Abstract
Background
Predictive models for mental disorders or behaviors (e.g., suicide) have been successfully developed at the level of populations, yet current demographic and clinical variables are neither sensitive nor specific enough for making individual clinical predictions. Forecasting episodes of illness is particularly relevant in bipolar disorder (BD), a mood disorder with high recurrence, disability, and suicide rates. Thus, to understand the dynamic changes involved in episode generation in BD, we propose to extract and interpret individual illness trajectories and patterns suggestive of relapse using passive sensing, nonlinear techniques, and deep anomaly detection. Here we describe the study we have designed to test this hypothesis and the rationale for its design.
Method
This is a protocol for a contactless cohort study in 200 adult BD patients. Participants will be followed for up to 2 years during which they will be monitored continuously using passive sensing, a wearable that collects multimodal physiological (heart rate variability) and objective (sleep, activity) data. Participants will complete (i) a comprehensive baseline assessment; (ii) weekly assessments; (iii) daily assessments using electronic rating scales. Data will be analyzed using nonlinear techniques and deep anomaly detection to forecast episodes of illness.
Discussion
This proposed contactless, large cohort study aims to obtain and combine high-dimensional, multimodal physiological, objective, and subjective data. Our work, by conceptualizing mood as a dynamic property of biological systems, will demonstrate the feasibility of incorporating individual variability in a model informing clinical trajectories and predicting relapse in BD.
Similar content being viewed by others
Background
Bipolar disorder (BD) is a mood disorder with high recurrence and disability rates [1]. It is estimated that the suicide risk in patients with BD is 20 times higher than in the general population [2]. A key feature of BD is its varying severity over time, in some patients manifested as distinct mood episodes, in others as fluctuating intensity of mood symptoms. As a result, interventions for long-term treatment and relapse prevention are crucial for the clinical management of patients with BD [3]. However, these strategies have not decreased the incidence of relapse or adverse outcomes [4]. One of the reasons relapse prevention may not be successful is because traditional clinical monitoring is not frequent enough to manage the capricious nature of the illness [5].
Similarly, predictive models for mental disorders or behaviors (e.g., suicide) have been successfully developed at the level of populations, yet current demographic and clinical variables are neither sensitive nor specific enough for making individual clinical predictions. For instance, a recent meta-analysis concluded that predictive ability for dying from suicide has not improved over 50 years of research [6]. New analytical approaches based on nonlinear mathematical methods (i.e., time-series analyses and entropy calculations) and machine learning (anomaly detection), coupled with passive sensing (i.e., wearables) may allow us to address this problem, which has been intractable so far.
The wide availability of low-cost wearables enables us to measure how the environment and our experiences impact our physiology. Recent studies in electronic (e-)monitoring in BD have typically focused on group differences rather than improving outcomes [7]. Also, these studies have faced two major challenges: (i) a high proportion of missing data; (ii) and inadequate statistical tools to process large amounts of data. Moreover, participants identify forgetfulness as the top barrier for engagement with e-monitoring [8]. Thus, passive sensing, which requires minimal effort from participants, may lead to more effective data collection.
While most studies concur on the difficulty of finding an overarching prediction algorithm that is applicable to a wide variety of patients, individual variability is a concept that has not been taken into account, even though it is the most important factor in individualized risk estimation [9, 10]. We hypothesize that we will not be able to make actionable individual predictions for patients with mental disorders without characterizing individual variability [10]. Thus, to understand the dynamic changes involved in episode generation in BD, we propose to integrate physiological, objective, and subjective clinical variables relevant for mood regulation using nonlinear techniques. We hypothesize that integrating these variables will allow us to assess the impact of individual variability on illness trajectories and predict the onset of episodes in BD. Here we describe the study we have designed to test this hypothesis and the rationale for its design. We also illustrate the analytical process involved in deep anomaly detection, which is the technique we will use to forecast episodes of illness.
Methods
Study overview
The study will recruit 200 participants diagnosed with BD I or BD II. After completing a comprehensive baseline assessment, they will be followed for up to 2 years during which they will be monitored continuously with a wearable Oura ring (Oura Health Oy, Generation 2, Oulu, Finland) and will complete questionnaires regularly. The study will be contactless, i.e., all the research procedures will be conducted by phone or virtually. Oura Health Oy did not sponsor the study nor has access to collected data stemming from this study.
