Abstract
Background
Measuring arterial partial pressure of carbon dioxide (PaCO2) is crucial for proper mechanical ventilation, but the current sampling method is invasive. End-tidal carbon dioxide (EtCO2) has been used as a surrogate, which can be measured non-invasively, but its limited accuracy is due to ventilation-perfusion mismatch. This study aimed to develop a non-invasive PaCO2 estimation model using machine learning.
Methods
This retrospective observational study included pediatric patients (< 18 years) admitted to the pediatric intensive care unit of a tertiary children’s hospital and received mechanical ventilation between January 2021 and June 2022. Clinical information, including mechanical ventilation parameters and laboratory test results, was used for machine learning. Linear regression, multilayer perceptron, and extreme gradient boosting were implemented. The dataset was divided into 7:3 ratios for training and testing. Model performance was assessed using the R2 value.
Results
We analyzed total 2,427 measurements from 32 patients. The median (interquartile range) age was 16 (12−19.5) months, and 74.1% were female. The PaCO2 and EtCO2 were 63 (50−83) mmHg and 43 (35−54) mmHg, respectively. A significant discrepancy of 19 (12–31) mmHg existed between EtCO2 and the measured PaCO2. The R2 coefficient of determination for the developed models was 0.799 for the linear regression model, 0.851 for the multilayer perceptron model, and 0.877 for the extreme gradient boosting model. The correlations with PaCO2 were higher in all three models compared to EtCO2.
Conclusions
We developed machine learning models to non-invasively estimate PaCO2 in pediatric patients receiving mechanical ventilation, demonstrating acceptable performance. Further research is needed to improve reliability and external validation.
Similar content being viewed by others
Background
Evaluating adequate oxygen and carbon dioxide (CO2) exchange is crucial for managing patients receiving mechanical ventilation. The arterial partial pressure of CO2 (PaCO2) is commonly used for evaluation, but its invasive measurement via arterial blood sampling can be painful and cause iatrogenic complications such as mechanical damage of arteries, anemia, embolism, thrombosis, or nerve injury. In general, as the difficulty of the procedure increases in smaller infant, the degree of complications may become more severe [1]. Therefore, this highlights the need for a non-invasive method to estimate PaCO2 in children.
End-tidal CO2 (EtCO2) and transcutaneous CO2 (TCO2) monitoring are feasible alternatives. EtCO2 reflects exhaled CO2, but ventilation-perfusion mismatch limits its accuracy as a surrogate for PaCO2. Factors like dead-space ventilation, severe atelectasis, and intrapulmonary shunts can influence its reliability in various clinical settings. Therefore, bedside practice often utilizes the EtCO2 trend to estimate PaCO2 trends rather than relying on a single value [2,3,4]. TCO2, measuring the partial pressure of CO2 in arteriolarized capillaries through skin warming, also has limitations that make it difficult to use it widely in clinical practice, especially in acute medical settings [5, 6].
Few studies have attempted to estimate the dead space fraction using the difference between EtCO2 and PaCO2, which was even described as an independent risk factor associated with mortality [7, 8]. However, developing a robust formula for this purpose remains challenging due to the influence of various physiological factors [7,8,9,10,11,12]. Recent research has demonstrated the successful application of artificial intelligence for individualized estimation and prediction of complex factors, overcoming limitations of conventional methods [13,14,15].
This study aimed to develop machine learning models based on non-invasively collected data and compare their performance to estimate PaCO2 more accurately in mechanically ventilated children.
Methods
Study setting and data collection
This retrospective cross-sectional observational study was conducted in the pediatric intensive care unit (PICU) of a university-affiliated children’s hospital. Patients aged less than 18 years who were admitted to the PICU and received mechanical ventilation between January 2021 and June 2022 were eligible for inclusion. Patients who underwent extracorporeal membrane oxygenation, which allows oxygenation and CO2 elimination using methods other than mechanical ventilation, were excluded. Additionally, patients who did not have PaCO2 measured during mechanical ventilation were excluded.
