Abstract
Background
Recent epidemics have entailed global discussions on revamping epidemic control and prevention approaches. A general consensus is that all sources of data should be embraced to improve epidemic preparedness. As a disease transmission is inherently governed by individual-level responses, pathogen dynamics within infected hosts posit high potentials to inform population-level phenomena. We propose a multiscale approach showing that individual dynamics were able to reproduce population-level observations.
Methods
Using experimental data, we formulated mathematical models of pathogen infection dynamics from which we simulated mechanistically its transmission parameters. The models were then embedded in our implementation of an age-specific contact network that allows to express individual differences relevant to the transmission processes. This approach is illustrated with an example of Ebola virus (EBOV).
Results
The results showed that a within-host infection model can reproduce EBOV’s transmission parameters obtained from population data. At the same time, population age-structure, contact distribution and patterns can be expressed using network generating algorithm. This framework opens a vast opportunity to investigate individual roles of factors involved in the epidemic processes. Estimating EBOV’s reproduction number revealed a heterogeneous pattern among age-groups, prompting cautions on estimates unadjusted for contact pattern. Assessments of mass vaccination strategies showed that vaccination conducted in a time window from five months before to one week after the start of an epidemic appeared to strongly reduce epidemic size. Noticeably, compared to a non-intervention scenario, a low critical vaccination coverage of 33% cannot ensure epidemic extinction but could reduce the number of cases by ten to hundred times as well as lessen the case-fatality rate.
Conclusions
Experimental data on the within-host infection have been able to capture upfront key transmission parameters of a pathogen; the applications of this approach will give us more time to prepare for potential epidemics. The population of interest in epidemic assessments could be modelled with an age-specific contact network without exhaustive amount of data. Further assessments and adaptations for different pathogens and scenarios to explore multilevel aspects in infectious diseases epidemics are underway.
Similar content being viewed by others
Background
Epidemics of infectious diseases are currently listed among the potential catastrophes that can set the world back in the next decades [1]. Overwhelming research efforts have been developed to predict the danger of the epidemics but their crisis nature often left scientists no better option than learning from the past [1, 2]. Confronting outbreaks of emerging infections, however, requires a swift response and thus the ability to evaluate quickly and early possible outcomes [1]. As such, computer simulations of epidemic models undoubtedly hold the potential as the first-aid toolbox for decision making amid the crisis [1, 3, 4].
Computational approaches in epidemic modelling date back a few centuries ago [5]. Since then, an overwhelming amount of research has been conducted and contributed profoundly to understanding of epidemic control and prevention [5, 6]. The aims of epidemic modelling are to address questions such as whether or not a substantial population fraction is getting infected? how large would the outbreak spread? or how can the outbreak be mitigated with intervention approaches at hand? among others [7, 8]. Answering these questions requires the quantification of these models using disease data on a population level [7, 9–11], which are often delayed [12, 13], under-reported [14], or not readily available [9]. As a result, epidemic models using population data, while progress understanding on diseases, might have limited applications to an ongoing epidemic [15]. A potentially useful way to predict future disease dynamics is using within-host processes [11].
In reality, the within-host infection process determines key parameters in a disease transmission ([16–18], Fig. 1). In an infected subject, interactions between the pathogen and immune responses shape the pathogen dynamics which, ultimately, define the incubation period, the transmission potential, and the recovery rate [11, 18]. It is also evident that susceptibility to infection is not the same for all the susceptible but, among others, it is highly correlated with a subject’s age due to age-related changes of the immune system [19, 20]. These observations together point towards some inherent biological processes where individual dynamics can help to predict population-level epidemics. Differences in the within-host infection profile as well as the susceptibility to infection complicate greatly epidemic models but at the same time underline their influential roles in determining epidemics features and intervention effects [21–23].
Schematic presentation of the within-host infections processes and their relation to transmission parameters. At the within-host level, viral replication and immune responses race with each other that eventually determines an individual infectivity, for example, his symptoms and possibly behaviours. At the between hosts level, infected individuals make contact(s) with susceptible individual(s) that eventually lead to a transmission, depending on both the infectivity of the infectious and the susceptibility of the susceptible. Noting that while the contact network can be fixed, the portion actively partake in epidemic spreading dynamically change over time [75]
The interplays between within-host infection and between hosts transmission led to arising attempts connecting the two levels [4, 17, 18, 24–28], but the approach is still at its infancy [16]. On one hand, most of these models were conceptual and theoretical [16, 28] or relied on assumptive and previously obtained parameter estimates [10, 11, 29]. We propose that this limitation can be overcomed by using explicitly within-host infection model as a unit in epidemic simulations. This allows not only a high level of heterogeneity to be expressed, but also the study of stochasticity effects in epidemic spreading.
On the other hand, implementations of population level models were either a general representation using probabilistic assumptions [30, 31] or a computationally demanding implementation of a particular population [11, 29, 32]. These approaches, while able to recover valuable insights, may not be representative and accessible for another population of interest, because either none or large amount of data were needed. In this case, an implementation of an epidemic model with social mixing attributes, i.e., number of contacts per day and to whom the contact occurs, could be less computationaly intensive [33], consistent across populations [34, 35], and more representative for a realistic disease spreading process [36, 37].
Improving understanding on EBOV transmission is crucial: EBOV can cause a large scale outbreak with a high fatality rate [14] and it re-emerges continuously [38]; as of May 2018, a total of 32 EBOV disease cases have been reported from Democratic Republic of the Congo, including 18 deaths [38]. Based on our previous studies of the within-host EBOV infection [39–41], we brought forward a within-host infection model to study the transmission fitness of EBOV at the population level. We built a network model based on social contact data [34] and the respective epidemic simulation algorithm that embedded the within-host model into the contact network. Thus, this multiscale model was data-informed at each level of the epidemic process.
