Abstract
Background
Respiratory syncytial virus (RSV) is a major cause of acute lower respiratory infections in children, especially bronchiolitis. Our study aimed to identify the key genes and upstream transcription factors in RSV.
Methods
To screen for RSV pathogenic genes, an integrated analysis was performed using the RSV microarray dataset in GEO. Functional annotation and potential pathways for differentially expressed genes (DEGs) were further explored by GO and KEGG enrichment analysis. We constructed the RSV-specific transcriptional regulatory network to identify key transcription factors for DEGs in RSV.
Results
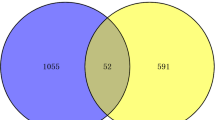
From three GEO datasets, we identified 1059 DEGs (493 up-regulated and 566 down-regulated genes, FDR < 0.05 and |Combined.ES| > 0.8) between RSV patients and normal controls. GO and KEGG analysis revealed that ‘response to virus’ (FDR = 7.13E-15), ‘mitochondrion’ (FDR = 1.39E-14) and ‘Asthma’ (FDR = 1.28E-06) were significantly enriched pathways for DEGs. The expression of IFI27, IFI44, IFITM3, FCER1A, and ISG15 were shown to be involved in the pathogenesis of RSV.
Conclusions
We concluded that IFI27, IFI44, IFITM3, FCER1A, and ISG15 may play a role in RSV. Our finding may contribute to the development of new potential biomarkers, reveal the underlying pathogenesis and also identify novel therapeutic targets for RSV.
Similar content being viewed by others
Background
Globally, respiratory syncytial virus (RSV) is the major pathogen of acute lower respiratory infections in children under 5 years old [1,2,3]. RSV also is an severe virus in adults, causing 25% of lethal respiratory infections in cold seasons, similar to the rate for seasonal influenza [4]. Morbidity caused by RSV may also be long-term, children hospitalized with RSV are more likely to suffer from asthma, sometimes for years, after acute infection [5].
Similar to other respiratory viruses, RSV infection is absorbed by airway epithelial cells, alveolar macrophages, and intraepithelial dendritic cells, induces direct antiviral responses through cytokines and chemokines, and initiates adaptive immune responses [6]. The severity of RSV infection is partly due to currently known risk factors, including medical complications and young age [7]. However, most infants hospitalized for respiratory syncytial virus infection have reportedly been previously healthy and have no risk factors for serious illness [8, 9]. Therefore, currently known risk factors do not fully explain the significant variability of the severity of the disease. Thus, it is important to find biomarkers related to the diagnosis of RSV.
In our study, we performed an integrated analysis of three gene expression datasets to identify the DEGs and transcription factors (TFs) associated with RSV. We identified the differentially expressed genes (DEGs) and TFs of RSV in this integrated analysis. Functional annotation and PPI network construction were performed to explore the biological function of DEGs. Our purpose is to provide clues to reveal the underlying mechanism of RSV and further develop potential new diagnosis and treatment for RSV.
Methods
Microarray expression profiling in GEO and identification of DEGs in RSV
The gene expression profiles of children RSV were gained from GEO database with following key search terms: (“respiratory syncytial viruses”[MeSH Terms] OR Respiratory syncytial virus [All Fields]) AND “Homo sapiens”[porgn] AND “gse”[Filter]. Datasets meet the following criteria would be included in our study: (1) selected datasets should be whole-genome mRNA expression profile by array; (2) these data were derived from blood samples of patients with RSV and normal controls; (3) datasets were normalized or original.
After downloading the selected datasets, we deleted the undetectable gene (ie, the genes whose expression value was less than 0 was more than 20% of the total sample size). There were 8834 genes in the intersection of the three datasets. For each dataset, log2 is converted to scale standardization. MetaMA was applied to obtain the DEGs. Genes with FDR < 0.05 and |Combined.ES| > 0.8 were selected as DEGs.
Functional annotation of DEGs and PPI network construction
GeneCoDis3 was employed to perform GO and KEGG pathway enrichment analysis. The threshold of FDR < 0.05 was considered as significant. Top 50 up- and down-regulated DEGs were searched with the BioGrid, and PPI network was constructed with Cytoscape software.
