Abstract
Background
The ubiquitin conjugating enzyme E2L3 (UBE2L3) gene is associated with susceptibility to many autoimmune diseases. The aim of this study was to investigate the association between UBE2L3 gene and autoimmune thyroid diseases (AITDs) and their clinical phenotypes.
Methods
We genotyped five single-nucleotide polymorphisms (SNPs) rs131654, rs5754217, rs2298428, rs140489 and rs5998672 of UBE2L3 gene in case groups including 1028 patients with AITDs [676 cases of Graves’ disease (GD) and 352 cases of Hashimoto’s thyroiditis (HT)] and control group including 897 healthy individuals.
The genotyping was performed with the method of polymerase chain reaction-ligase detection reaction (PCR-LDR).
Results
The frequencies of allele and genotype of five SNPs in gene UBE2L3 showed no statistically significant difference between case groups and control group, respectively. Moreover, no significant differences in frequencies of allele and genotype of five SNPs of the gene were found between clinical subphenotypes of AITDs and control group. Such subphenotypes included GD, HT, and thyroid associated ophthalmopathy (TAO). The negative results were also found in the frequency of other haplotypes of the gene except the haplotype of TCGGC, which was significantly higher in HT group than in control group (P = 0.031, OR = 1.441).
Conclusions
The present findings indicate that TCGGC haplotype is associated with an increased risk of HT and UBE2L3 gene is likely to be a susceptibility factor to HT in a Chinese Han population.
Similar content being viewed by others
Background
Autoimmune thyroid diseases (AITDs) are a set of organ specific endocrine autoimmune diseases mediated by T lymphocyte. They mainly consist of Graves’ disease (GD), Hashimoto’s thyroiditis (HT), and thyroid associated ophthalmopathy (TAO). GD and HT are the most common phenotypes of AITDs. Now the pathogenesis of AITDs is thought to be immune dysfunction due to the interaction between genetic susceptibility and environmental factors. The recent studies have identified more than ten susceptibility loci for AITDs, which include cytotoxic T lymphocyte associated protein 4 (CTLA4) gene [1], thyroid stimulating hormone receptor (TSHR) gene [2], protein tyrosine phosphatase, non receptor type 22 (PTPN22) gene [3], interleukin (IL17) gene [4], thyroglobulin (TG) gene [5], human leukocyte antigen (HLA) gene [6], and FCRL3 gene [7]. The majority of susceptibility genes belong to immunity-related genes.
Ubiquintin proteasome pathway (UPP) is an important signal transduction system which involves many physiological processes. In this pathway, UBE2L3 is an ubiquitin conjugating enzyme (E2) participating in ubiquitination of substrate through sequential steps catalyzed together with activating (E1) and ligase (E3) enzymes, and exerts its degradation effects on targeting molecules [8]. These targeting molecules encompass p53 [9], c-Fos [10] and some immune or inflammatory reaction related molecules such as nuclear factor-κB (NF-κB) [11] and Toll-like receptors (TLR) [12].
Genetic variation in the UBE2L3 has been demonstrated in many autoimmune diseases such as systemic lupus erythematosus (SLE) [13–15], rheumatoid arthritis (RA) [15, 16] and Crohn’s disease (CD) [17, 18]. As we all know, many different autoimmune diseases often co-exist in one individual or aggregate in different members of one family. More importantly, accumulated evidence indicates that different autoimmune diseases may share common susceptible genes. Therefore, exploring the UBE2L3 gene mutation in the etiology of AITDs is a pivotal issue. In the present case–control study, we analyzed the SNPs in UBE2L3 region, rs131654, rs5754217, rs2298428, rs140489 and rs5998672 in AITDs in a Chinese Han population.
