC) in GLIS3 gene which is likely to result in transcript degradation by nonsense mediated decay [8]. These patients died from infection in infancy. Later on, 12 patients with mutations within the GLIS3 gene have presented with a wider phenotype consisting mainly of neonatal diabetes and congenital hypothyroidism, in addition to multiple features involving different organs. Dimitri et al described the genetic and clinical features of those 12 patients who had GLIS3 mutation [9]. The variation in the GLIS3 phenotype is attributed to the tissue expression of variable length transcripts derived from the 11 exon GLIS3 gene [10]. GLIS3 is expressed in a tissue-specific manner with the highest levels of expression observed in multiple tissues including testis, although genital abnormalities have not previously manifested in those patients. We describe a new case in which mutation in GLIS3 have resulted in severely affected patient with an extended phenotype including abnormalities in external genitalia that have not been previously described.
Case presentation
Our patient is a male, who was born with a weight of 1.3 kg, length of 42 cm and head circumference of 29.4 cm after 36 weeks of gestation, to consanguineous Saudi parents (1st degree cousins). He developed diabetes on day 2 after birth, requiring continuous intravenous insulin (0.03 IU/kg/h) and was subsequently treated with subcutaneous insulin (glargine once daily and Aspart as needed). Target blood glucoses have been difficult to achieve due to labile glucose level. His current daily dose of insulin is 0.4 IU/kg/day. Glycosylated haemoglobin was difficult to interpret due to persistently high levels of fetal haemoglobin.
On day 3 of life, hypothyroidism was identified. His cord thyroid stimulating hormone (TSH) level was extremely high. Serum TSH was 270μIU/L (normal 0.27–4.2) and free thyroxine (FT4) of 0.4pmol/L (normal 12–22). Thyroglobulin (TG) level was 427 ng/ml (normal 3.5–77). Maternal thyroid functions were normal. He was initially managed by oral Thyroxine20 mcg/kg per day of (25 mcg daily). Suppression of TSH proved difficult despite consistently normal FT4 measurements. Serum TSH concentration remains at above 100 μIU/L despite high dose of oral thyroxine (50 mcg/kg per day). Thyroid anatomy was normal on ultrasound scan while radioiodine scan was not done due to technical difficulties.
Patient developed early cholestasis. His total bilirubin was 289 umol/L (normal 2–193) with high conjugated bilirubin 103 umol/L (normal 0–5) and high gamma-glutamyl-transpeptidase 432 U/L (normal 8–61). His alanine aminotransferase and aspartate aminotransferase were normal. TORCH screen was negative. Ultrasound abdomen showed normal liver size and echogenicity. hepatobiliary scintigraphy showed patent biliary to bowel transit excluding biliary atresia with preserved hepatocyte function. Liver biopsy was refused by his parents but his liver function had improved gradually after starting of ursodeoxycholic acid orally.
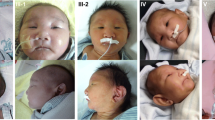
Ophthalmic examination showed bilateral severe glaucoma so; he underwent goniotomy and trabeculotomy in early neonatal period. He has subtle dysmorphic features in form of depressed nasal bridge, pointed chin and ocular hypertelorism. Cranial and renal ultrasound scan were normal. Skeletal survey showed no skeletal abnormalities. He has normal hearing test.
His genital examination showed stretched phallus length of 1.8 cm with scrotal hypospadias, ventral chordae and bifid scrotum. Gonads were palpable in the inguinal canal. Karyotyping showed 46XY. Ultrasound of the pelvis and inguinal region showed no mullerian system, both testicles are in the distal inguinal region at the external ring with normal echogenicity and normal vascular perfusion.
Cortisol level was 395 nmol/L and he was maintaining normal electrolytes and serum glucose. Adrenocorticotropic hormone (ACTH) level was normal; 5.1pmol/L. 17-hydroxy-progesterone (17-OHP) was 2.3 nmol/L. Human chorionic gonadotropin (hCG) stimulation test was done at age of 5 months. Patient was given daily intramuscular 1500 IU/m2 of hCG for 3 days. Androstenedione, testosterone and dihydrotestosterone pre and post hCG stimulation were shown in Table 1. According to the laboratory results showed in Table 1, 5-alfa reductase deficiency, partial androgen insensitivity, 17-beta-hydroxysteroid dehydrogenase deficiency and congenital adrenal hyperplasia are unlikely to be the cause of genital abnormalities in our patient.