Data used in this study were retrieved from our institution’s data warehouse. Demographic information, clinical characteristics (systolic blood pressure [BP], diastolic BP, heart rate [HR], respiratory rate [RR], peripheral oxygen saturation [SpO2], height, weight, and EtCO2), and ventilator parameters (minute ventilation, peak inspiratory pressure, fraction of inspired oxygen [FiO2], positive end-expiratory pressure [PEEP], and mean airway pressure [MAP]) were collected. Blood gas analysis results, representing the only invasive data in this study, were collected for training the machine learning model.
This study was conducted in accordance with the principles of the Declaration of Helsinki. The protocol of this study was reviewed by the Institutional Review Board of Seoul National University Hospital. This study was recognized as a minimal risk study by the above committee because it used only pseudonymous information and did not collect personally identifiable information. Therefore, for the above reasons, the above committee waived the need for written informed consent and approved the conduct of this study (approval no. H-2307-160-1452).
Data preprocessing
Among the collected data, cases containing duplicates or missing values were excluded. A limitation of retrospectively collected blood gas analysis results is the inability to definitively differentiate between arterial and venous blood samples. Therefore, we assumed arterial blood when the arterial partial pressure of oxygen (PaO2) was equal to or greater than 40 mmHg, thereby including cases with hypoxemia. Additionally, measurements outside the physiological range, suggesting errors in recording or measurement, were excluded. These criteria included: HR < 30 beats/minute or > 300 beats/minute, RR < 5 breaths/minute or > 120 breaths/minute, EtCO2 < 20 mmHg, and FiO2 < 21%. For clinical information and ventilator parameters, time values were converted to hours by discarding minutes and seconds. Laboratory findings were integrated into a one-hour window. To improve model performance, FiO2, which exhibits a known non-linear relationship with PaCO2, was log-transformed. Patients’ BPs were converted to percentiles based on age, sex, and height [16]. Pulse pressure and oxygenation saturation index (OSI) were calculated using the collected variables following the method outlined [17]. Data preprocessing was performed using R statistical packages R 4.3.1.
Strategy of analysis
The primary outcome of this study was the estimation performance of the developed models. Performance was compared with the R2 values and visualized through the Bland-Altman analysis with 95% limits of agreement and scatter plots. The measured PaCO2 was referred to as the gold standard, and the machine learning-derived estimated CO2 was served as the comparator. The correlation coefficients between EtCO2 and the measured PaCO2 were presented with P-values as a secondary outcome.
Demographics and characteristics were presented as median (interquartile range) and compared using the Wilcoxon signed-rank test for non-normally distributed categorical variables and the chi-square test for other categorical variables. To account for the discrepancy between PaCO2 and EtCO2, the absolute values of PaCO2-EtCO2 were calculated. Correlations among the values were evaluated using Spearman’s correlation coefficient. Linear regression analysis of PaCO2 and EtCO2 was performed to determine R2 values. The normality of residuals was assessed using the Shapiro-Wilk test before conducting the Bland-Altman analysis. Statistical analyses were performed using R software version 4.3.1., P -values less than 0.05 were considered statistically significant, and a confidence level of 95% was chosen for analysis.
Model development and validation
Using collinearity analysis of the classical linear regression and feature importance analysis in the models, the following variables that can be non-invasively retrievable in clinical settings were selected for machine learning: weight, percentile of systolic and diastolic BP, RR, HR, EtCO2, minute ventilation, log-transformed FiO2, SpO2, MAP and PEEP. To minimize the outliers’ impact, the variables were scaled according to the quantile range.
To mitigate potential overfitting and selection bias from the varying numbers of individual measurements per patient, each patient’s data was split into a 7:3 ratio for training and validation. Machine-learning models were developed using three algorithms: linear regression, multi-layer perceptron (MLP), and extreme gradient boosting (XGB). The final model of multivariate linear regression was built based on the Akaike information criterion, and multicolinearity was assessed using the variance inflation factor. Optimal hyperparameters for MLP and XGB models were extracted through a grid search with five-fold cross-validation [18, 19]. The MLP model was designed with two hidden layers: 50 nodes in the first and 10 in the second. ReLU activation and Adam solver were used. The XGB model had a maximum decision tree depth of seven, a learning rate of 0.01, and 500 estimators. Feature importance in the XGB model was determined using Shapley additive explanations (SHAP) values, reflecting the impact of various features on the model [20]. Machine learning was implemented using Python version 3.10.4 (Python Software Foundation, Beaverton, OR, USA; https://www.python.org) and open libraries such as Pandas, Keras, Pytorch, Numpy and Scikit-learn.