Our multi-scale model aimed to express the link between pathogen load and transmission potential of an infected host, the immune responses to the infection and vaccination, and the heterogeneity of the contact frequency distribution in a population. The parameters obtained from model simulations were compared to those estimated from population-level data and empirical observations. The results showed that within-host infection model reproduced estimates of the transmission parameters and allowed detailed evaluations of the effects of intervention timing on the course of the epidemic. Implementations of the network model from social contact data was straightforward and scalable for large simulations on high-performance computer clusters. In that capacity, epidemic assessments and preparations can be conducted quickly, ahead of time, and with high-resolution requirements.
Methods
In an EBOV-infected subject, the immune system components dynamically evolve in response to the virus replication dynamic [42]. As a result, a series of events is triggered determining the infection outcomes such as infectious status, symptoms, recovery, or death [42–44]. In this paper, the EBOV replication dynamic within a host was used to infer its transmission parameters.
Within-host model
Using virus dynamics and the immune response data within a host, mathematical relations can be defined to test hypothesized infection mechanisms [39, 45]. In this context, non-human primates (NHPs) are the standard animal model for developing EBOV’s therapeutics and vaccines in humans [46–48]. Epidemiological and pharmacological studies reported that a viral load level higher than 106 copies/mL [47, 49] was associated with a higher mortality rate, whereas observations based on experimental data in NHPs showed that a viral load level higher than 106 TCID50 was fatal [43, 44]. Here the viral load dynamics were simulated based on a model as follows [41]:
where rV,KV, and In denote the viral replication rate, the carrying capacity of the infected host, and a threshold expressing a lag-phase of viral replication. In particular, a logistic growth was assumed for EBOV, with a short delay when the virus level was low. The parameter KAb represents the strength of the immune system at which the antibody titre (Ab) completely inhibits the viral net growth rate [41, 44], i.e., it was assumed that the higher antibody level required to inhibit the viral replication the weaker the immune strength. An antibody titre level of 104 appeared to be protective in NHPs and it required approximately one week after vaccination to reach this threshold [44]. The model parameters were obtained previously [41] using two experimental datasets on NHPs [43, 44]. The antibody dynamic (Ab) was also fitted in [41] to the data of NHPs vaccinated with a recombinant vesicular stomatitis virus vaccine (rVSV-EBOV) [44]. This vaccine had shown a high efficacy in human [48]. Details of model fitting, data, and the parameter set can be found in [41] and “Availability of data and materials” section.
Simulated subject-specific infection course
To simulate subject-specific infection course, the antibody response strength KAb was varied from a normal level of approximately 102.5 [44, 50] to the highest observed level of 104.5 [44]. This parameter was assumed to follow a U-shaped function of an individual’s age, where infants and elderly have a higher susceptibility ([19], Fig. 1) (the extracted function is presented in “Availability of data and materials” section). As the infective dose can alter the course of infection [51], the initial condition V(0) of the model Eq. 1 was varied depending on from whom a subject acquired the infection, i.e., the infection dose was assumed as equal to the lethal dose (Vc=100.15 [41]) times the transmission potential of a subject transmitting the infection. Here we assumed a direct relation between the transmission potential and the viral load at the time of infection [16], i.e., the transmission potential pTrans(t)=V(t)/KV. Note that pTrans(t)=1 does not guarantee a successful transmission, but it was considered collectively with its contact’s susceptibility and with the existence of such a contact’s (details in “Availability of data and materials” section).
Infection outcomes definitions
Empirical observations from EBOV infected human and NHPs showed that the time from symptom onset to death was approximately one week [43, 44, 52]. Based on this, we used the total area under the viral load curve (AUC) seven days post-infection obtained from the subjects that died as a threshold above which the infection is lethal, i.e., \(\text {AUC}_{7} = \int _{0}^{7} V(t)dt\). Otherwise, infected subjects were assumed to have recovered once the viral load was no longer detectable (Fig. 2). Depending on the infective dose and the adaptive immune response strength, the infection model manifests different viral dynamics and consequently the infection outcomes. Based on that, we defined the transmission parameters as in Table 1A-C (detailed implementations can be seen in “Availability of data and materials” section).
Simulated infection course using within-host infection dynamics. The viral replication, the antibody dynamics, and their interaction were modelled to define epidemiological parameters. It is assumed that when the EBOV-specific antibody concentration reaches a certain threshold, it can inhibit the viral replication [76]. The total viral load under the curve (AUC) in lethal cases is used to define infection outcomes [77]
Network model
The European’s contact patterns survey data [34] were used to generate a network model reflecting the number of contacts and the mixing patterns among age-groups. This dataset is currently the largest collection available on human contact patterns; moreover, a similar pattern has also been observed outside Europe [35]. To make the contact pattern more specific to an EBOV-affected population, the age distribution of Freetown city in Sierra Leon [53] was used to weight the contact pattern towards this area. In particular, while the contact frequency and pattern remain the same, these data were imposed on a network that has more young and less elderly subjects. A detailed description of the network implementation can be found in Availability of data and materials.
Because EBOV spreads through direct contacts with infectious subjects [51], and the highest risk of infection is contacting with blood, faeces, and vomit [54], we used only the data of physical contacts and excluded those contacts with a duration of less than five minutes. In the 2014 EBOV epidemic, an important transmission route was through contacting with the deceased who had not been buried [2]. To account for this we considered deceased EBOV-infected subjects as infectious until they were buried. During the 2015 EBOV epidemic, the time from death to burial was from one to two days on average, but it can be a week [55]. This information was used to formulate a truncated normal distribution for the time from death to burial, i.e., the distribution was truncated at zero and seven and had unit mean and variance (detailed implementations can be seen in Availability of data and materials).