Methods
Subjects
We studied samples from Department of Endocrinology, the First Affiliated Hospital of Xi’an Jiaotong University Health Science Center, and Department of Endocrinology, Jinshan Hospital of Fudan University. The case groups including 1028 patients with AITDs cohort consisted of 676 in GD subgroup (208 males and 468 females, mean age of 36.918 years) and 352 in HT subgroup (44 males and 308 females, mean age of 34.774 years). The subjects in our study were not related to each other. Diagnostic basis for GD was typical high metabolic syndrome, different degrees of diffuse goiter, laboratory examination indicating hyperthyroidism and positive antibody against TSH receptor (TRAb). HT was confirmed by goiter, thyroid antibody against Tg (TgAb) or antibody against thyroid peroxidase (TPOAb) or even thyroid puncture pathological examination. The control group included 897 healthy controls (300 males and 597 females, mean age of 38.734 years), who were all screened for absence of thyroid goiter and personal or family history of thyroid diseases and any autoimmune diseases. The thyroid function was in the normal range. All subjects were ethnic Chinese Han and signed informed consent at each site. The study was approved by the Ethics Committee of Jinshan Hospital of Fudan University.
Single-nucleotide polymorphism (SNP) genotyped of UBE2L3
We selected five SNPs rs131654, rs5754217, rs2298428, rs5998672 and rs140489 of UBE2L3 region because of their strong association with the other autoimmune diseases such as SLE, RA, Crohn’s disease, and Sjögren’s syndrome [13–18].
DNA extraction
The genomic DNA was extracted from 2 ml of peripheral blood from each subject using RelaxGene Blood DNA System (Tiangen biotech, Beijing, China) according to the manufacture’ s protocol.
Genotyping
UBE2L3 SNPs were genotyped using polymerase chain reaction-ligase detection reaction (PCR-LDR) and analyzed with polymerase chain reaction (PCR) machine (PRISM 3730, ABI). Five pairs of primers and the target gene LDR-probe sequences for SNPs of UBE2L3 region are shown as Additional file 1: Table S1.
Clinical phenotype analysis
We analyzed the case groups’ clinical characteristics and correlation between frequencies of allele, genotype and haplotype and subphenotypes on the basis of [1] the onset age of disease (≤18 years versus ≥19 years); [2] presence or absence of AITDs family history (defined as the subjects’ first-degree relatives including parents, children and siblings or second-degree relatives such as grandparents, uncles and aunts who had AITDs occurrence); [3] presence or absence of ophthalmopathy (defined as a distinctive disorder characterized by inflammation and swelling of the extraocular muscles, eyelid retraction, periorbital edema, episcleral vascular injection, conjunctive swelling and proptosis); [4] euthyroid or hypothyroid HT (defined as serum TSH over 10 U/l) (Roche, China). The clinical characteristic data of the case groups are shown in Table 1.
Statistical analysis
Allele and genotype frequencies in case and control groups were implemented using SPSS17.0 (SPSS Inc., Chicago, IL, USA). Comparison of genotype and allele frequencies between case and control groups was made by Chi-square test. P value of < 0.05 was considered significant. Differences between groups were assessed by the odd ratio (OR) and 95 % confidence interval (95%CI). Haplotype frenquencies and linkage disequilibrium (LD) test were analyzed using Haploview 4.2 [19]. LD test was carried out using the pairwise LD analyze D’ and r2.
Results
Allele and genotype frequency distribution
There were no differences in allele or genotype frequency distribution in any SNP between patients (GD and HT) and control group (Additional file 1: Table S2). The same results were also found in subphenotypes such as GD patients with ophthalmopathy, family history and different thyroid function of HT patients (the data not shown).
Haplotype frequencies analysis
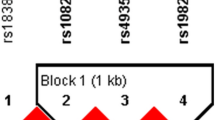
We found that five SNPs rs131654, rs5754217, rs2298428, rs5998672, and rs140489 of UBE2L3 region were in the same LD block (65 kilobases) using Hapmap CHB data (corresponding to samples of China ancestry from Beijing) and Haploview software. Strong linkage disequilibrium was shown for five SNPs (Fig. 1).
We obtained four haplotypes GCGGC, TTTAT, TCGGC and TTTAC in the UBE2L3 LD block using Haploview software, with frequencies all above 0.05. Unfortunately, compared with control group, the frequencies of haplotypes of GCGGC, TTTAT and TTTAC revealed no statistical difference with GD, HT or integrated AITD group respectively. However, the frequency of TCGGC haplotype in HT patients was significantly higher than that in healthy controls (P = 0.031, OR = 1.441), which suggested that the haplotype TCGGC increased the risk of HT (Table 2).