Construction of TF regulatory network
With UCSC Genome Bioinformatics (http://genome.ucsc.edu), the corresponding promoters of the top 20 up-regulated or down-regulated DEGs were acquired. Transcription factors (TFs) involved in regulating these DEGs were collected from the match tools in TRANSFAC. The transcriptional regulatory network was visualized by using Cytoscape software.
QRT-PCR confirmation
We collected blood samples from three RSV patients and three healthy children, and RNA samples were isolated from which to verify the expression level of candidate genes using qRT-PCR. The clinical characteristics of individuals included in this study were displayed in Table S1. We obtained the written informed consent from every participant and the approval from the ethics committee of The Affiliated Hospital of Qingdao University (QYFYW2LL25724). The human 18srRNA was used as endogenous control in analysis.
Validation in the GEO dataset and receiver operating characteristic (ROC) analysis
GSE34205, GSE38900, GSE42026 and GSE105450 were downloaded from GEO database. GSE34205 performed on GPL570, including 22 healthy controls and 51 RSV. GSE38900 performed on GPL10558, including 8 healthy controls and 28 RSV. GSE42026 performed on GPL6947, including 33 healthy controls and 22 RSV. GSE105450 performed on GPL10558, including 38 healthy controls and 89 RSV. The same data processing was performed for these four datasets as for the integration analysis. The expression levels of selected DEGs were validated with these four datasets. Then, by using pROC package in R language, we performed the ROC analysis to assess the diagnostic value of DEGs. The area under the curve (AUC) was further calculated.
Results
Differential expression analysis of genes in RSV
After filtering, a total of three datasets (GSE103842, GSE80179 and GSE77087) were retained for the analysis, the details of these three datasets were shown in the Table 1. A principal component analysis (PCA) of these three datasets was performed (Figure S1). By integrated analysis, 1059 DEGs (493 up- and 566 down-regulated) were obtained in RSV with FDR < 0.05 and |Combined.ES| > 0.8. Among them, IFI27 and MEGF6 was the most up- and down-regulated genes, respectively (Table 2). The heatmap of top 100 up- and down-regulated DEGs produced by cluster analysis is shown in Fig. 1.
Functional annotation
In Fig. 2a-c, GO enrichment revealed that the DEGs were significantly enriched in the biological processes of ‘cytokine-mediated signaling pathway’ (FDR = 1.17E-25), ‘innate immune response’ (FDR = 1.02E-15), ‘response to virus’ (FDR = 7.13E-15), the cellular components of ‘cytoplasm’ (FDR = 4.29E-61), ‘nucleus’ (FDR = 1.35E-44), ‘cytosol’ (FDR = 3.12E-41), ‘mitochondrion’ (FDR = 1.39E-14) and molecular functions of ‘protein binding’ (FDR = 3.90E-48), ‘metal ion binding’ (FDR = 4.42E-23) and ‘zinc ion binding’ (FDR = 2.34E-17). Furthermore, as shown in Fig. 2d, the results of KEGG pathway enrichment analysis revealed that DEGs were enriched in ‘Tuberculosis’ (FDR = 2.38E-09), ‘Measles’ (FDR = 2.29E-07), ‘Leishmaniasis’ (FDR = 5.62E-07) and ‘Asthma’ (FDR = 1.28E-06, Fig. 3).
PPI network construction
In Fig. 4, the PPI network consists of 229 nodes and 226 edges. Among them, the genes with higher degrees were FBXO6 (degree = 66), ISG15 (degree = 30), EIF2AK2 (degree = 19), CRY2 (degree = 13), TRAF1 (degree = 13), GLTSCR2 (degree = 9), TXN (degree = 7), TCTN1 (degree = 7), SRPK2 (degree = 7), EEF2 (degree = 6), LMNB1 (degree = 6), EIF4B (degree = 6), FBL (degree = 6), LUC7L (degree = 6), PLSCR1 (degree = 6). Among which, the three proteins of FBXO6, ISG15 and EIF2AK2 were hub proteins.
