Abstract
Background
Clinical treatment of osteosarcoma suffers from high recurrence rate. Therefore, is of great clinical values to develop predictive markers for recurrent osteosarcoma. Cervical carcinoma high-expressed lncRNA 1 (lncRNA CCHE1) participates in several types of malignancies, while its functionality in osteosarcoma is unknown. This study was therefore carried out to explore the involvement of lncRNA CCHE1 in recurrent osteosarcoma.
Methods
A total of 87 osteosarcoma patients received surgical resection and 38 healthy volunteers were included in this study. The 87 osteosarcoma patients were followed up for 5 years to record the recurrence of osteosarcoma. Plasma levels of lncRNA CCHE1 and ROCK1 on the day of discharge and during follow-up were measured by real-time quantitative PCR and ELISA, respectively. The effects of CCHE1 siRNA silencing on ROCK1 expression were analyzed by real-time quantitative PCR and western blot. Transwell assay was performed to analyze the role of lncRNA CCHE1 and ROCK1 in regulating cell invasion and migration.
Results
We observed that, on the day of discharge, plasma lncRNA CCHE1 was upregulated in osteosarcoma patients who developed distant recurrence (DR) during follow-up than in osteosarcoma patients who developed local recurrence (LR), patients with non-recurrence (NR) and healthy controls. On the day of discharge, plasma levels of ROCK1 were higher in DR, LR and NR groups in comparison to healthy controls. On the day of discharge, plasma levels of lncRNA CCHE1 were positively correlated with plasma levels of ROCK1 only in patients who developed DR during follow-up, but not in patients who developed LR, NR and control groups. During follow-up, plasma levels of lncRNA CCHE1 were further increased in DR group but slightly decreased in LR and NR groups. LncRNA CCHE1 siRNA silencing inhibited, while ROCK1 overexpression promoted osteosarcoma cell invasion and migration. ROCK1 overexpression attenuated the role of CCHE1. LncRNA CCHE1 siRNA silencing led to inhibited ROCK1 expression in cancer cells.
Conclusion
Therefore, lncRNA CCHE1 may participate in postoperative distant recurrence of osteosarcoma caner possibly by interacting with ROCK1 to promote cancer cell invasion and migration.
Similar content being viewed by others
Background
With the development of early diagnosis programs and surgical resection techniques, treatment outcomes of cancer at early stages have been significantly improved [1, 2]. However, postoperative survival of cancer patients is still challenged by the higher recurrence rate, which is a major cause of poor survival [3, 4]. As the most common histological type of primary bone cancer, osteosarcoma is a rare malignancy that only affect less than 1 per 100,000 [5]. However, increasing incidence rate of this disease was observed during the past several decades [6]. Although surgical resection can reach radical treatment in many cases of osteosarcoma patients, recurrence inevitably occur in a considerable portion of osteosarcoma patients, lead to poor survival [6]. Therefore, how to further improve the treatment and prevent postoperative recurrence of osteosarcoma is still a focus of the clinical treatment of osteosarcoma.
Rho-associated protein kinase 1 (ROCK1) is a major downstream effector of the small GTPase RhoA that plays pivotal roles in the regulation of cell motility [7]. In cancer biology, ROCK1 promotes cancer development and progression by mediating tumor metastasis [8]. At present, inhibition of ROCK1 has shown promising application values in the treatment of different types of malignancies [9, 10]. However, studies on the functions and clinical applications of ROCK1 in osteosarcoma are rare. LncRNA CCHE1 has been reported to be an oncogenic lncRNA that participates in several types of malignancies [11,12,13,14], while its involvement in osteosarcoma is unknown. Therefore, it will be interesting to investigate the roles of lncRNA CCHE1 in osteosarcoma and to explore its interactions with ROCK1. In the present study we showed that lncRNA CCHE1 could participate in postoperative distant recurrence of osteosarcoma possibly by interacting with ROCK1.
Methods
Cell lines and human plasma samples
Two osteosarcoma cell lines MG-63 and U2OS from ATCC (USA) were used. EMEM medium supplemented with 10% FBS was used to cultivate cancer cells under normal conditions (37 °C, 5% CO2).