Discussion
Ubiquitin-mediated specific cellular protein degradation is involved in the regulation of many cellular processes, including cell cycle progression, differentiation, transcriptional adjustment, antigen presenting, receptor signal transduction, tumorigenesis and cell apoptosis [20, 21]. Increasing evidence suggests that UPP is closely related to the occurrence and progression of many autoimmune diseases. UPP’s influence on the immune system is embodied in both innate and adaptive immunities [22]. Under normal circumstances, the body’s innate immune system identifies and combines with intrusive pathogens by such receptors as toll-like receptor (TLR), RIG-1 receptor (RLR), and NOD receptors (NLR), and activates certain transcription factors such as NF-κB, interferon regulatory factor (IRF) family, subsequently inducing the release of various cytokines and inflammatory chemokines to get rid of pathogen from the body. It has been already demonstrated that innate immunity related TLR gene is one of genetic factors for AITDs [23, 24]. The influence of UPP on innate immunity is also controlled negatively by deubiquitylating enzymes (DUBs). As one of the DUBs, TNFAIP3 gene was also showed an association with the AITDs in our previous study [25]. So, studying the relationship between the polymorphisms of UBE2L3 and AITDs is pivotal and interesting. In our study, the frequency of haplotype TCGGC was higher in HT patients than in normal group, This suggests that the genotype which influences the pathogenesis of HT may exist in the vicinity of or in the haplotype itself. That is to say, UBE2L3 gene polymorphism is a predisposing factor for HT occurrence.
Adaptive immunity is generally mediated by T and B lymphocytes. UPP is involved in the regulation of T and B cell functions by adjusting T cell receptor (TCR) and B cell receptor (BCR) signaling transduction. There have been reports on the correlation of five SNPs in UBE2L3 with many T cell or B cell mediated autoimmune diseases, such as SLE [14, 15, 26], RA [15, 16], Crohn’s disease [16, 18], celiac disease [16], Sjögren’s syndrome (SS) and neonatal lupus erythematosus [27]. As a set of complex autoimmune diseases, not only T cell and B cell infiltrate in the lesioned thyroid tissues of AITDs, but also both innate and adaptive immunities may be implicated in its development. It did not come singly but in pairs in the present study, which revealed the relation between UBE2L3 gene variation and the pathogenesis of AITDs.
Although various subtypes of AITDs share common genetic factors and biomarkers, they indeed have their own specific manifestations and probably specific pathophysiolgic mechanisms. We speculated that different clinical types of AITDs may have different prominent alleles and genotypes. But we did not get the predicted result after stratifying the AITDs patients into GD, HT, and TAO ones, cases with positive family history or not, and HT in euthyroid or hypothyroid state.
Herein, our study provide an important clue that SNPs in UBE2L3 are involving in the pathogenesis of HT. Nevertheless, we have to admit this result may be influenced by sample size, different ethnic groups or even different research methods. Further research on UBE2L3 gene in AITDs of other races or even on UBE2L3 gene expression in AITDs is expected in the future.
Conclusions
Thus, we may conclude that UBE2L3 gene variant may be implicated in HT in a Chinese Han population. Further investigations with other ethnic cohorts await to confirm the implication.
Availability of data and materials
The dataset supporting the conclusions of this article is included within the article and its additional file.
Abbreviations
- AITDs:
-
autoimmune thyroid diseases
- CD:
-
crohn’s disease
- CTLA4:
-
cytotoxic t lymphocyte associated protein 4
- E1:
-
ubiquitin activating enzyme
- E3:
-
ubiquitin ligase enzymes
- GD:
-
graves’ disease
- HLA:
-
human leukocyte antigen
- HT:
-
hashimoto’s thyroiditis
- IL17:
-
interleukin 17
- NF-κB:
-
nuclear factor-κB
- PCR-LDR:
-
polymerase chain reaction-ligase detection reaction
- PTPN22:
-
protein tyrosine phosphatase, non receptor type 22
- RA:
-
rheumatoid arthritis
- SLE:
-
systemic lupus erythematosus
- SNPs:
-
single-nucleotide polymorphisms
- TAO:
-
thyroid associated ophthalmopathy
- TG:
-
thyroglobulin
- TgAb:
-
thyroid antibody against Tg
- TPOAb:
-
antibody against thyroid peroxidase
- TRAb:
-
antibody against TSH receptor
- TSHR:
-
thyroid stimulating hormone receptor
- UBE2L3:
-
ubiquitin conjugating enzyme E2L3
- UPP:
-
ubiquintin proteasome pathway
References
Zhao SX, Pan CM, Cao HM, Han B, Shi JY, Liang J, et al. Association of the CTLA4 gene with Graves’ disease in the Chinese Han population. PLoS One. 2010;5(3):e9821.