The PPI network of top 100 significantly DEGs in RSV. The green ellipses were represented the proteins encoded by down-regulated DEGs and the red ellipses were represented the proteins encoded by up-regulated DEGs. Among which, ellipses with black border were derived from the top 20 down-regulated DEGs in RSV
TF regulatory network
TF regulatory network, which consists of 103 nodes and 287 edges, involving in 64 TFs and 39 DEGs, were obtained (Fig. 5). Among them, Pax-4, 1-Oct, Nkx2–5, HNF-4, COMP1, and Pax-6 were top 6 TFs with the most downstream genes (Table 3). The highest degree of 10 target genes were EPSTI1 (degree = 16), GYG1 (degree = 14), SHISA5 (degree = 14), PPP1R13B (degree = 14), MEF2D (degree = 13), RALGPS1 (degree = 12), MEGF6 (degree = 10), FCER1A (degree = 10), LMNB1 (degree = 10), EEF2 (degree = 10).
QRT-PCR confirmation
Six genes, including IFI27, IFI44, IFITM3, FCER1A, EEF2 and ISG15, were selected to test by qRT-PCR. In Fig. 6, except for IFI27 and IFI44, the expression of IFITM3, FCER1A, EEF2 and ISG15 in qRT-PCR was consistent with our integrated analysis.
Validation in the GEO dataset and ROC analysis
The expression patterns of six DEGs, including IFI27, IFI44, IFITM3, FCER1A, EEF2 and ISG15, were verified with GSE34205, GSE38900, GSE42026 and GSE105450. As shown in Fig. 7, IFI27, IFI44, IFITM3 and ISG15 were up-regulated, and FCER1A and EEF2 were down-regulated in RSV, which were consistent with our integrated analysis.
We performed ROC curve analyses and calculated the AUC to assess the diagnostic value of these six DEGs. The AUC of these six DEGs, including IFI27 (0.935), IFI44 (0.903), IFITM3 (0.872), FCER1A (0.852), EEF2 (0.803) and ISG15 (0.889), was more than 0.80, which indicated that these six DEGs were with diagnostic value (Fig. 8).
The ROC curves of DEGs in RSV. The ROC curves were used to show the diagnostic ability of these selected DEGs with sensitivity (the proportion of true positive) and 1-specificity (the proportion of false positive). The x-axis shows 1-specificity and y-axis shows sensitivity. a IFI27, b IFI44, c IFITM3, d FCER1A, e EEF2, f ISG15
Discussion
RSV is the most common viral pathogen causing acute lower respiratory tract infections in infants, children and older people [10]. In this study, we performed an integrated analysis using data obtained from the GEO database. KEGG, GO and other biological information databases, and R analysis tools were used to analyze the DEGs. We obtained 1059 DEGs in RSV (493 genes were up-regulated, 566 genes were down-regulated). We also identified important signaling pathways that affect the pathogenesis of RSV such as ‘response to virus’ and ‘Asthma’. In addition, based on the promoter sequence of DEGs obtained from UCSC, a TF regulatory network was constructed using the match tool of the TRANFAC website to obtain the corresponding TFs.
IFI27 is a hydrophobic mitochondrial protein composed of 122 amino acid [11]. IFI27 belong to a group of small interferon stimulated genes (ISGs) [12, 13]. Rosebeck and Leaman et al. reported that IFI27 maintains a low background expression in various mammalian cells and participates in a variety of biological processes, including apoptosis and congenital immunity [14, 15]. IFI27 expression was elevated in the psoriatic lesions and uterine fibroids, ovarian cancer, and other diseases [16, 17]. It has also been shown to have a direct antiviral effect against certain viruses [18]. Hans-Olav Fjaerli et al. reported the gene IFI27 is up-regulated in whole blood of infants hospitalised with RSV [19]. According to our study, up-regulated IFI27 was among the top 20 differentially expressed mRNAs and was enriched in the GO item mitochondrion (FDR = 1.39E-14).