Plasma samples were derived from 87 osteosarcoma patients and 38 healthy volunteers, who were admitted by Liaocheng People’s Hospital from January 2011 to May 2013. Patients’ inclusion criteria: 1) patients diagnosed as osteosarcoma at stage I and II for the first time by pathological examinations; 2) patients signed informed consent. Patients’ exclusion criteria: 1) patients with initiated therapy; 2) patients who were suffering from multiple diseases. All patients received surgical resection. Ethics committee of Liaocheng People’s Hospital approved this study.
Follow-up and grouping
A 5-year follow-up was performed after discharge. Distant recurrence (DR) occurred in 18 cases, local recurrence (LR) in 21 cases and non-recurrence (NR) in 48 cases.
Blood extractions on the day of discharge and during follow-up
Blood was extracted on the day of discharge, on the day of the diagnosis of DR or LR during follow-up or on the last day of follow-up in cases of NR. See Table 1 for patients’ information.
Real-time quantitative PCR
Monarch® Total RNA Miniprep Kit (NEB) was used to isolate RNA. RT-qPCR was performed using BlazeTaq™ One-Step SYBR Green RT-qPCR Kit (Genecopoeia). Primers used in PCR reactions were: 5′-AAGGTCCCAGGATACTCGC-3′ (forward) and 5′-GTGTCGTGGACTGGCAAAAT-3′ (reverse) for lncRNA CCHE1; 5′-ACCTGTAACCCAAGGAGATGTG-3′ (forward) and 5′-CACAATTGGCAGGAAAGTGG-3′ (reverse) for ROCK1; 5′-CAGGAGGCATTGCTGATGAT-3′ (forward) and 5′-GAAGGCTGGGGCTCATTT-3′ (reverse) for GAPDH. Thermal conditions for PCR reactions were: 95 °C for 45 s, then 95 °C for 16 s and 58.5 °C for 42 s for 40 cycles. Ct values were processed using 2-ΔΔCT methods.
ELISA
Plasma levels of ROCK1 were measured using ROCK1 ELISA Kit (Human) (OKEH06554, Aviva Systems Biology). All steps were completed according to manufacturer’s instructions. Plasma levels of ROCK1 were expressed as ng/ml.
Cell transfection
ROCK1 expression vector, lncRNA CCHE1 siRNA (5′-GCTTCTGACCAGCGACGCTAGGAGTAGCTG-3′) and negative control (NC) siRNA were from GenePharma (Shanghai, China). Cancer cells were cultivated to reach 80–90% confluence and transfection was carried out using lipofectamine 2000 (11668–019, Invitrogen, Carlsbad, USA) reagent with vectors and siRNAs at 10 nM and 50 nM, respectively. All steps were completed according to manufacturer’s instructions. Control group included untransfected cells and NC cells were empty vector- or NC siRNA-transfected cells. Cells were harvested at 24 h after transfection.
Transwell migration and invasion assay
Overexpression rate of ROCK1 above 200% and knockdown rate of lncRNA CCHE1 below 50% were reached before this experiment. Briefly, 3000 cells in 0.1 ml medium were transferred to the upper chamber and medium containing 20% FBS was used to fill the lower chamber. After 24 h, membranes were cleaned and 0.5% crystal violet (Sigma-Aldrich, USA) staining was performed in dark for 18 min at room temperature. This protocol was used for both migration and invasion assay except that the upper chamber was not pre-coated before invasion assay. Similar results, in terms of cell migration and invasion were obtained from two negative controls. Therefore, only one negative control is shown in results section for simplification.
Western-blot
Knockdown rate of lncRNA CCHE1 were reached before this experiment. RIPA (Bio-Rad) was used to isolate proteins. Proteins were separated using 12% SDS-PAGE gel. Western blotting was performed using conventional method. Primary antibodies included rabbit anti-human ROCK1 (1: 1200, ab97592, Abcam) and rabbit anti-human GAPDH antibody (1: 1200, ab37168, Abcam). IgG-HRP (1:1000, MBS435036, MyBioSource) was used as the secondary antibody. ECL (Sigma-Aldrich, USA) was used for signal development and data normalization was performed using Image J software.