Transmission outcomes definitions
To obtain EBOV’s epidemic metrics, the within-host infection model was embedded into the network model. Simulations of EBOV epidemic are detailed in Fig. 3. In short, a network of ten thousand nodes was generated. Scenarios in which the population was randomly vaccinated during one-week vaccination program were tested and compared to a control simulation without vaccination. To isolate the effect of vaccination, we assumed that no other interventions were in place, e.g., no treatments were provided and no quarantine or isolation programs occurred. For each scenario, one thousand simulations were performed, each of which started with a single random index case. Each time when a contact occurred, the viral load at that time point was extracted to determine the transmission potential. Next, the susceptibility of the contact persons was computed as a function of their age [19]. A Bernoulli trial was then used to determine if the contact results in an infection given the overall transmission probability. If the transmission succeeded, for the newly infected subject his/her own infection profile was computed. Based on simulation outputs, the epidemic outcomes were determined as in Table 1D-F (detailed implementations can be seen in Availability of data and materials).
Potential weaknesses and remedial approaches
The following assumptions were used given the lack of specific experimental data, but further efforts to produce the data can be done to address the issues listed here:
-
(i)
Secondary antibody responses were assumed to be similar to primary responses. This underestimates the effect of vaccination strategies conducted before an epidemic. Experimental studies on secondary immune responses to EBOV infection are needed, especially those with a longer follow-up period.
-
(ii)
The transmission potential was assumed as directly related to viral load. While this is reasonable, non-linear relationships might exist [16, 28]. Dedicated animal experiments to define the exact relationship between the viral load and the ability to transmit the virus are needed.
-
(iii)
The contact pattern was assumed similar for Ebola affected regions as in European countries. Although the contact patterns seemed similar across countries [34], a more sociable population would have higher contact rate and thus increase R0. As collecting this data for all countries can be laborious, simulation studies addressing the effect of contact patterns on the connectivity in network models are needed.
-
(iv)
Infection status was assumed to have no influence on the network structure, except that those buried were removed from the network. This could overestimate R0 [56]. Taking people’s behaviour changes into epidemic modelling remains a grand challenge [56].
-
(v)
Susceptibility to EBOV infection was assumed similar to a general viral infection disease. Studies on susceptibility functions are lacking and require more attention of the infection research community.
Computational implementation
The simulations were written in vectorized R language [57]. Ordinary differential equations (ODEs) were solved with deSolve package [58]. The network was generated as an adjacency matrix and was visualized with the package igraph [59]. Computations of infection dynamics of the newly infected nodes were done during the epidemic simulation after obtaining its infective dose and immunization status. For nodes with identical conditions, their infection courses were copied instead of recomputing the ODEs for speed (Availability of data and materials). Repeated runs of epidemic simulations to obtain uncertainty estimates were done on computer clusters of the Center for Scientific Computing (CSC) of the Goethe University Frankfurt. Distribution of computation resources was sent from within R to SLURM Workload Manager.
Results
Basic transmission characteristics
Table 2 shows that the within-host infection model captured well the population-level transmission parameters. The results suggest that in contrary to using outbreak data, employing within-host infection model can provide this information prior to outbreaks, and even for a scenario where a pathogen X has never caused an epidemic before [60].
The network model
Figure 4 shows an example of the generated networks and its required data. In particular, given a network of size \(N \in \mathbb {N}\), each node was assigned an age such that the network’s age-distribution resembled that of the target population. Subsequently, nodes were assigned a number of contacts per day following a defined contact distribution of interest. Finally, the algorithm visited each node to generate the defined number of contacts, not at random but following a defined contact matrix. The network was returned as an adjacent matrix that is compatible to available network analyses algorithms, e.g., igraph, graph-tool [61, 62]. Storing data as a sparse matrix, a regular installation of R could generate reliably networks of 10-20 thousand nodes with the generation time 6-10 minutes on a single thread Intel Core i7, 8GB RAM. Note that R theoretically can only handle a maximum square matrix ≈44721 rows and columns.
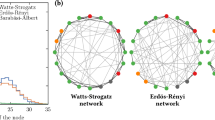
Required data and an example of an age-structure network model. (a) Generated network of one hundred individuals that mimics distribution of physical contact, contact matrix, and population age structure. The node’s size reflects its number of contacts. Nodes in the same age-group have the same colour; (b) Distribution of number of physical contacts shows a majority of individuals have a few physical contact per day [34]; (c) A heat map of contact matrix shows higher contact frequencies in darker shades. The matrix reflects the assortative pattern of human contacts, that is people contact mainly with their peers, follow by their children or parents. The age-group with the highest contacts are teenagers and young adults [34]; (d) Reconstructed age-structure of Sierra Leon population based on Statistics Sierra Leone and ICF International data [53]
Calculating basic reproductive number (R0)
After each simulation, the uninfected nodes were removed from the initial network. Then the basic reproduction number was calculated as the average network degree, considering the network as a directed and without loop network (Fig. 5-Left). Simulation results showed that the overall estimate of R0 was 1.43 (Fig. 5-Right). However, the estimates differed across age-groups with the highest of 4.7 for the group of 10-14 years of age. Intuitively, the age-groups with a higher contact rate had also a higher R0. Simulations of epidemics with varied intervention strategies showed that Re can be reduced below unity if a vaccination program with 85% coverage was deployed at the time as far as five months before the introduction of the index case (time zero) or as late as one week after that (Additional file 1: Figure S1). This coverage threshold was tested as it is the highest vaccine coverage currently achieved worldwide for some diseases, e.g., Hepatitis B, measles, and polio [63]. Late initiations of a similar intervention from one to five months after the time zero gradually shifted the Re to the outbreak domain.