Settings
Participants will be recruited at two academic centers in Canada: the Adult Psychiatry Division, Centre for Addiction and Mental Health (CAMH), Toronto, Ontario; and the Mood Disorders Program, Queen Elizabeth II Health Sciences Centre, Halifax, Nova Scotia. The study has been approved by the local REBs with extensive feedback from the Information Technology (IT) and Privacy leaders.
Recruitment and consent
We will include adult men or women of any race and ethnicity, aged 18 and older (with no upper age limit), with a primary diagnosis of BD I or II, according to DSM-5 criteria [11] based on the SCID-5 [12], in any phase of the illness (i.e., euthymic, depressive, (hypo)manic, or mixed). Treating physicians at the two academic centers will refer their potentially eligible patients who express an interest in participating in the study. All participants will provide written informed consent using a form approved by the local REB and will be advised of their right to withdraw from the study.
Baseline assessment and clinical follow-up
After providing informed consent, participants will complete a comprehensive baseline assessment as summarized in Table 1. This baseline assessment includes six domains: socio-demographic, diagnosis, clinical course, cardiovascular screening, chronotype determination, and pharmacotherapy. Collecting this information will allow us to characterize the sample and assess variables that will be included in the predictive model, including Illness Burden Index [13]. Participants will receive standard (“as usual”) medical treatment.
A trained rater will administer the Young Mania Rating Scale (YMRS) [14] and the Montgomery-Asberg Depression Rating Scale (MADRS) [15] to determine polarity at study entrance. Euthymia will be operationalized as a score ≤ 10 on both scales. MADRS scores will be used to define the presence and severity of depressive episodes as per established guidelines [16] (mildly ill: 11–18; moderately ill: 19–23; moderate to severely ill: 24–36; severely ill: 37–39; or extremely ill: ≥ 40). Similarly, YMRS scores will be used to define episodes of hypomania, mania, or mania with psychosis (hypomania: 11–19; mania; ≥ 20, with or without psychotic symptoms). Both scales will be used in all participants because mixed episodes may be difficult to detect in the early stages. Cardiovascular (functional) capacity will be assessed with the Duke Activity Status Index Scale (DASI) [17], a 12-item questionnaire that assesses a person’s ability to perform a set of activities (personal care, ambulation, household tasks, sexual activity, recreational activity). The DASI scores integrates weighted answers based on the known metabolic cost for each activity in Metabolic-Equivalent Task (MET) units [18]; it ranges from 0 to 58.2, with higher values indicative of higher (better) functional capacity. Chronotype will be assessed with the 19-item Morningness-Eveningness Questionnaire (MEQ) [19], the responses of which are combined into a composite score that indicates participant’s chronotype (morning, intermediate or evening). Finally, we will classify medications (and changes in medications), as we have done in previous studies, based on class, dosage, and duration of treatment [13].
Passive sensing
Participants will use the Oura ring (ouraring.com), a titanium-made ring weighing 4–6 g that collects data automatically and transfers them via Bluetooth to a smartphone on which a specialized app has been loaded; in turn, that app transfers the data to a centralized database maintained by the manufacturer and accessible to the research team. The ring assesses activity and sleep with a 3-D accelerometer and gyroscope, and heart rate (HR) and heart rate variability (HRV) with an infrared optical pulse measurement. Total sleep time and sleep latency determined by the ring are correlated 0.93 with polysomnography [20].
Participants will be sent a sizing kit to determine their ring size; they will be encouraged to try several sizes for a day or two, including during their sleep, to ensure a comfortable fit. Once participants confirm their size, we will mail them the actual wearable and they will be asked to wear it continuously for the duration of the study. As summarized in Table 2, we will use this wearable to objectively and continuously track some measures related to mood regulation: HR, HRV; parameters related to sleep (e.g., total sleep and REM-sleep duration), activity (e.g., number of steps during the day and of active and inactive minutes), and energy (e.g., energy consumption in MET during the day).