Statistical analysis
Mean ± standard deviation values were used to express the data of 3 replicates. Comparisons of plasma levels of CCHE1 and ROCK1 among different groups as well as cell migration and invasion and ROCK1 expression among cells with different treatments were performed using one-way ANOVA and Tukey test. Comparisons of plasma levels of CCHE1 measure on the discharge and during follow-up levels were performed by paired test. Correlation analyses were performed using Pearson correlation coefficient. p < 0.05 was statistically significant.
Results
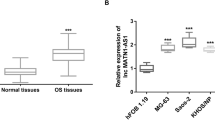
Differential expression of plasma lncRNA CCHE1 in different groups
QRT-PCR results revealed that, on the day of discharge, plasma lncRNA CCHE1 was upregulated in osteosarcoma patients who developed DR during follow-up than in osteosarcoma patients who developed LR, patients with NR and controls (Fig. 1, p < 0.05). Levels of plasma lncRNA CCHE1 were also higher in osteosarcoma patients with LR and patients with NR than in healthy controls (Fig. 1, p < 0.05). No significant differences in plasma levels of lncRNA CCHE1 were found between LR and NR groups (Fig. 1, p > 0.05).
Differential expression of plasma ROCK1 in different groups
ELISA results revealed that, on the day of discharge, plasma ROCK1 was upregulated in DR, LR and NR groups than in controls (Fig. 2, p < 0.05). No significant differences in plasma levels of ROCK1 were found among DR, LR and NR groups (Fig. 2, p > 0.05).
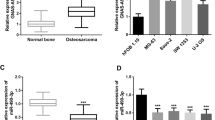
Plasma levels of lncRNA CCHE1 were positively correlated with plasma levels of ROCK1 only in DR group
On the day of discharge, Pearson correlation coefficient analysis showed that plasma levels of lncRNA CCHE1 were positively correlated with plasma levels of ROCK1 in DR group (Fig. 3a). In contrast, the correlation between plasma levels of ROCK1 and lncRNA CCHE1 were not significant in LR (Fig. 3b), NR (Fig. 3c) and control group (Fig. 3d).
Plasma levels of lncRNA CCHE1 were further increased during follow-up only in DR group
Compared with plasma levels of lncRNA CCHE1 on the day of discharge, significantly upregulated plasma lncRNA CCHE1 during follow-up was observed in DR group (Fig. 4a, p < 0.05). In contrast, slightly decreased plasma levels of lncRNA CCHE1 were observed in LR (Fig. 4b) and NR (Fig. 4c) groups.
LncRNA CCHE1 silencing inhibited migration and invasion of cells of MG-63 and U2OS human osteosarcoma cell lines
LcRNA CCHE1 silencing and OCK1 overexpression were reached at 24 h after transfection (Fig. 5a, p < 0.05). Compared with control group (C) and negative control group (NC), lncRNA CCHE1 silencing significant inhibited, while ROCK1 overexpression significantly promoted the migration (Fig. 5b) and invasion (Fig. 5c) of cells of both MG-63 and U2OS human osteosarcoma cell lines (p < 0.05). In addition, ROCK1 overexpression attenuated the inhibitory effects of lncRNA CCHE1 on cancer cell migration (Fig. 5b) and invasion (Fig. 5c) (p < 0.05).
LncRNA CCHE1 silencing inhibited migration and invasion of cells of MG-63 and U2OS human osteosarcoma cell lines. LcRNA CCHE1 silencing and OCK1 overexpression were reached at 24 h after transfection (a). LncRNA CCHE1 silencing significant inhibited, while ROCK1 overexpression significantly promoted the migration (b) and invasion (c) of cells of both MG-63 and U2OS human osteosarcoma cell lines. In addition, ROCK1 overexpression attenuated the inhibitory effects of lncRNA CCHE1 on cancer cell migration and invasion (*, p < 0.05)
LncRNA CCHE1 silencing mediated the inhibition of ROCK1 expression
Western-blot results showed that, compared with control group (C) and negative control group (NC), lncRNA CCHE1 silencing led to significantly inhibited expression of ROCK1 in cells of both MG-63 and U2OS human osteosarcoma cell lines (Fig. 6, p < 0.05).
Discussion
Postoperative recurrence of osteosarcoma is common [15]. The major finding of our study is that lncRNA CCHE1 may participate in distant recurrence but not in local recurrence of early stage osteosarcoma after surgical resection. We also provided experimental evidences that the actions of lncRNA CCHE1 are likely achieved through the interactions with ROCK1.