Genetic analysis
Methods
Consent was obtained from his parents to perform genetic analysis. The 10 coding exons (exons2–11) of the GLIS3 gene on chromosome 9p24.2 (OMIM 610192) mutations were amplified by polymerase chain reaction (PCR) and sequenced directly. The resulting sequence data were compared with the reference sequence NM_001042413.
Results
Detection of homozygous mutation c.23113_2314dupTC (p.Pro772Leufs*35) in the GLIS3 gene.
Interpretation
Sequence analysis revealed a homozygous duplication of two nucleotides at position c.2313_2314 in exon 9 of the GLIS3 gene (c.2313_2314dupTC). This leads to a frameshift, resulting in premature termination codon (p.Pro772leufs*35) and subsequently in degradation of the mRNA (nonsense-mediated decay) or in a truncation of the protein. To best of our knowledge, this mutation has not been described in the literature so far and can be regarded pathogenic. Furthermore, the silent variant c.176G > T (p.Arg589Arg) in the axon 5 of GLIS3 gene could be detected in the homozygous state. Analysis by different bioiformatic tools did not predict splicing. This variant can most likely be regarded a pathogenic. The result was confirmed by sequencing of an independent PCR product. Although sequencing analysis cannot exclude a large heterozygous deletion in the GLIS3 gene in trans to c.2313_2314dupTC (i.e hemizygosity), homozygosity for c.2313_2314dupTC is most likely. Both homozygosity and hemizygosity for c.2313_2314dupTC in the GLIS3 gene would be compatible with the clinical diagnosis of our patient. In order to distinguish between homozygosity and compound heterozygosity for c.2313_2314dupTC and large deletion comprising this part on the allele we did analysis for the mutation in both parents through the same methods in the same genetic laboratory. Sequence analysis for both parents revealed a heterozygous duplication of two nucleotides at position c.2313_2314 in exon 9 of the GLIS3 gene (c.2313_2314dupTC), resulting in premature termination codon (p.Pro772leufs*35) and subsequently in degradation of the mRNA (nonsense-mediated decay) or in a truncation of the protein. Furthermore, the silent variant c.176G > T (p.Arg589Arg) in the axon 5 of GLIS3 gene was detected in the heterozygous state. Analysis by different bioiformatic tools did not predict significant scores for aberrant splicing. This variant can most likely be regarded a pathogenic.
Discussion
We herein describe a new patient who has a novel homozygous mutation of GLIS3 gene with duplication of two nucleotides at position c.2313_2314 in exon 9 of the GLIS3 gene (c.2313_2314dupTC).
In agreement with all previous reports of GLIS3 mutations, our patient had neonatal diabetes, congenital hypothyroidism, congenital glaucoma, hepatic cholestasis, intrauterine growth retardation, developmental delay and characteristic facial dysmorphism but there was absent renal, hearing and skeletal involvement.
In this patient, we have described congenital hypothyroidism that has not responded to conventional treatment. Eleven out of twelve patients described with GLIS3 mutation, had high TSH and low FT4. In most cases, patients have not responded to conventional treatment and maintained elevated levels of TSH despite normalization of T4. However, abnormalities in thyroid anatomy and/or T4 uptake are not sufficient to explain this [10]. Three patients described by Senee et al responded to daily T4 treatment, but subsequent TSH levels were not reported. Thyroid ultrasound and scintigraphy results also suggested athyreosis or hypoplasia with absent radioiodine uptake [8]. Patients described by Dimitri and Taha had high daily T4 requirements with persistently elevated TSH and increased TG levels despite normal thyroid anatomy which are similar to our patient [7, 11]. One patient of GLIS3 mutation with a deletion of exons 1–2, hypothyroidism was identified on day four of life with TSH levels >150μIU/l and T4 at 4.3pmol/l. The patient was treated with 20 μg/kg of T4 daily with sufficient TSH suppression. However, at 2 months of age, TSH levels exceeded 150mIU/l and remained high despite and increased dose of 75 μg/kg daily [11].
Although it is clear that almost all patients with a GLIS3 mutation present with thyroid dysfunction, the absence of consistent pathological features in patients makes it difficult to ascertain a unifying causative mechanism. It is possible that the markedly elevated levels of TSH observed in our patient and those described previously, combined with the variation in thyroid anatomy are a result of partial to complete TSH resistance. However, this does not explain the variable reduction in TSH, on occasions, values within the normal range following initial T4 supplementation, and the need thereafter to administer T4 three times daily to normalize free T4 [7].