Follow-up ratings
As shown in Table 2, participants will rate their mood, anxiety, and energy level daily using electronic visual analog scales (e-VAS) accessed via an e-mailed link. The scale ranges from ‘1’ to ‘9’, with ‘5’ being “your usual”, 1 “your lowest”, and 9 “your highest” and participants describe with a swipe of their finger on the e-VAS their mood, anxiety, or energy level throughout the day, according to the Day Reconstruction Method [21]. These e-VAS, which we used in our previous studies with a paper-version, use a densely-sampled scale (change interval is 0.1), which allows us to generate continuous, fine-grain data. We will use interpolation methods when data are missing or incomplete for 1 or 2 days in a row. When data are missing for 3 days, we will contact participants by text or email and remind them about the importance of filling out the scale on a daily basis. In addition, every week, participants will complete the Patient Health Questionnaire (PHQ-9) [22] and the Altman Self-Rating Mania Scale (ASRS) [23]. Participants will get a secure email with a link to both scales and complete self-ratings, which will be uploaded into REDCap.
Data collection
All study data will be managed using REDCap electronic data capture tools hosted at CAMH. REDCap (Research Electronic Data Capture) is a secure, web-based software platform designed to support data capture for research studies, providing: an intuitive interface for validated data capture; audit trails for tracking data manipulation and export procedures; automated export procedures for seamless data downloads to common statistical packages; and procedures for data integration and interoperability with external sources [24].
Statistical analyses
First, a descriptive analysis will characterize the participants at baseline and summarize missing data. Then we will compare the baseline characteristics based on the outcome groups: “becoming ill” or “not becoming ill”. We will also use graphs to present the data and individual trajectories. We will evaluate the association among time-varying variables (i.e., sleep, mood, activity). Traditional statistical techniques will be used to model the data for comparative purposes. Cox Regression models adjusted for baseline and time-varying variables will be used to model the time until events (e.g., relapse). We will use time-dependent covariates and the occurrence of the event as censoring. We will also use dynamic time-series to fit independent models for each participant. Finally, because negative correlations have been found between age or socioeconomic status, and use of health apps [25], we will incorporate self-reported gender, age, and other relevant factors (e.g., years of education) in all our analytical models and assess whether any of these influence our results. Statistical analyses will be done using MATLAB®.
Sample size calculation
Previous studies report that up to 44–48% of BD patients relapse in the first 2 years of follow-up, and that depressive episodes are more common than hypomanic or manic ones [26]. With 200 participants with BD receiving standard treatment and followed for 12 months, we expect at least 60 participants to experience the onset of a new depressive episode and 10 to experience the onset of a new hypomanic/manic episode throughout the study duration (i.e., 35% of the sample). In our previous study, entropy calculations for the mood series showed a normal distribution with mean (SD) of 1.04 (0.6) in 30 BD patients and 1.47 (0.3) in 30 healthy comparators. Thus, if we replicate these findings in our 200 participants (H1a), we will have a power of 0.90 to differentiate highly correlated series (i.e., equivalent to low entropy levels). While this project has not been powered for H2, if the true difference in entropy levels between euthymia and illness is 0.22, we will have a power of 0.80 to reject the null hypothesis for H2 (with a Type I error probability of 0.05).
Nonlinear analyses: time-series analyses and entropy calculations
For all the time-series generated (e.g., sleep), we will compute auto-correlation for each series (i.e., how one point in the series correlates to the next point in the same series); and cross-correlation between all series (i.e., how one point in one series correlates with a corresponding point in a different series). Furthermore, we will analyze whether these variables: (i) differentially contribute to the model to identify clinical trajectories; (ii) differentially predict events (e.g., relapse). Entropy calculations for each variable will be continuously recomputed from each of the time-series. In turn, as described below, time series-data from each of the above-mentioned variables will be fed into our machine learning technique one step at a time.
Outlier and anomaly detection
The use of digital analytics for precision health is now possible given advances in machine learning models that can extract complex patterns from multiple sources of high-dimensional data over time. However, understanding how some machine-learning techniques work remains a challenging task, as it has an inherent “black box” character. Thus, we propose to use deep learning anomaly detection, a machine learning technique that doesn’t have this limitation. The principle behind outlier detection is simple and well established [27]. While this approach works robustly for single variables, once multidimensional data is considered, the deviation from the mean is not as easily computed. Scaling, distribution, and, most importantly for this study, the temporal sequence of values matters greatly. Thus, by using deep anomaly detection, instead of computing a mean and the deviation from the mean, a deep neural network can be trained on the data. The model makes predictions and the deviation of the model’s predictions from the observed participant identifies anomalous behavior. To date, deep anomaly detection has been mostly applied in risk management, security and financial surveillance [28].