Liu L, Wu HQ, Wang Q, Zhu YF, Zhang W, Guan LJ, et al. Association between thyroid stimulating hormone receptor gene intron polymorphisms and autoimmune thyroid disease in a Chinese Han population. Endocr J. 2012;59(8):717–23.
Heward JM, Brand OJ, Barrett JC, Carr-Smith JD, Franklyn JA, Gough SC. Association of PTPN22 haplotypes with Graves’ disease. J Clin Endocrinol Metab. 2007;92(2):685–90.
Yan N, Yu YL, Yang J, Qin Q, Zhu YF, Wang X, et al. Association of interleukin-17A and -17 F gene single-nucleotide polymorphisms with autoimmune thyroid diseases. Autoimmunity. 2012;45(7):533–9.
Maierhaba M, Zhang JA, Yu ZY, Wang Y, Xiao WX, Quan Y, et al. Association of the thyroglobulin gene polymorphism with autoimmune thyroid disease in Chinese population. Endocr J. 2008;33(3):294–9.
Zeitlin AA, Heward JM, Newby PR, Carr-Smith JD, Franklyn JA, Gough SC, et al. Simmonds MJ. Analysis of HLA class II genes in Hashimoto’s thyroiditis reveals differences compared to Graves’ disease. Genes Immun. 2008;9(4):358–63.
Chu X, Pan CM, Zhao SX, Liang J, Gao GQ, Zhang XM, et al. A genome-wide association study identifies two new risk loci for Graves’ disease. Nat Genet. 2011;43(9):897–901.
Weissman AM. Themes and variations on ubiquitylation. Nat Rev Mol Cell Biol. 2001;2(3):169–78.
Ciechanover A, Shkedy D, Oren M, Bercovich B. Degradation of the tumor suppressor protein p53 by the ubiquitin-mediated proteolytic system requires a novel species of ubiquitin-carrier protein, E2. J Biol Chem. 1994;269(13):9582–9.
Nuber U, Schwarz S, Kaiser P, Schneider R, Scheffner M. Cloning of human ubiquitin-conjugating enzymes UbcH6 and UbcH7 (E2-F1) and characterization of their interaction with E6-AP and RSP5. J Biol Chem. 1996;271(5):2795–800.
Orian A, Whiteside S, Israël A, Stancovski I, Schwartz AL, Ciechanover A. Ubiquitin-mediated processing of NF-kappa B transcriptional activator precursor p105. Reconstitution of a cell-free system and identification of the ubiquitin-carrier protein, E2, and a novel ubiquitin-protein ligase, E3, involved in conjugation. J Biol Chem. 1995;270(37):21707–14.
Chuang TH, Ulevitch RJ. Triad3A, an E3 ubiquitin-protein ligase regulating Toll-like receptors. Nat Immunol. 2004;5(5):495–502.
Agik S, Franek BS, Kumar AA, Kumabe M, Utset TO, Mikolaitis RA, et al. The autoimmune disease risk allele of UBE2L3 in African American patients with systemic lupus erythematosus: a recessive effect upon subphenotypes. J Rheumatol. 2012;39(1):73–8.
Wang S, Adrianto I, Wiley GB, Lessard CJ, Kelly JA, Adler AJ, et al. A functional haplotype of UBE2L3 confers risk for systemic lupus erythematosus. Genes Immun. 2012;13(5):380–7.
Orozco G, Eyre S, Hinks A, Bowes J, Morgan AW, Wilson AG, et al. Study of the common genetic background for rheumatoid arthritis and systemic lupus erythematosus. Ann Rheum Dis. 2011;70(3):463–8.