Results
Baseline characteristics
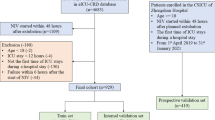
From 32 pediatric patients, we collected a total of 2,427 measurements. After applying the exclusion criteria, 1,546 measurements from 30 patients remained for analysis (Fig. 1). Patient demographics and baseline characteristics were presented in Table 1.
Continuous variables are expressed as medians (interquartile ranges), and categorical variables are expressed as numbers (percentages). BP, blood pressure; EtCO2, end-tidal carbon dioxide; SpO2, peripheral oxygen saturation; FiO2, fraction of inspired oxygen; MAP, mean airway pressure; PIP, peak inspiratory pressure; PEEP, positive end-expiratory pressure; PaCO2, arterial partial pressure of carbon dioxide; PaO2, arterial partial pressure of oxygen; (PaCO2 - EtCO2), discrepancy between PaCO2 and EtCO2.
The age at admission was 16 (12−19.5) months and female predominance was observed. The OSI indicated mild to moderate acute respiratory distress syndrome (ARDS), and the proportion of patients with OSI ≥ 12, corresponding to severe ARDS, was 31.46% [17]. The percentiles of systolic and diastolic BP suggested that most patients were not hypotensive and had BP above the fifth percentile. The majority of patients were hypercapnic and the discrepancy between PaCO2 and EtCO2 was 19 (12−31) mmHg. The PaO2/FiO2 ratio was 119.4 (82.6−202.2) mmHg.
Main outcomes
The R2 values and correlation coefficients for the linear, MLP, and XGB models were summarized in Table 2. Bland-Altman analyses were performed for the models using scaled data. The linear regression model exhibited a mean difference of -0.574 with a 95% limit of agreement (LOA) ranging from − 18.449 to 17.301. The MLP model improved upon this, showing a mean difference of -0.108 with a tighter 95% LOA of -14.107 to 13.892. The XGB model achieved the best performance, with a mean difference of -0.340 and a narrow 95% LOA of -15.716 to 15.036. This is further confirmed by the scatter plot, which demonstrates minimal bias (Fig. 2).
Performance and bias of the machine learning models. The Bland-Altman analysis for the linear regression model, the multi-layer perceptron (MLP) model, and the extreme gradient boosting (XGB) model are shown in (A), (B), and (C), respectively. The scatter plots for the linear regression model, the MLP model, and the XGB model are exhibited in (D), (E), and (F), respectively. CO2: carbon dioxide
Correlations were estimated using the Spearman correlation coefficient, and R2 values were calculated to compare the developed models. MLP, multi-layer perceptron; XGB, extreme gradient boosting.
The Shapiro-Wilk test performed on residuals yielded P-values of 0.042, < 0.001, and < 0.001 for the linear regression, MLP, and XGB models, respectively. The developed models presented high correlations with PaCO2. In comparison, EtCO2 and PaCO2 displayed a correlation coefficient of 0.78 (P < 0.001) and an R2 value of 0.58, highlighting the superior performance of machine learning models. Notably, the discrepancy between EtCO2 and PaCO2 increased with higher PaCO2 values, and the group with OSI ≥ 12 was more prevalent at a greater discrepancy between EtCO2 and PaCO2 (Fig. 3). The feature importance of the XGB model, which exhibited the highest performance, is visualized in Fig. 4.