Left. Three examples of infected networks. The three networks were randomly chosen from the simulated epidemics. Uninfected nodes were removed and the network is plotted. Based on these, the R0 was calculated based on the edges assuming a directed network, i.e., each edge counted in only one direction. Right. Estimates of the basic reproductive number without any intervention, overall and by age-groups. Simulations of a network of size ten thousand during a period of one year. One thousand simulations were run, each time with a random index case. At the end of each simulation, networks of infected nodes were extracted to compute the average number of secondary infections
A lower vaccination coverage of 33%, while substantially reduced the epidemic size, still posed a potential of a large outbreak regardless the timing of vaccination program (Additional file 1: Figure S1). This coverage was tested as it is a theoretically protective threshold in stochastic and heterogeneous mixing models, i.e., 1-1/R0 [7, 64]. Note that the tested time window of five months before the appearance of the index case was chosen based on the windows of opportunity for EBOV vaccination [41]. As of now, no detailed data are available on the secondary antibody responses to EBOV; it was therefore assumed that secondary responses are similar to the primary responses.
Case-fatality rate
Simulations showed that the case-fatality rate in the absence of intervention was about 91% (Additional file 2: Figure S2), which falls within the range of literature estimates of 0.4 to 0.91 [79]. Note that the simulations assumed worst case scenarios where no other inverventions were done except the vaccination. Furthermore, simulation results showed that all the intervention strategies mentioned previously reduced the case-fatality rate. These results highlighted a benefit of vaccination programs even if they were late: reducing the disease severity in newly infected subjects after the introduction of the vaccination program. As such, relying on R0 as the solely indicator for evaluating intervention programs would overlook this life-saving aspect.
Epidemic final size
Theoretical analyses of stochastic epidemic models showed that when R0 is larger than unity, the final size of an epidemic converges to a bimodal distribution: either the epidemic dies out with a small number of infected cases or the epidemic takes off to a normal distribution with a high number of cases [7]. Our simulation results recreated this epidemic behavior (Fig. 6). Without intervention, EBOV had approximately 50% probability to infect more than half the population. The introduction of vaccination programs at the two previously mentioned coverages and at any vaccination time points under assessment scaled down the epidemic size (Fig. 6). The earlier the vaccination programs were deployed, the closer the epidemics size distribution resemble to a uni-modal distribution centered at a low infected fraction. The high vaccine coverage strategy effectively eliminated the possibility of having a major outbreak infecting a large proportion of the population. This was achieved when the vaccination programs were deployed at any time point from one week to five months before time zero.
Distribution of the final infected fraction in different timing and coverage of vaccination strategies. A synthetic population of ten thousand individuals was generated. One thousand simulations were run to simulate the epidemic in the time course of one year. Each time, one individual was chosen randomly as the index case. Circles, diamonds, and connected lines are median. Filled areas are the corresponding non-parametric densities estimates [78]. Two median values are presented for multi-modal density estimates, determining by inflection points
Figure 6 also shows that a random vaccination program covering 33% of the population one week before the time zero reduced the final size by more than 100 times compared to a no intervention scenario. However, the low coverage strategy still showed a small probability that the epidemic becomes major, whereas the high coverage strategies did not. Vaccination programs deployed during the epidemics also substantially reduced the epidemic’s size: the vaccination program conducted one month after time zero still reduced the final size by more than ten times. Furthermore, these interventions were not only able to reduce the final size, but they could also increase the epidemics extinction probability (Fig. 6).
Discussion
In estimating parameters and designing prevention strategies for infectious diseases, it is important to take into account mechanisms underlying the heterogeneity in immune responses [65] and in the population [34]. This paper combined a within-host immunological model of Ebola virus (EBOV) infection and an age- and demographic-specfic contact network to study EBOV epidemic. The multi-scale model reproduced major characteristics of EBOV epidemics and allowed finer assessments of the timing of vaccination strategies.
Estimates of the EBOV’s incubation period suggested a contact tracing period of three weeks for Ebola epidemics, matching the current WHO’s recommendation of 21 days [66]. Estimates of the delay distributions agreed with information that EBOV infected subjects can be infectious from day 3 up to three weeks post infection [79]. Understanding of these delay distributions is critical in clinical and epidemiological perspectives [67]. These distributions, however, are often only partially observed in practice: it is difficult to know the exact time of exposure to the pathogen or to have complete outbreak data [68, 69]. As such, parameter estimation of these distributions have been relied on testing and comparing distributional assumptions [69]. The mechanistically generated transmission characteristics using virus dynamics remarkably resemble literature estimates. This suggests some inherent biological processes where individual dynamics accumulate to produce population-level epidemics. This approach is thus promising and practical given the accumulating experimental data on varieties of pathogens, such as a disease X [60] that as yet unknown in epidemic contexts.
To determine infection outcomes, the threshold AUC7 was chosen based on suggestions derived from empirical data in humans [52] and non-human primates [43, 44]. Simulations of the epidemics using this threshold reproduced estimate of EBOV case-fatality rate (Additional file 2: Figure S2), suggesting that the use of total viral load (AUC) as a criterion for determining infection outcomes is appropriate. It is worth noting that the calculated case fatality rate was based on the assumption of worst case scenarios where no treatments were provided. In practice there were medical approaches to reduce disease severity. Scientific literature has shown that for severe cases of EBOV infection (equivalent to unvaccinated subjects or those vaccinated too late in our model), supportive treatments increased substantially the survival chance [70]. Although a more precise threshold criterion is desirable, it might not be feasible to obtain it in practice considering the inherent ethical reasons. Thus a similar criterion as used here could be considered when adapting this approach to other diseases, but ideally derived from dedicated experimental data.
Different classes of network models have been proposed, but they cannot reproduce properties observed in real world networks [71]. In addition, choices of theoretical network structure used for simulation can alter epidemic outcomes [30]. Thus, a network model built from empirical data would provide a more solid ground for epidemic simulations. Apart from mimicking the contact data properties, our network model can express age-related infection traits via the assigned age attribute. It was used in this paper to express individual differences in the susceptibility and immune response to viral infection—the crucial elements in a realistic disease transmission. Although contact data might not be available for a certain target area, the assortative patterns of human contacts and the highly skewed distribution of the number of contacts might hold true across regions [34, 35]. Thus, this paper presents a simple way to bring empirical contact data into epidemic modelling studies.