Zhernakova A, Stahl EA, Trynka G, Raychaudhuri S, Festen EA, Franke L, et al. Meta-analysis of genome-wide association studies in celiac disease and rheumatoid arthritis identifies fourteen non-HLA shared loci. PLoS Genet. 2011;7(2):e1002004.
Franke A, McGovern DP, Barrett JC, Wang K, Radford-Smith GL, Ahmad T, et al. Genome-wide meta-analysis increases to 71 the number of confirmed Crohn’s disease susceptibility loci. Nat Genet. 2010;42(12):1118–25.
Fransen K, Visschedijk MC, van Sommeren S, Fu JY, Franke L, Festen EA, et al. Analysis of SNPs with an effect on gene expression identifies UBE2L3 and BCL3 as potential new risk genes for Crohn’s disease. Hum Mol Genet. 2010;19(17):3482–8.
Haploview, Broad Institute. http://www.broadinstitute.org/haploview. Accessed 18 Jan 2015.
Kornitzer D, Ciechanover A. Modes of regulation of ubiquitin-mediated protein degradation. J Cell Physiol. 2000;182(1):1–11.
Ciechanover A. The ubiquitin–proteasome pathway: on protein death and cell life. EMBO J. 1998;17(24):7151–60.
Bhoj VG, Chen ZJ. Ubiquitylation in innate and adaptive. Nature. 2009;458(7237):430–7.
Cho WK, Jang JP, Choi EJ, Jeon YJ, Jung IA, Kim SH, et al. Association of Toll-Like Receptor 10 Polymorphisms with Autoimmune Thyroid Disease in Korean Children. Thyroid. 2015;25(2):250–5.
Liao WL, Chen RH, Lin HJ, Liu YH, Chen WC, Tsai Y, et al. Toll-like receptor gene polymorphisms are associated with susceptibility to Graves’ ophthalmopathy in Taiwan males. BMC Med Genet. 2010;11:154.
Song RH, Yu ZY, Wang Q, Muhali FS, Jiang WJ, Xiao L, et al. Polymorphisms of the TNFAIP3 region and Graves’ disease. Autoimmunity. 2014;47(7):459–65.
Han JW, Zheng HF, Cui Y, Sun LD, Ye DQ, Hu Z, et al. Genome-wide association study in a Chinese Han population identifies nine new susceptibility loci for systemic lupus erythematosus. Nat Genet. 2009;41(11):1234–7.
Chung SA, Taylor KE, Graham RR, Nititham J, Lee AT, Ortmann WA, et al. Differential genetic associations for systemic lupus erythematosus based on anti-dsDNA autoantibody production. PLoS Genet. 2011;7(3):e1001323.
Acknowledgements
The present work was supported by grants from the National Natural Science Foundation of China (No. 81270871 and 81471004) and the Key Disciplines Development of Shanghai Jinshan District (No. 2012–23). The authors thank all of the participants in the present study.
Author information
Authors and Affiliations
Corresponding author
Additional information
Competing interests
No competing interest was stated from all the authors. The authors declared they are responsible for writing of the paper.
Authors’ contributions
YW conceived of the study and participated in its design, data collection, manuscript writing. YFZ collaborated on patients recruitment, data collection and statistical analysis. QW, JX (Jing Xu) and NY carried out laboratory tests. LFS, JX (Jian Xu) and STH performed statistical analysis and helped to draft the manuscript. JAZ conceived of the study and participated in its design, statistical analysis and revision of the manuscript for important intellectual content. All authors read and approved the final manuscript.
Additional file
Additional file 1:
Table S1. Annealing temperature, primer and probe sequences for the sequences analysis of UBE2L3 SNPs. Table S2. Allele and genotype frequencies in GD and HT patients. (DOC 103 kb)
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Wang, Y., Zhu, Yf., Wang, Q. et al. The haplotype of UBE2L3 gene is associated with Hashimoto’s thyroiditis in a Chinese Han population. BMC Endocr Disord 16, 18 (2016). https://doi.org/10.1186/s12902-016-0098-6
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12902-016-0098-6