We will use two different types of anomaly detection techniques: autoencoders and recurrent neural networks (RNN) for time-series prediction. We will use both methods because they differ in what they predict: an autoencoder takes a time-series (or pattern) as an input and tries to generate the same series or pattern as an output. While this sounds trivial, if the autoencoder is large enough, it becomes challenging, because the autoencoder has an information bottleneck. This bottleneck consists of very few nodes, and requires the data to be compressed, losing information, and only allowing major trends to pass through. Conversely, when using RNN or their more advanced variants, such as Long Short-Term Memory (LSTM) [29], the data is fed into the model one time step after the other, while the model continuously predicts the following time point.
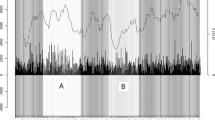
Figure 1 illustrates this process with a simulated dataset including three different signals (e.g., heart rate, activity, and sleep). A RNN was trained on this dataset (including the outlier period) to predict the next data points based on the last 20 time points. After successful training, the difference between the model’s prediction and the actual data was computed for all time points. The distribution of that deviation allows us to determine a threshold, i.e., 95% confidence interval. All data points for which the prediction is aberrant from the observation successfully identified the period in which the anomaly is expected (figure not shown). The RNN was trained on the data containing the outlier period, as this would be the case with participants’ data, which will also contain outlying periods.
Using deep learning models with data containing different signals. An example of using deep learning models with data containing three different signals. This data, while following a sinus rhythm with low noise (red, green, and blue lines), experiences high noise at the end of the series (solid black line for noise and dashed lines identifying outlier period)
Discussion
This proposed contactless, large cohort study aims to obtain and combine high-dimensional, multimodal physiological (HRV), objective (sleep, activity), and subjective data using passive sensing and e-rating scales (self-ratings and clinician-rating scales). The overall objective of the study is to extract and interpret individual illness trajectories and patterns suggestive of relapse using deep anomaly detection. Our Methods are predicated on the following: (i) models do not generalize from one participant to another; (ii) model hyper-parameters that optimally predict one participant’s trajectory (including outliers) may be different from participant to participant; (iii) a single model trained on all participants’ data cannot be used to identify outliers. Moreover, unlike many models generated by other machine learning techniques, the resulting model would be understandable and interpretable (i.e., it would not be a “black-box”). The integration of densely-sampled (i.e., “deep” data) objective and autonomic data with each participant’s unique contextual information will further enhance the performance of the model.
Potential impediments to the success of this study include (i) some participants will not be willing or able to use the wearable; (ii) relapse rates may be too low over the length of the study; (iii) participant dropout. To minimize the impact of these potential impediments, we will monitor adherence and contact participants who have not uploaded data for 3 days. In addition, participants will have their usual, regular clinical follow-ups, which will give us an additional opportunity to encourage adherence with study procedures. However, participants are informed through the consent form that we are not able to monitor their illness in real time; and they are advised to obtain medical advice if they feel their mood is deteriorating. In these instances, we expect that most psychiatrists will promptly assess their patients to confirm whether they are indeed experiencing a relapse and will implement appropriate actions when warranted clinically.
The major strengths of our study include its foundation on a solid work of an interdisciplinary team on clinical trajectories in BD; nonlinear properties of mood regulation in healthy controls, BD patients, and their unaffected first-degree relatives [10, 30]; feasibility of using passive sensing in adults with BD during different types of episodes [13]; and a simulation using machine learning for outcome prediction [31]. By conceptualizing mood as a dynamic property of biological systems, we plan to demonstrate the feasibility of incorporating individual variability in a model informing clinical trajectories and predicting relapse in BD. Ultimately, if we succeed in predicting relapses, we should be able to prevent them.
Availability of data and materials
Not applicable.