Our network model currently can only simulate epidemics in a population of size 10–20 thousand. This is because of the limit in R with the theoretical maximum square matrix size of approximately 45000 [57]. A more efficient storing of the network could extend the network size, such as a lazy evaluation used in igraph [61]. However, it could be more realistic to have several communities amount to a large population size instead of a large single network. This can be implemented by generating different communities across computers and allow them to communicate, speeding also the computation processes [72]. In this case, additional data are needed to model the communication among the communities, such as transportation network and immigration flow.
Regarding EBOV epidemics, previous R0 estimates based on epidemic data varied strongly, depending on model choices and assumptions [79]. Our estimate of R0 was 1.4 which is within the range of the previous estimates, ranging from 1.2 to 2.6, with some exceptional estimates up to 4.7 and 8.3 [79]. Notably, we showed that the estimates differed by age-groups with the highest of 4.7 for the group of 10–14-years of age. Although these estimates depend on Sierra Leon’s age-structure, the differences of R0 estimate stress the role of the age-structure and contact patterns in the estimation of R0, prompting that age-specific intervention strategies should be considered. The estimates by sub-groups also single out the effort required to control the epidemic [73]. With the assumptions used in our models, targeting interventions to the group 5–20-years of age would be the most effective strategy. Note that the differences of R0 by age-group also provide us an explanation of the wide variation of the previous estimates of R0 where different samples were used [79]. As the R0 estimates were larger in sub-groups, our results confirmed that the critical vaccine coverage also needed to be larger to ensure eradication of the epidemic [64, 74]. Using our approach, we have shown further that while a low coverage could not completely eradicate the epidemic, it could largely reduce both the size and severity of an epidemic—which is worth pursuing in cases of lacking resources to reach an optimal threshold.
Conclusion
Throughout this paper, we showed the possibilities to investigate practical and intriguing questions using a within-host viral dynamic model and an age-structured network model. The advantages of using explicitly within-host dynamics are the availability of experimental data, the possibility of conducting experiments to characterize transmission parameters, and the ability to provide high-resolution subject-specific responses to infection. The advantages of using an age-structured network model are its simple implementation, its representativeness for disease transmission, and the availability of the age-structured data. Therefore, immunological studies of infectious agents could be seamlessly integrated into studies of between hosts transmission, promoting evidence-based public health practices.
Abbreviations
- AUC:
-
Area under the curve
- EBOV:
-
Ebola virus
- NHPs:
-
Non-human primates
- R0:
-
Basic reproduction number
- Re:
-
Effective reproduction number
- VSV:
-
Vesicular stomatitis virus
- WHO:
-
World health organization
References
Gates B. The next epidemic–lessons from Ebola. N Engl J Med. 2015; 372(15):1381–4.
Piot P. Public health: Beating Ebola. Nature. 2016; 537(7621):484–5.
Lofgren ET, Halloran ME, Rivers CM, Drake JM, Porco TC, Lewis B, Yang W, Vespignani A, Shaman J, Eisenberg JN, Eisenberg MC, Marathe M, Scarpino SV, Alexander KA, Meza R, Ferrari MJ, Hyman JM, Meyers LA, Eubank S. Opinion: Mathematical models: a key tool for outbreak response. Proc Natl Acad Sci USA. 2014; 111(51):18095–6.
Willem L, Verelst F, Bilcke J, Hens N, Beutels P. Lessons from a decade of individual-based models for infectious disease transmission: a systematic review (2006-2015). BMC Infect Dis. 2017; 17(1):612. https://doi.org/10.1186/s12879-017-2699-8.
Anderson RM, May RM. Infectious Diseases of Humans: Dynamics and Control. Oxford: OUP Oxford; 1992.
Heesterbeek H, Anderson RM, Andreasen V, Bansal S, De Angelis D, Dye C, Eames KTD, Edmunds WJ, Frost SDW, Funk S, Hollingsworth TD, House T, Isham V, Klepac P, Lessler J, Lloyd-Smith JO, Metcalf CJE, Mollison D, Pellis L, Pulliam JRC, Roberts MG, Viboud C. Modeling infectious disease dynamics in the complex landscape of global health. Science. 2015; 347(6227):4339–9. https://doi.org/10.1126/science.aaa4339.
Britton T. Stochastic epidemic models: A survey. Math Biosci. 2010; 225(1):24–35. 0910.4443.
Hens N, Shkedy Z, Aerts M, Faes C, Van Damme P, Beutels P. Modeling Infectious Disease Parameters Based on Serological and Social Contact Data: A Modern Statistical Perspective. Statistics for Biology and Health. New York: Springer; 2012. https://books.google.de/books?id=IH08pTAoe6QC.
Pomeroy LW, Bansal S, Tildesley M, Torres KIM, Moritz M, Xiao N, Carpenter TE, Garabed RB. Data-Driven Models of Foot-and-Mouth Disease Dynamics: A Review. Transboundary Emerg Dis. 2017; 64(3):716–28. https://doi.org/10.1111/tbed.12437.
Merler S, Ajelli M, Fumanelli L, Parlamento S, Pastore Y Piontti A, Dean NE, Putoto G, Carraro D, Longini IM, Halloran ME, Vespignani A. Containing Ebola at the Source with Ring Vaccination. PLoS Negl Trop Dis. 2016; 10(11):0005093.
Lukens S, DePasse J, Rosenfeld R, Ghedin E, Mochan E, Brown ST, Grefenstette J, Burke DS, Swigon D, Clermont G. A large-scale immuno-epidemiological simulation of influenza A epidemics. BMC Public Health. 2014; 14:1019.
World Health Organization. Outbreak communication: Best practices for communicating with the public during an outbreak. Technical report.
Nguyen VK, Parra-Rojas C, Hernandez-Vargas EA. The 2017 plague outbreak in Madagascar: data descriptions and epidemic modelling. Epidemics. 2018. pii: S1755-4365(18)30007-0. https://doi.org/10.1016/j.epidem.2018.05.001.