GLIS3 interacts with key regulatory genes in pancreatic embryogenesis including ONECUT1 and NEUROGENIN3 (NEUROG3) [11–13]. GLIS3 expression also persists beyond the embryonic period promoting beta cell proliferation and regulating insulin gene expression through binding to GLI-RE on the INS gene [14, 15]. Therefore, the variation in insulin sensitivity between patients with GLIS3 mutations may relate to the impact of the mutation on the nuclear localization, GLI-binding element activity, transactivation, pancreatic development, subsequent cell proliferation, and remnant endogenous insulin production. In humans GLIS3 has been identified as a susceptibility locus for the risk of type 1 and 2 diabetes [15, 16].
As previously reported, most of the patients who had GLIS3 mutations presented with renal parenchymal disease, primarily renal cystic dysplasia [8]. However, some patients (including our patient) with mutations in GLIS3 did not develop renal disease. The variable presentation of renal disease therefore may be related to the relative qualitative and quantitative expression of tissue transcripts and the encoded proteins in individual patients, or alternatively the variation in the expression of regulatory transcripts.
Our patient has abnormalities in the external genitalia that have not been previously described. We believe that this is probably a coincidental finding. Whether or not GLIS3 mutations would be the cause of these abnormalities needs extensive genetic testing and work up which are beyond the scope of our report which aims to report the recently discovered novel mutation of GLIS3. If any other reports are released in future literature this might raise the need for more work up of these patient but at the level of our report we can’t raise this suspicion as being a coincidental finding is much more logic specially with the base line relatively high incidence of these findings in male neo borns with 5 in 1000 and 1–5 in 100 newborns for scrotal hypospedius with bifid scrotum [17, 18] and undescended testis [19] respectively.
Conclusions
In summary, we have described a patient with novel deletions in GLIS3, whose clinical phenotype includes more features than previously described and whose hypothyroidism was relatively resistant to conventional interventions as well as neonatal diabetes with labile blood glucose level. The extension of the phenotype, we have described, is probably coincidental.
Abbreviations
- 17-OHP:
-
17-hydroxy-progesterone
- ACTH:
-
Adrenocorticotropic hormone
- DHT:
-
Dihydrotestosterone
- FT4:
-
Free thyroxine
- GLIS3:
-
GLI-similar 3
- hCG:
-
Human chorionic gonadotropin
- PCR:
-
Polymerase chain reaction
- RORγ:
-
Retinoic acid-related orphan receptor γ
- TG:
-
Thyroglobulin
- TSH:
-
Thyroid stimulating hormone
References
Kim YS, Lewandoski M, Perantoni AO, Kurebayashi S, Nakanishi G, Jetten AM. Identification of Glis, a novelGli-related, Kruppel-like zincfinger protein containing transactivation and repressor functions. JBiol Chem. 2002;277:30901–13.
Lamar E, Kintner C, Goulding M. Identification of NKL, a novel GliKruppel zinc-finger protein that promotes neuronal differentiation. Development. 2001;128:1335–46.
Zhang F, Nakanishi G, Kurebayashi S, Yoshino K, Perantoni A, Kim YS, Jetten AM. Characterization of Glis2, a novel gene encoding a Glirelated, Kruppel-like transcription factor with transactivation and repressor functions. Roles in kidney development and neurogenesis. J Biol Chem. 2002;277:10139–49.
Lichti-Kaiser K, ZeRuth G, Jetten AM. transcription factor GLI-SIMILAR 3 (GLIS3): implications for the development of congenital hypothyroidism. J Endocrinol Diabet Obesity. 2014;2(2):1024.
Kim YS, Nakanishi G, Lewandoski M, Jetten AM. GLIS3, a novel member of the GLIS subfamily of Kruppel-like zinc finger proteins with repressor and activation functions. Nucleic Acids Res. 2003;31:5513–25.
Beak JY, Kang HS, Kim YS, Jetten AM. Functional analysis of the zinc finger and activation domains of Glis3 and mutant Glis3(NDH1). Nucleic Acids Res. 2008;36:1690–702.
Taha D, Barbar M, Kanaan H, Williamson Balfe J. Neonatal diabetes mellitus, congenital hypothyroidism, hepatic fibrosis, polycystic kidneys, and congenital glaucoma: a new autosomal recessive syndrome? Am J Med Genet A. 2003;122A(3):269–73.