Abbreviations
- ASRS:
-
Altman Self-Rating Mania Scale
- BD:
-
Bipolar disorder
- BMI:
-
Body mass index
- DASI:
-
Duke Activity Status Index Scale
- HR:
-
Heart rate
- HRV:
-
Heart rate variability
- IBI:
-
Illness Burden Index
- MADRS:
-
Montgomery-Asberg Depression Rating Scale
- MET:
-
Metabolic Equivalent of a Task
- MEQ:
-
Morningness-Eveningness Questionnaire
- PHQ-9:
-
Patient Health Questionnaire, 9 items
- REM:
-
Rapid Eye Movement
- RMSSD:
-
Root Mean Square of Successive Differences
- SCID: 5:
-
Structured Clinical Interview for DSM-5
- YMRS:
-
Young Mania Rating Scale
References
Deckersbach T, Nierenberg AA, McInnis MG, Salcedo S, Bernstein EE, Kemp DE, et al. Baseline disability and poor functioning in bipolar disorder predict worse outcomes: results from the bipolar CHOICE study. J Clin Psychiatry. 2016;77(1):100–8.
Simon GE, Hunkeler E, Fireman B, Lee JY, Savarino J. Risk of suicide attempt and suicide death in patients treated for bipolar disorder. Bipolar Disord. 2007;9(5):526–30.
Morriss RK, Faizal MA, Jones AP, Williamson PR, Bolton C, McCarthy JP. Interventions for helping people recognise early signs of recurrence in bipolar disorder. Cochrane Database Syst Rev. 2007;2007(1):Cd004854.
Kessing LV, Andersen PK, Vinberg M. Risk of recurrence after a single manic or mixed episode - a systematic review and meta-analysis. Bipolar Disord. 2018;20(1):9–17.
Judd LL, Akiskal HS, Schettler PJ, Endicott J, Maser J, Solomon DA, et al. The long-term natural history of the weekly symptomatic status of bipolar I disorder. Arch Gen Psychiatry. 2002;59(6):530–7.
Franklin JC, Ribeiro JD, Fox KR, Bentley KH, Kleiman EM, Huang X, et al. Risk factors for suicidal thoughts and behaviors: a meta-analysis of 50 years of research. Psychol Bull. 2017;143(2):187–232.
Ortiz A, Maslej MM, Husain I, Daskalakis J, Mulsant BH. Apps and gaps in bipolar disorder: a systematic review on electronic monitoring for episode prediction. J Affect Disord. 2021;295:1190–200.
Van Til K, McInnis MG, Cochran A. A comparative study of engagement in mobile and wearable health monitoring for bipolar disorder. Bipolar Disord. 2020;22(2):182–90.
Nelson B, McGorry PD, Wichers M, Wigman JTW, Hartmann JA. Moving from static to dynamic models of the onset of mental disorder: a review. JAMA Psychiatry. 2017;74(5):528–34.
Ortiz A, Bradler K, Mowete M, MacLean S, Garnham J, Slaney C, et al. The futility of long-term predictions in bipolar disorder: mood fluctuations are the result of deterministic chaotic processes. Int J Bipolar Disord. 2021; In press.
Diagnostic and statistical manual of mental disorders. 5th ed. American Psychiatric Publishing; 2013. DSM-V, http://repository.poltekkes-kaltim.ac.id/657/1/Diagnostic%20and%20statistical%20manual%20of%20mental%20disorders%20_%20DSM-5%20%28%20PDFDrive.com%20%29.pdf.
First M, Williams J, Karg R, RL S. Structured clinical interview for DSM-5, research version (SCID-5). Arlington: American Psychiatric Association; 2015.
Ortiz A, Bradler K, Moorti P, MacLean S, Husain MI, Sanches M, et al. Reduced heart rate variability is associated with higher illness burden in bipolar disorder. J Psychosom Res. 2021;145:110478.
Young RC, Biggs JT, Ziegler VE, Meyer DA. A rating scale for mania: reliability, validity and sensitivity. Br J Psychiatry. 1978;133:429–35.
Montgomery SA, Asberg M. A new depression scale designed to be sensitive to change. Br J Psychiatry. 1979;134:382–9.
Thase ME, Harrington A, Calabrese J, Montgomery S, Niu X, Patel MD. Evaluation of MADRS severity thresholds in patients with bipolar depression. J Affect Disord. 2021;286:58–63.
Hlatky MA, Boineau RE, Higginbotham MB, Lee KL, Mark DB. A brief self-administered questionnaire to determine functional capacity. Am J Cardiol. 1989;64(10):651–4.
Passmore R, Durnin JV. Human energy expenditure. Physiol Rev. 1955;35(4):801–40.