Scatter plot showing the correlation of PaCO2 and EtCO2. The Spearman correlation coefficient was 0.78, and the R2 value was 0.58. The plot reveals a poor correlation between PaCO2 and EtCO2 as PaCO2 increases. The values of those who belong to severe acute respiratory distress syndrome, defined by the oxygenation saturation index ≥ 12, were represented with red dots. OSI: oxygenation saturation index; (PaCO2 - EtCO2): discrepancy between PaCO2 and EtCO2
The feature importance in the XGB model. The relative importance of each feature was measured by Shapley additive explanations values. etco2: end-tidal carbon dioxide; log_fio2: log-transformed fraction of inspired oxygen; wt: weight; peep: positive end-expiratory pressure; ht: height; map: mean airway pressure, rr: respiratory rate; spo2: peripheral oxygen saturation; ve: minute ventilation; p_dbp: percentile of diastolic blood pressure; pip: peak inspiratory pressure; hr: heart rate; p_sbp: percentile of systolic blood pressure; |SHAP|: the absolute values of Shapley additive explanations; SHAP: Shapley additive explanations
Discussion
This study aimed to develop a non-invasive machine learning model for estimating PaCO2 in critically ill children receiving mechanical ventilation. We focused on minimizing the difference between PaCO2 and EtCO2 through machine learning, recognizing the significant impact that dead space ventilation or perfusion status can have on their correlation [7, 21, 22].
In the study cohort, the correlation coefficient between EtCO2 and PaCO2 was 0.78 (P < 0.001), with an R2 value of 0.58. This was in with previous reporting R2 values ranging from 0.4 to 0.9, depending on disease entities and ventilator modes [3, 4, 23].
Cross-sectional studies and clinical trials conducted in both adults and pediatric patients have investigated factors influencing the difference between EtCO2 and PaCO2. These studies suggest a negative relationship with PaO2/FiO2 [3, 23]. However, these studies primarily included patients with relatively small discrepancies between PaCO2 and EtCO2, and limited representation of patients with PaO2/FiO2 ≤ 200 mmHg. McDonald et al. [23] observed a discrepancy of 6.8 mmHg with a standard deviation of 6.4 mmHg, while Wang et al. [3] reported an even smaller discrepancy of 1.86 ± 7.42 mmHg in patients undergoing synchronized intermittent mandatory ventilation.
In contrast to these previous studies, our study included patients with a larger discrepancy between PaCO2 and EtCO2 of 19 (12−31) mmHg and a lower PaO2/FiO2 of 119.4 (82.6−202.2) mmHg. Even though the median OSI was in accordance with mild to moderate ARDS, the proportion of the group with OSI ≥ 12 was considerable. In our cohort, severely ill children with OSI ≥ 12 were observed more frequently at a higher discrepancy between EtCO2 and PaCO2, especially in hypercapnic groups.
Additionally, PaCO2 levels in previous studies were reported to be lower than those observed in this study. Razi et al. [4] reported high accuracy (R2 > 0.8) for patients with a mean of PaCO2 of 45.8 ± 17.1 mmHg in synchronized intermittent mandatory ventilation mode. In our study, the PaCO2 was hypercapnic at 63 (50−83) mmHg, with an OSI of 7.2 (4.4−8.6). As respiratory failure and significant dead space are relatively common in critically ill patients, the performance of the models estimating PaCO2 may depend on their reliability in hypercapnic and hypoxemic patient groups. Utilizing artificial intelligence, we built more customized estimations compared to the traditional risk stratification methods [13,14,15]. The XGB model achieved the highest R2 value of 0.877 among the three models, all of which demonstrated better correlations with PaCO2 than EtCO2, even in patients with hypercapnia and hypoxemia.
Nevertheless, several limitations need to be acknowledged. Firstly, utilizing PaO2 as a cutoff to identify arterial blood in blood gas analysis might have led to the inclusion of venous blood results and potential exclusion of true arterial blood samples. This limitation is inherent to retrospective studies and arises due to the lack of explicit and clear labeling. Secondly, this study was conducted within a single-institution PICU. Consequently, external validation by other institutions or utilizing different measurement equipment was performed. Thirdly, despite separating training and testing data for each patient, repeated sampling from the individuals might have resulted in overfitting. Further studies with external validation and prospective cohorts are required. At last, given that our models were developed based on data retrieved in a 1-hour window, future analyses based on real-time data would prompt the clinical application of the developed model.
One of the key elements influencing the difference between EtCO2 and PaCO2 is the heterogeneous distribution of ventilation-perfusion mismatch among alveoli [21]. Including capnographic waveforms as a variable is expected to improve model performance [22]. At last, although interpreting relevant parameters is crucial for understanding gas exchange and dead space ventilation, the “black box” nature of machine learning makes it difficult to access detailed information regarding the developed model’s mechanisms. Regarding this limitation, we tried to incorporate an explanation using SHAP values; however, it remains still challenging to provide an intuitive interpretability.