In spite of the development of early prediction approaches for cancer postoperative recurrence [16, 17], the accuracy is still low. In a recent study, Ying et al. reported multiple lncRNAs, such as RP1-261G23.7, RP11-69E11.4 and SATB2-AS1, can be used to predict recurrent osteosarcoma [18]. However, this study failed to distinguish local and distant recurrence. In our study we observed that lncRNA CCHE1 levels in plasma on the day of discharge were significantly higher in patients who developed distant recurrence than in patients with local recurrence and patients without recurrence. Therefore, detecting plasma lncRNA CCHE1 may provide guidance of follow-up chemotherapy or radiation therapy to prevent distant recurrence, which is a more malignant form compared with local recurrence.
As an oncogenic gene, ROCK1 is usually upregulated in the development of different types of cancers [7, 19]. Consistently, our study also observed significantly upregulated plasma ROCK1 in osteosarcoma patients than in healthy controls on the day of discharger. However, plasma ROCK1 was not differentially expressed in patients with different types of recurrence. Therefore, ROCK1 itself may not specifically participate in postoperative recurrence of osteosarcoma or at least have no predictive values for its recurrence.
ROCK1 participate in cancer biology not only through the interactions with proteins, but also by the cross-talk with non-coding RNAs, such as lncRNA [20, 21]. Interesting, our study observed positive correlation between plasma levels of ROCK1 and lncRNA CCHE1 on the day of discharge in patients who developed distant recurrence during follow-up. Our in vitro cell experiments data also revealed that lncRNA CCHE1 silencing can mediated the inhibited expression of ROCK1 in osteosarcoma cells, and lncRNA CCHE1 silencing may inhibit osteosarcoma cell migration and invasion through the downregulation of ROCK1. Therefore, inhibition of lncRNA CCHE1 expression may potentially be a therapeutic target for the treatment of osteosarcoma.
It is worth noting that we in this study only included a small number of patients and controls. To further analyze the reliability of the prediction of NSCLC distant recurrence by CCHE1, more clinical studies with bigger sample size are needed to further confirm the conclusion. Moreover, animal model experiments are also needed to confirm the functions of CCHE1.
Conclusion
In conclusion, overexpression of lncRNA CCHE1 is involved in the distant recurrence of early stage osteosarcoma after surgical resection. The actions of lncRNA CCHE1 are at least partially achieved through the interactions with ROCK1.
Availability of data and materials
The analyzed data sets generated during the study are available from the corresponding author on reasonable request.
Abbreviations
- DR:
-
Distant recurrence
- lncRNA:
-
Long non-coding RNA
- LR:
-
Local recurrence
- NR:
-
Non-recurrence
- ROCK1:
-
Rho-associated, coiled-coil-containing protein kinase 1
References
Hussain T, Nguyen QT. Molecular imaging for cancer diagnosis and surgery. Adv Drug Deliv Rev. 2014;66:90–100.
Kraft M, Fostiropoulos K, Gürtler N, Arnoux A, Davaris N, Arens C. Value of narrow band imaging in the early diagnosis of laryngeal cancer. Head Neck. 2016;38(1):15–20.
Ansari D, Gustafsson A, Andersson R. Update on the management of pancreatic cancer: surgery is not enough. World J Gastroenterol. 2015;21(11):3157–65.
Anderson ME. Update on survival in osteosarcoma. Orthop Clin North Am. 2016;47(1):283–92.
Durfee RA, Mohammed M, Luu HH. Review of osteosarcoma and current management. Rheumatol Ther. 2016;3(2):221–43.
Wu J, Sun H, Li J, Guo Y, Zhang K, Lang C, Zou C, Ma H. Increased survival of patients aged 0-29 years with osteosarcoma: a period analysis, 1984-2013. Cancer Med. 2018;7(8):3652–61.
Rath N, Olson MF. Rho-associated kinases in tumorigenesis: re-considering ROCK inhibition for cancer therapy. EMBO Rep. 2012;13(10):900–8.
Su SC, Hsieh MJ, Yang WE, Chung WH, Reiter RJ, Yang SF. Cancer metastasis: mechanisms of inhibition by melatonin. J Pineal Res. 2017;62(1):e12370.