Senee V, Chelala C, Duchatelet S, Feng D, Blanc H, Cossec JC, et al. Mutations in GLIS3 are responsible for a rare syndrome with neonatal diabetes mellitus and congenital hypothyroidism. Nat Genet. 2006;38(6):682–7.
Dimitri P, Habeb AM, Garbuz F, Millward A, Wallis S, Moussa K, Akcay T, Taha D, Hogue J, Slavotinek A, Wales JKH, Shetty A, Hawkes D, Hattersley AT, Ellard S, De Franco E. Expanding the clinical spectrum associated with GLIS3 mutations. J Clin Endocrinol Metab. 2015;100:0000.
Dimitri P, Warner JT, Minton JAL, Patch AM, Ellard S, Hattersley AT, Barr S, Hawkes D, Wales JK, Gregory JW. Novel GLIS3 mutations demonstrate an extended multisystemphenotype. Eur J Endocrinol. 2011;164:437–43.
Kim YS, Kang HS, Takeda Y, Hom L, Song HY, Jensen J, et al. Glis3 regulates neurogenin 3 expression in pancreatic beta-cells and interacts with its activator, Hnf6. Mol Cells. 2012;34(2):193–200.
Poll AV, Pierreux CE, Lokmane L, Haumaitre C, Achouri Y, Jacquemin P, et al. A vHNF1/TCF2-HNF6 cascade regulates the transcription factor network that controls generation of pancreatic precursor cells. Diabetes. 2006;55(1):61–9.
Kang HS, Kim YS, ZeRuth G, Beak JY, Gerrish K, Kilic G, et al. Transcription factor Glis3, a novel critical player in the regulation of pancreatic beta-cell development and insulin gene expression. Mol Cell Biol. 2009;29(24):6366–79.
Yang Y, Chang BH, Chan L. Sustained expression of the transcription factor GLIS3 is required for normal beta cell function in adults. EMBO Mol Med. 2013;5(1):92–104.
Cho YS, Chen CH, Hu C, Long J, Ong RT, Sim X, et al. Metaanalysisof genome-wide association studies identifies eight new locifor type 2 diabetes in east Asians. Nat Genet. 2011;44(1):67–72.
Santin I, Eizirik DL. Candidate genes for type 1 diabetes modulatepancreatic islet inflammation and beta-cell apoptosis. Diabet Obes Metab. 2013;15 Suppl 3:71–81.
Paulozzi LJ, Erickson JD, Jackson RJ. Hypospadias trends in two US surveillance systems. Pediatrics. 1997;100:831–4.
Mai CT, Isenberg J, Langlois PH, Alverson CJ, Gilboa SM, Rickard R, Canfield M, Anjohrin SB, Lupo PJ, Jackson DR, Stallings EB, Scheuerle AE, Kirby RS for the National Birth Defects Prevention Network. Brief report, population-based birth defects data in the United States, 2008 to 2012: presentation of state-specific data and descriptive brief on variability of prevalence. Birth Def Res (Part A). 2015;103:972–94.
Abacı A, Çatlı G, Anık A, Böber E. Epidemiology, classification and management of undescended testes: does medication have value in its treatment? J Clin Res Pediatr Endocrinol. 2013;5(2):65–72.
Funding
This research did not receive any grant from any funding agency.
Availability of data and materials
All data generated or analysed during this study are included in this published article.
Authors’ contributions
KA, AAlsaedi, AAljasser, A Altawil; diagnosed the patient, did investigations, followed up him, drafted the manuscript. NMK, KA: reviewed literature, drafted the manuscript, and reviewed the manuscript for final publication.
Competing interests
The authors declare that they have no competing interests.
Consent for publication
Written informed consent was obtained from the patient’s father for publication of details of the case report. Personal information were not mentioned in a way which can lead to identification of the patient or his family.
Ethics approval and consent to participate
The study was approved by the alhada armed forces hospital research and ethical committee. Written informed consent was obtained from the patient’s father for contribution of his child in the study.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Alghamdi, K.A., Alsaedi, A.B., Aljasser, A. et al. Extended clinical features associated with novel Glis3 mutation: a case report. BMC Endocr Disord 17, 14 (2017). https://doi.org/10.1186/s12902-017-0160-z
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12902-017-0160-z