In conclusion, we successfully developed a machine learning-based model capable of non-invasively estimating PaCO2 in critically ill pediatric patients receiving mechanical ventilation. The model demonstrated acceptable performance; however, external validation has not yet been performed, and further improvement in performance through follow-up studies is promising.
Data availability
No datasets were generated or analysed during the current study.
Change history
25 April 2024
A Correction to this paper has been published: https://doi.org/10.1186/s12887-024-04772-5
Abbreviations
- ARDS:
-
Acute respiratory distress syndrome
- BP:
-
Blood pressure
- CO2 :
-
Carbon dioxide
- EtCO2 :
-
End-tidal carbon dioxide
- FiO2 :
-
Fraction of inspired oxygen
- HR:
-
Heart rate
- LOA:
-
Limit of agreement
- MAP:
-
Mean airway pressure
- MLP:
-
Multi-layer perceptron
- OSI:
-
Oxygenation saturation index
- PaCO2 :
-
Arterial partial pressure of carbon dioxide
- PaO2 :
-
Arterial partial pressure of oxygen
- PEEP:
-
Positive end-expiratory pressure
- PICU:
-
Pediatric intensive care unit
- RR:
-
Respiratory rate
- SHAP:
-
Shapley additive explanations values
- SpO2 :
-
Peripheral oxygen saturation
- TCO2 :
-
Transcutaneous carbon dioxide
- XGB:
-
Extreme gradient boosting
References
Rowling SC, Flojstrup M, Henriksen DP, Viberg B, Hallenberg C, Lindholt JS, Alberg-Flojborg A, Nanayakkara PWB, Brabrand M. Arterial blood gas analysis: as safe as we think? A multicentre historical cohort study. ERJ Open Res. 2022;8(1):00535–2021.
Siobal MS. Monitoring exhaled carbon dioxide. Respir Care. 2016;61(10):1397–416.
Wang J, Zhang J, Liu Y, Shang H, Peng L, Cui Z. Relationship between end-tidal carbon dioxide and arterial carbon dioxide in critically ill patients on mechanical ventilation: a cross-sectional study. Med (Baltim). 2021;100(33):e26973.
Razi E, Moosavi GA, Omidi K, Khakpour Saebi A, Razi A. Correlation of end-tidal carbon dioxide with arterial carbon dioxide in mechanically ventilated patients. Arch Trauma Res. 2012;1(2):58–62.
Lermuzeaux M, Meric H, Sauneuf B, Girard S, Normand H, Lofaso F, Terzi N. Superiority of transcutaneous CO2 over end-tidal CO2 measurement for monitoring respiratory failure in nonintubated patients: a pilot study. J Crit Care. 2016;31(1):150–6.
Mummery V, Rogers E, Padmanaban V, Matthew D, Woodcock T, Bloch S. Transcutaneous carbon dioxide measurement is not a reliable alternative to arterial blood gas sampling in the acute medical setting. Eur Respir J. 2019;53(4):1801726.
Lecompte-Osorio P, Pearson SD, Pieroni CH, Stutz MR, Pohlman AS, Lin J, Hall JB, Htwe YM, Belvitch PG, Dudek SM, et al. Bedside estimates of dead space using end-tidal CO(2) are independently associated with mortality in ARDS. Crit Care. 2021;25(1):333.
Siddiki H, Kojicic M, Li G, Yilmaz M, Thompson TB, Hubmayr RD, Gajic O. Bedside quantification of dead-space fraction using routine clinical data in patients with acute lung injury: secondary analysis of two prospective trials. Crit Care. 2010;14(4):R141.
Hardman JG, Aitkenhead AR. Estimation of alveolar deadspace fraction using arterial and end-tidal CO2: a factor analysis using a physiological simulation. Anaesth Intensive Care. 1999;27(5):452–8.
Doppmann P, Meuli L, Sollid SJM, Filipovic M, Knapp J, Exadaktylos A, Albrecht R, Pietsch U. End-tidal to arterial carbon dioxide gradient is associated with increased mortality in patients with traumatic brain injury: a retrospective observational study. Sci Rep. 2021;11(1):10391.