Horne JA, Ostberg O. A self-assessment questionnaire to determine morningness-eveningness in human circadian rhythms. Int J Chronobiol. 1976;4(2):97–110.
de Zambotti M, Rosas L, Colrain IM, Baker FC. The sleep of the ring: comparison of the OURA sleep tracker against Polysomnography. Behav Sleep Med. 2017:1–15.
Kahneman D, Krueger AB, Schkade DA, Schwarz N, Stone AA. A survey method for characterizing daily life experience: the day reconstruction method. Science. 2004;306(5702):1776–80.
Kroenke K, Spitzer RL, Williams JB. The PHQ-9: validity of a brief depression severity measure. J Gen Intern Med. 2001;16(9):606–13.
Altman EG, Hedeker D, Peterson JL, Davis JM. The Altman self-rating mania scale. Biol Psychiatry. 1997;42(10):948–55.
Harris PA, Taylor R, Thielke R, Payne J, Gonzalez N, Conde JG. Research electronic data capture (REDCap)--a metadata-driven methodology and workflow process for providing translational research informatics support. J Biomed Inform. 2009;42(2):377–81.
Carroll JK, Moorhead A, Bond R, LeBlanc WG, Petrella RJ, Fiscella K. Who uses Mobile phone health apps and does use matter? A secondary data analytics approach. J Med Internet Res. 2017;19(4):e125.
Perlis RH, Ostacher MJ, Patel JK, Marangell LB, Zhang H, Wisniewski SR, et al. Predictors of recurrence in bipolar disorder: primary outcomes from the systematic treatment enhancement program for bipolar disorder (STEP-BD). Am J Psychiatry. 2006;163(2):217–24.
Bullen RJ, Cornford D, Nabney IT. Outlier detection in scatterometer data: neural network approaches. Neural Netw. 2003;16(3–4):419–26.
Pang G, Shen C, Cao L, Hengel AVD. Deep learning for anomaly detection: a review. ACM Comput Surv. 2021;54(2):1–38.
Hochreiter S, Schmidhuber J. Long short-term memory. Neural Comput. 1997;9(8):1735–80.
Ortiz A, Bradler K, Garnham J, Slaney C, Alda M. Nonlinear dynamics of mood regulation in bipolar disorder. Bipolar Disord. 2015;17(2):139–49.
Ortiz A, Bradler K, Hintze A. Episode forecasting in bipolar disorder: is energy better than mood? Bipolar Disord. 2018;20(5):470–6.
Acknowledgements
Not applicable.
Funding
This work has been supported by the National Institute of Mental Health (NIMH) grant 1R21MH123849-01A1 (Principal Investigator: Dr. Ortiz) and by the Canadian Institutes of Health Research (CIHR) grant 02010PJT-450770-BSB-CEAH-188794 (Principal Investigator: Dr. Ortiz). Funding agencies had no role in the design and conduct of the study. The research team had full autonomy in all aspects of the study.
Author information
Authors and Affiliations
Contributions
AO conceptualized the study, managed, and coordinated responsibility for the grant funding and activity planning; wrote the original draft, reviewed, and edited the final draft. AH performed data curation, implemented the computer code and supporting algorithms for data analysis; and verified the overall reproducibility of the results. RB contributed to the original draft. CGT contributed to the original draft. JM, SU, and DY contributed to the original draft and to the statistical analyses of the data. MA reviewed and edited the draft. BM provided mentorship external to the core team, critically reviewed and edited drafts. The author(s) read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Participants will be recruited at two academic centers in Canada: the Adult Psychiatry Division, Centre for Addiction and Mental Health (CAMH), Toronto, Ontario; and the Mood Disorders Program, Queen Elizabeth II Health Sciences Centre, Halifax, Nova Scotia. The study has been approved by both ethics committees: the Research Ethics Board at CAMH (#059–2019), with extensive feedback from the Information Technology (IT) and Privacy leaders; as well as the Research Ethics Board at the Queen Elizabeth II Health Sciences Centre in Halifax, NS. All participants will provide written informed consent using a form approved by the local REB and will be advised of their right to withdraw from the study.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Ortiz, A., Hintze, A., Burnett, R. et al. Identifying patient-specific behaviors to understand illness trajectories and predict relapses in bipolar disorder using passive sensing and deep anomaly detection: protocol for a contactless cohort study. BMC Psychiatry 22, 288 (2022). https://doi.org/10.1186/s12888-022-03923-1
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12888-022-03923-1