World Health Organization. Ebola Situation Report - 7 January 2015 | Ebola. Technical report. http://apps.who.int/ebola/en/status-outbreak/situation-reports/ebola-situation-report-7-january-2015.
Lloyd-Smith JO, Funk S, McLean AR, Riley S, Wood JLN. Nine challenges in modelling the emergence of novel pathogens. Epidemics. 2014:1–5. https://doi.org/10.1016/j.epidem.2014.09.002.
Handel A, Rohani P. Crossing the scale from within-host infection dynamics to between-host transmission fitness: a discussion of current assumptions and knowledge. Philos Trans R Soc Lond B Biol Sci. 2015;370(1675).
Alizon S, Luciani F, Regoes RR. Epidemiological and clinical consequences of within-host evolution. Trends Microbiol. 2011; 19(1):24–32.
Chen SC, Chio CP, Jou LJ, Liao CM. Viral kinetics and exhaled droplet size affect indoor transmission dynamics of influenza infection. Indoor Air. 2009; 19(5):401–13.
Farber DL, Yudanin NA, Restifo NP. Human memory T cells: generation, compartmentalization and homeostasis. Nat Rev Immunol. 2014; 14(1):24–35.
Hernandez-Vargas EA, Wilk E, Canini L, Toapanta FR, Binder SC, Uvarovskii A, Ross TM, Guzman CA, Perelson AS, Meyer-Hermann M. Effects of aging on influenza virus infection dynamics. J. Virol. 2014; 88(8):4123–31.
Rozhnova G, Metcalf CJE, Grenfell BT. Characterizing the dynamics of rubella relative to measles: the role of stochasticity. J R Soc Interface. 2013; 10(88):20130643–20130643. https://doi.org/10.1098/rsif.2013.0643.
Caudron Q, Mahmud AS, Metcalf CJE, Gottfrethsson M, Viboud C, Cliff AD, Grenfell BT. Predictability in a highly stochastic system: final size of measles epidemics in small populations. J R Soc Interface. 2014; 12(102):20141125. https://doi.org/10.1098/rsif.2014.1125.
Finkenstadt BF. A stochastic model for extinction and recurrence of epidemics: estimation and inference for measles outbreaks. Biostatistics. 2002; 3(4):493–510. https://doi.org/10.1093/biostatistics/3.4.493.
Murillo LN, Murillo MS, Perelson AS. Towards multiscale modeling of influenza infection. J Theor Biol. 2013; 332:267–90.
Day T, Alizon S, Mideo N. Bridging scales in the evolution of infectious disease life histories: theory. Evolution. 2011; 65(12):3448–61.
Longini IM, Nizam A, Xu S, Ungchusak K, Hanshaoworakul W, Cummings DA, Halloran ME. Containing pandemic influenza at the source. Science. 2005; 309(5737):1083–7.
Cen X, Feng Z, Zhao Y. Emerging disease dynamics in a model coupling within-host and between-host systems. J Theor Biol. 2014; 361:141–51.
Almocera AES, Nguyen VK, Hernandez-Vargas EA. Multiscale model within-host and between-host for viral infectious diseases. J Math Biol. 2018; 19(1):1–23. https://doi.org/10.1007/s00285-018-1241-y.
Lee BY, Brown ST, Korch G, Cooley PC, Zimmerman RK, Wheaton WD, Zimmer SM, Grefenstette JJ, Bailey RR, Assi T-M, Burke DS. A Computer Simulation of Vaccine Prioritization, Allocation, and Rationing During the 2009 H1N1 Influenza Pandemic. Vaccine. 2010; 28(31):4875–9. https://doi.org/10.1016/j.vaccine.2010.05.002.
Rahmandad H, Hu K, Tebbens RJD, Thompson KM. Development of an individual-based model for polioviruses: implications of the selection of network type and outcome metrics. Epidemiol Infect. 2011; 139(6):836–848. https://doi.org/10.2307/27975665.
Pastor-Satorras R, Vespignani A. Epidemic spreading in scale-free networks. Phys Rev Let. 2001; 86:3200–3. 0010317.
Brown ST, Tai JH, Bailey RR, Cooley PC, Wheaton WD, Potter MA, Voorhees RE, LeJeune M, Grefenstette JJ, Burke DS, McGlone SM, Lee BY. Would school closure for the 2009 H1N1 influenza epidemic have been worth the cost?: a computational simulation of Pennsylvania. BMC Public Health. 2011; 11(1):353. https://doi.org/10.1186/1471-2458-11-353.
Eames KTD, Read JM, Edmunds WJ. Epidemic prediction and control in weighted networks. Epidemics. 2009; 1(1):70–76. https://doi.org/10.1016/j.epidem.2008.12.001.
Mossong J, Hens N, Jit M, Beutels P, Auranen K, Mikolajczyk R, Massari M, Salmaso S, Tomba GS, Wallinga J, Heijne J, Sadkowska-Todys M, Rosinska M, Edmunds WJ. Social contacts and mixing patterns relevant to the spread of infectious diseases. PLoS Med. 2008; 5(3):74.
Horby P, Pham QT, Hens N, Nguyen TT, Le QM, Dang DT, Nguyen ML, Nguyen TH, Alexander N, Edmunds WJ, Tran ND, Fox A, Nguyen TH. Social contact patterns in Vietnam and implications for the control of infectious diseases. PLoS ONE. 2011; 6(2):16965.
Pastor-satorras R, Castellano C, Mieghem PV, Vespignani A. Epidemic processes in complex networks. 2015:1–62. arXiv:1408.2701v2.
Pellis L, Ball F, Bansal S, Eames K, House T, Isham V, Trapman P. Eight challenges for network epidemic models. Epidemics. 2015; 10:58–62.
World Health Organization. Ebola Virus Disease – Democratic Republic of the Congo. http://www.who.int/csr/don/10-may-2018-ebola-drc/en/.