Robertson HT. Dead space: the physiology of wasted ventilation. Eur Respir J. 2015;45(6):1704–16.
McSwain SD, Hamel DS, Smith PB, Gentile MA, Srinivasan S, Meliones JN, Cheifetz IM. End-tidal and arterial carbon dioxide measurements correlate across all levels of physiologic dead space. Respir Care. 2010;55(3):288–93.
Rajpurkar P, Chen E, Banerjee O, Topol EJ. AI in health and medicine. Nat Med. 2022;28(1):31–8.
Tsai WC, Liu CF, Ma YS, Chen CJ, Lin HJ, Hsu CC, Chow JC, Chien YW, Huang CC. Real-time artificial intelligence system for bacteremia prediction in adult febrile emergency department patients. Int J Med Inf. 2023;178:105176.
Seitz KA-O, Spicer AB, Casey JA-OX, Buell KG, Qian ET, Graham Linck EJ, Driver BE, Self WH, Ginde AA, Trent SA, et al. Individualized treatment effects of Bougie versus Stylet for tracheal intubation in critical illness. Am J Respir Crit Care Med. 2023;207(12):1602–11.
Martin B, DeWitt PE, Albers D, Bennett TD. Development of a pediatric blood pressure percentile tool for clinical decision support. JAMA Netw Open. 2022;5(10):e2236918.
Yehya N, Smith L, Thomas NJ, Steffen KM, Zimmerman J, Lee JH, Erickson SJ, Shein SL, Second Pediatric Acute Lung Injury Consensus Conference of the Pediatric Acute Lung I, Sepsis Investigators N. Definition, incidence, and epidemiology of pediatric acute respiratory distress syndrome: from the second pediatric acute lung injury consensus conference. Pediatr Crit Care Med. 2023;24(12 Suppl 2):S87–S98.
Pedregosa F, Varoquaux G, Gramfort A, Michel V, Thirion B, Grisel O, Blondel M, Prettenhofer P, Weiss R, Dubourg V, et al. Scikit-learn: machine learning in python. J Mach Learn Res. 2011;12(null):2825–30.
Park DJ, Park MW, Lee H, Kim Y-J, Kim Y, Park YH. Development of machine learning model for diagnostic disease prediction based on laboratory tests. Sci Rep. 2021;11(1):7567.
Lundberg SM, Lee S-I. A unified approach to interpreting model predictions. Adv Neural Inf Process Syst. 2017;30.
Nassar BS, Schmidt GA. Capnography during critical illness. Chest. 2016;149(2):576–85.
Thompson JE, Jaffe MB. Capnographic waveforms in the mechanically ventilated patient. Respir Care. 2005;50(1):100–8. discussion 108–109.
McDonald MJ, Montgomery VL, Cerrito PB, Parrish CJ, Boland KA, Sullivan JE. Comparison of end-tidal CO2 and Paco2 in children receiving mechanical ventilation. Pediatr Crit Care Med. 2002;3(3):244–9.
Acknowledgements
Not applicable.
Funding
This study was conducted with financial support from Seoul National University Hospital (grant number: 03-2022-0370) and the National Research Foundation of Korea grant funded by the Korea government (Ministry of Science and ICT) (grant number: 2023R1A2C1007095).
Author information
Authors and Affiliations
Contributions
Conceptualization: BL. Data curation: HJH and BL. Formal analysis: HLH. Methodology: HJH, BL. Project administration: BL. Visualization: HJH. Writing – original draft: HJH. Writing – review & editing: HJH, BL, and JDP.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
This study was conducted in accordance with the principles of the Declaration of Helsinki. The protocol of this study was reviewed by the Institutional Review Board of Seoul National University Hospital. This study was recognized as a minimal risk study by the above committee because it used only pseudonymous information and did not collect personally identifiable information. Therefore, for the above reasons, the above committee waived the need for written informed consent and approved the conduct of this study (approval no. H-2307-160-1452).
Consent for publication
Not applicable as mentioned above.
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Han, HJ., Lee, B. & Park, J.D. Individualized estimation of arterial carbon dioxide partial pressure using machine learning in children receiving mechanical ventilation. BMC Pediatr 24, 149 (2024). https://doi.org/10.1186/s12887-024-04642-0
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12887-024-04642-0