Matsubara M, Bissell MJ. Inhibitors of rho kinase (ROCK) signaling revert the malignant phenotype of breast cancer cells in 3D context. Oncotarget. 2016;7(22):31602–22.
Datta A, Sandilands E, Mostov KE, Bryant DM. Fibroblast-derived HGF drives acinar lung cancer cell polarization through integrin-dependent RhoA-ROCK1 inhibition. Cell Signal. 2017;40:91–8.
Yang M, Zhai X, Xia B, Wang Y, Lou G. Long noncoding RNA CCHE1 promotes cervical cancer cell proliferation via upregulating PCNA. Tumuor Biol. 2015;36(10):7615–22.
Xu G, Zhang Y, Li N, Zhang JB, Xu R. LncRNA CCHE1 in the proliferation and apoptosis of gastric cancer cells. Eur Rev Med Pharmacol Sci. 2018;22(9):2631–7.
Liao Y, Cheng S, Xiang J, Luo C. lncRNA CCHE1 increased proliferation, metastasis and invasion of non-small lung cancer cells and predicted poor survival in non-small lung cancer patients. Eur Rev Med Pharmacol Sci. 2018;22(6):1686–92.
Peng W, Fan H. Long noncoding RNA CCHE1 indicates a poor prognosis of hepatocellular carcinoma and promotes carcinogenesis via activation of the ERK/MAPK pathway. Biomed Pharmacother. 2016;83:450–5.
Aponte-Tinao L, Ayerza MA, Muscolo DL, Farfalli GL. Survival, recurrence, and function after epiphyseal preservation and allograft reconstruction in osteosarcoma of the knee. Clin Orthop Relat Res. 2015;473(5):1789–96.
Mattonen SA, Palma DA, Haasbeek CJA, Senan S, Ward AD. Early prediction of tumor recurrence based on CT texture changes after stereotactic ablative radiotherapy (SABR) for lung cancer. Med Phys. 2014;41(3):033502.
Mallidi S, Watanabe K, Timerman D, Choenfeld D, Hasan T. Prediction of tumor recurrence and therapy monitoring using ultrasound-guided photoacoustic imaging. Theranostics. 2015;5(3):289–301.
Ying T, Dong J, Yuan C, Li P, Guo Q. The lncRNAs RP1-261G23. 7, RP11-69E11. 4 and SATB2-AS1 are a novel clinical signature for predicting recurrent osteosarcoma. Biosci Rep. 2020;40(1):BSR20191251.
Gilkes DM, Xiang L, Lee SJ, Chaturvedi P, Hubbi ME, Wirtz D, Semenza GL. Hypoxia-inducible factors mediate coordinated RhoA-ROCK1 expression and signaling in breast cancer cells. Proc Natl Acad Sci U S A. 2014;111(3):E384–93.
Cui M, Wang J, Li Q, Zhang J, Jia J, Zhan X. Long non-coding RNA HOXA11-AS functions as a competing endogenous RNA to regulate ROCK1 expression by sponging miR-124-3p in osteosarcoma. Biomed Pharmacother. 2017;92:437–44.
Wang Y, Zeng X, Wang N, Zhao W, Zhang X, Teng S, Zhang Y, Lu Z. Long noncoding RNA DANCR, working as a competitive endogenous RNA, promotes ROCK1-mediated proliferation and metastasis via decoying of miR-335-5p and miR-1972 in osteosarcoma. Mol Cancer. 2018;17(1):89.
Acknowledgements
Not applicable.
Funding
Not applicable.
Author information
Authors and Affiliations
Contributions
ZZ literature review, data collection, data analysis, manuscript written. WG data collection, data analysis. TY supervision, data collection, data analysis, manuscript edition. The authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
The present study was approved by the Ethics Committee of Liaocheng People’s Hospital. The research has been carried out in accordance with the World Medical Association Declaration of Helsinki. All patients provided written informed consent prior to their inclusion within the study.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Zhang, Z., Yu, T. & Geng, W. Long non-coding RNA CCHE1 participates in postoperative distant recurrence but not local recurrence of osteosarcoma possibly by interacting with ROCK1. BMC Musculoskelet Disord 21, 462 (2020). https://doi.org/10.1186/s12891-020-3184-x
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12891-020-3184-x