Nguyen VK, Binder SC, Boianelli A, Meyer-Hermann M, Hernandez-Vargas EA. Ebola virus infection modeling and identifiability problems. Front Microbiol. 2015; 6:7590. https://doi.org/10.3389/fmicb.2015.00257.
Nguyen VK, Klawonn F, Mikolajczyk R, Hernandez-Vargas EA. Analysis of Practical Identifiability of a Viral Infection Model. PLoS ONE. 2016; 11(12):0167568. https://doi.org/10.1371/journal.pone.0167568.
Nguyen VK, Hernandez-Vargas EA. Windows of opportunity for Ebola virus infection treatment and vaccination. Sci Rep. 2017; 7(1):8975. https://doi.org/10.1038/s41598-017-08884-0.
Prescott JB, Marzi A, Safronetz D, Robertson SJ, Feldmann H, Best SM. Immunobiology of Ebola and Lassa virus infections. Nat Rev Immunol. 2017; 17(3):195–207.
Qiu X, Wong G, Audet J, Bello A, Fernando L, Alimonti JB, Fausther-Bovendo H, Wei H, Aviles J, Hiatt E, Johnson A, Morton J, Swope K, Bohorov O, Bohorova N, Goodman C, Kim D, Pauly MH, Velasco J, Pettitt J, Olinger GG, Whaley K, Xu B, Strong JE, Zeitlin L, Kobinger GP. Reversion of advanced Ebola virus disease in nonhuman primates with ZMapp. Nature. 2014; 514(7520):47–53.
Marzi A, Robertson SJ, Haddock E, Feldmann F, Hanley PW, Scott DP, Strong JE, Kobinger G, Best SM, Feldmann H. EBOLA VACCINE, VSV-EBOV rapidly protects macaques against infection with the 2014/15 Ebola virus outbreak strain. Science. 2015; 349(6249):739–42.
Nowak M, May RM. Virus Dynamics : Mathematical Principles of Immunology and Virology: Mathematical Principles of Immunology and Virology. Oxford: Oxford University Press; 2000.
Sullivan N, Yang Z, Nabel GJ. Ebola virus pathogenesis: implications for vaccines and therapies,. J Virol. 2003; 77(18):9733–7.
Madelain V, Nguyen TH, Olivo A, de Lamballerie X, Guedj J, Taburet AM, Mentre F. Ebola Virus Infection: Review of the Pharmacokinetic and Pharmacodynamic Properties of Drugs Considered for Testing in Human Efficacy Trials. Clin Pharmacokinet. 2016; 55(8):907–23.
Henao-Restrepo AM, Camacho A, Longini IM, Watson CH, Edmunds WJ, Egger M, Carroll MW, Dean NE, Diatta I, Doumbia M, Draguez B, Duraffour S, Enwere G, Grais R, Gunther S, Gsell PS, Hossmann S, Watle SV, Konde MK, Keita S, Kone S, Kuisma E, Levine MM, Mandal S, Mauget T, Norheim G, Riveros X, Soumah A, Trelle S, Vicari AS, Rottingen JA, Kieny MP. Efficacy and effectiveness of an rVSV-vectored vaccine in preventing Ebola virus disease: final results from the Guinea ring vaccination, open-label, cluster-randomised trial (Ebola Ça Suffit!). Lancet. 2017; 389(10068):505–18.
Li J, Duan HJ, Chen HY, Ji YJ, Zhang X, Rong YH, Xu Z, Sun LJ, Zhang JY, Liu LM, Jin B, Zhang J, Du N, Su HB, Teng GJ, Yuan Y, Qin EQ, Jia HJ, Wang S, Guo TS, Wang Y, Mu JS, Yan T, Li ZW, Dong Z, Nie WM, Jiang TJ, Li C, Gao XD, Ji D, Zhuang YJ, Li L, Wang LF, Li WG, Duan XZ, Lu YY, Sun ZQ, Kanu AB, Koroma SM, Zhao M, Ji JS, Wang FS. Age and Ebola viral load correlate with mortality and survival time in 288 Ebola virus disease patients. Int J Infect Dis. 2016; 42:34–9.
Agarwal S, Cunningham-Rundles C. Assessment and clinical interpretation of reduced IgG values. Ann Allergy Asthma Immunol. 2007; 99(3):281–3.
Feldmann H, Geisbert TW. Ebola haemorrhagic fever,. Lancet (London, England). 2011; 377(9768):849–62.
Akerlund E, Prescott J, Tampellini L. Shedding of Ebola virus in an asymptomatic pregnant woman,. N Engl J Med. 2015; 372(25):2467–9.
Statistics Sierra Leone (SSL) and ICF International. Sierra Leone Demographic and Health Survey 2013: Freetown, Sierra Leone and Rockville, Maryland, USA: SSL and ICF International; 2014.
World Health Organization. What We Know About Transmission of the Ebola Virus Among Humans: Ebola situation assessment; 2014. http://www.who.int/mediacentre/news/ebola/06-october-2014/en/.
Lipton J. Care and burial practices in urban Sierra Leone. 2014. http://www.ebola-anthropology.net/wp-content/uploads/2014/11/care-and-burial-practice.pdf.
Funk S, Bansal S, Bauch CT, Eames KT, Edmunds WJ, Galvani AP, Klepac P. Nine challenges in incorporating the dynamics of behaviour in infectious diseases models. Epidemics. 2015; 10:21–25.
R Core Team. R: A Language and Environment for Statistical Computing. Vienna, Austria: R Foundation for Statistical Computing; 2015. https://www.R-project.org/.
Soetaert K, Cash J, Mazzia F. Solving Differential Equations in R. Berlin, Heidelberg: Springer; 2012. https://doi.org/10.1007/978-3-642-28070-2. http://link.springer.com/10.1007/978-3-642-28070-2.
Csardi G, Nepusz T. The igraph software package for complex network research. InterJournal. 2006; Complex Systems:1695.
World Health Organization. List of Blueprint priority diseases. Cambridge: WHO; 2018.
Csardi G, Nepusz T. The igraph software package for complex network research. InterJournal. 2006; Complex Systems:1695.
Peixoto TP. The graph-tool python library. figshare. 2014. https://doi.org/10.6084/m9.figshare.1164194. Accessed 10 Sep 2014.
World Health Organization. Immunization Coverage: Geneva: World Health Organization; 2010.
Keeling MJ, Rohani P. Modeling Infectious Diseases in Humans and Animals. Princeton: Princeton University Press; 2011.
Metcalf CJE, Andreasen V, Bjørnstad ON, Eames K, Edmunds WJ, Funk S, Hollingsworth TD, Lessler J, Viboud C, Grenfell BT. Seven challenges in modeling vaccine preventable diseases. Epidemics. 2015; 10:11–15. https://doi.org/10.1016/j.epidem.2014.08.004.
Centers for Disease Control and Prevention and World Health Organization. Implementation and Management of Contact Tracing for Ebola Virus Disease: Emergency Guideline. 2015.
Nishiura H. Early efforts in modeling the incubation period of infectious diseases with an acute course of illness. Emerg Themes Epidemiol. 2007; 4(1):2. https://doi.org/10.1186/17622-4-2.
Obadia T, Haneef R, Boelle PY. The R0 package: a toolbox to estimate reproduction numbers for epidemic outbreaks. BMC Med Inform Decis Mak. 2012; 12:147. https://doi.org/10.1186/1472-6947-12-147.
Virlogeux V, Fang VJ, Park M, Wu JT, Cowling BJ. Comparison of incubation period distribution of human infections with MERS-CoV in South Korea and Saudi Arabia. Sci Rep. 2016; 6:35839.
Martínez MJ, Salim AM, Hurtado JC, Kilgore PE. Ebola Virus Infection: Overview and Update on Prevention and Treatment. Infect Dis Ther. 2015; 4(4):365–30. https://doi.org/10.1007/s40121-015-0079-5.
Pastor-Satorras R, Castellano C, Van Mieghem P, Vespignani A. Epidemic processes in complex networks. Rev Mod Phys. 2015; 87(3):925.
Bisset KR, Chen J, Feng X, Kumar VSA, Marathe MV. EpiFast: a Fast Algorithm for Large Scale Realistic Epidemic Simulations on Distributed Memory Systems. a fast algorithm for large scale realistic epidemic simulations on distributed memory systems. New York, New York, USA: ACM; 2009. https://doi.org/10.1145/1542275.1542336.
Heffernan JM, Smith RJ, Wahl LM. Perspectives on the basic reproductive ratio. Journal of The Royal Society Interface. 2005; 2(4):281–293.
Camacho A, Kucharski AJ, Funk S, Breman J, Piot P, Edmunds WJ. Potential for large outbreaks of Ebola virus disease. Epidemics. 2014; 9:70–78. https://doi.org/10.1016/j.epidem.2014.09.003.
Bansal S, Grenfell BT, Meyers LA. When individual behaviour matters: homogeneous and network models in epidemiology. J R Soc Interface. 2007; 4(16):879–91. https://doi.org/10.1098/rsif.2007.1100.
Abbas AK. B Cell Activation and Antibody Production. In: Cellular and Molecular Immunology. Elsevier Health Sciences: 2011. p. 1–26.
Vegvari C, Hadjichrysanthou C, Cauët E, Lawrence E, Cori A, de Wolf F, Anderson RM. How Can Viral Dynamics Models Inform Endpoint Measures in Clinical Trials of Therapies for Acute Viral Infections?PLoS ONE. 2016; 11(7):0158237–13. https://doi.org/10.1371/journal.pone.0158237.
Bowman A, Azzalini A. R Package sm: Nonparametric Smoothing Methods (version 2.2-5.4). 2014.
Van Kerkhove MD, Bento AI, Mills HL, Ferguson NM, Donnelly CA. A review of epidemiological parameters from Ebola outbreaks to inform early public health decision-making. Sci Data. 2015; 2:150019.
Funding
This work was supported by the Alfons und Gertrud Kassel-Stiftung. VKN was supported by the Presidents Initiative and Networking Funds of the Helmholtz Association of German Research Centres (HGF) under contract number VH-GS-202. The funders have no role in the design of the study and collection, analysis, and interpretation of data and in writing the manuscript.
Availability of data and materials
All data generated or analysed during this study are included in this published article and its supplementary information files.
Code and examples — Generating age-specific contact network R code for generating an example contact network can be found at http://doi.org/10.5281/zenodo.1037264.
Code and examples — Epidemic simulations R code for epidemic simulations can be found at https://doi.org/10.5281/zenodo.1045404.
Author information
Authors and Affiliations
Contributions
EAHV supervised the project. VKN designed the modelling and performed the simulations. VKN, EAHV and RM provided and analysed the data. VKN, EAHV and RM discussed and wrote the manuscript. All authors reviewed the manuscript. All authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Additional files
Additional file 1
Figure S1. Estimates of the reproductive number in different vaccination schemes. Simulations of a network of size ten thousand during a period of one year are performed. One thousand simulations were run, each time with a random index case. At the end of each simulation, the network of infected nodes was extracted to compute the average number of secondary infections. (PDF 27 kb)
Additional file 2
Figure S2. Case-fatality rate in different vaccination schemes. Simulations of a network of size ten thousand during a period of one year are performed. One thousand simulations were run, each time with a random index case. At the end of each simulation, the network of infected nodes was extracted to compute the case-fatality rate. (PDF 28 kb)
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License(http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver(http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Nguyen, V., Mikolajczyk, R. & Hernandez-Vargas, E. High-resolution epidemic simulation using within-host infection and contact data. BMC Public Health 18, 886 (2018). https://doi.org/10.1186/s12889-018-5709-x
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12889-018-5709-x