IFI44 is a member of the type I interferon-inducible gene family. Microtubule-associated protein 44 (IFI44) has been reported to be antiproliferative [20]. IFI44, also termed interferon-inducible protein 44 or p44 as it aggregates to form microtubular structures, is part of the type I IFN-inducible gene family. Its promoter region contains an IFN-α stimulation responsive elements, which can mediate type I IFN-inducible gene pathway [21]. Jacqueline U. McDonald et al. identified IFI44 gene serve as potential targets for future investigation in RSV disease [22]. In our study, IFI44 was up-regulated and among the top 20 differentially expressed mRNAs, which support the previous researches. Furthermore, IFI44 was enriched the GO term response to virus (FDR = 7.13E-15).
IFITM3 is a member of the interferon-inducible transmembrane protein family, which play a role in regulating antiviral signaling, inflammation, and somatogenesis [23]. In the IFITM3 knockout mouse model, IFITM3 has been reported to inhibit RSV cell infection and control the pathogenesis of the disease [24]. In our integrated analysis, IFITM3 was up-regulated and among the top 20 differentially expressed mRNAs.
The Fc fragment of IgE, a high affinity I, is a receptor for alpha polypeptides, also known as FCER1A, a protein encoded by the FCER1A gene in humans [25]. High-affinity IgE receptors play an important role in allergic diseases, coupled allergens, and mast cells, triggering inflammation and immediate allergic reactions, which are characteristic of diseases such as hay fever and asthma. Infants with severe RSV infections will subsequent develop asthma later during childhood [26]. In the KEGG analysis, the item of ‘Asthma’ (FDR = 1.28E-06) was significantly enriched and the down-regulated FCER1A was enriched in this pathway. In addition, FCER1A was among the top 20 differentially expressed mRNAs. Moreover, in the transcription factors regulation network, FCER1A (degree = 10) was among the top 10 targeted genes with high degree.
IFN-stimulated genes (ISGs) produce an antiviral state that plays an important role in determining host innate and adaptive immune responses [27]. One of the most highly induced genes in the IFN response is ISG15, which encodes a 17 kDa small UBL protein that forms a covalent conjugate with cellular proteins that mediate a large number of antiviral responses [28, 29]. Rubén González-San et al. found that ISG15 is up-regulated in respiratory pseudostratified epithelial cells and infant nasopharyngeal lavage fluids infected with RSV [30]. In our results, ISG15 was up-regulated and was the hub protein in the PPI network.
Conclusion
In conclusion, five DEGs (IFI27, IFI44, IFITM3, FCER1A, and ISG15) were identified to be involved in RSV. From the three GEO datasets analyzed, we identified 1059 DEGs (493 up-regulated and 566 down-regulated genes) between RSV and normal controls. Our findings may contribute to the elucidation of new potential biomarkers, reveal the underlying pathogenesis and identify novel therapeutic targets for the treatment of RSV. Our study also had limitations. The samples used for study was blood samples for a mucosa-limited infection disease, and no functional experiments was performed to validate the results. To confirm the exact function of the biomarkers found in this study, more samples will be collected and more in deep research on functional experiments will be included in our future work.
Availability of data and materials
The datasets used and/or analysed during the current study available from the corresponding author on reasonable request.
Abbreviations
- RSV:
-
Respiratory syncytial virus
- TF:
-
Transcription factor
- DEG:
-
Differentially expressed gene
- GEO:
-
Gene Expression Omnibus
- FDR:
-
False discovery rate
- PPI:
-
Protein-protein interaction
- UCSC:
-
University of California Santa Cruz
- ROC:
-
Receiver operating characteristic
- AUC:
-
Area under the curve
- PCA:
-
Principal component analysis
- ISG:
-
Interferon stimulated gene
References
Lee WJ, Kim YJ, Kim DW, Lee HS, Lee HY, Kim K. Complete genome sequence of human respiratory syncytial virus genotype a with a 72-nucleotide duplication in the attachment protein G gene. J Virol. 2012;86(24):13810–1.
Zhang Y, Zhuochao L. Analysis of clinical characteristics and therapy in respiratory syncytial virus acute lower respiratory infection. J Pediatr Pharmacy. 2015;21(12):15-9.
Arguedasflatts YN, Capistrán MA, Christen JA, Noyola DE. An analysis of the interaction between influenza and respiratory syncytial virus based on acute respiratory infection records. arXiv e-prints. 2013:arXiv:1312.0594.
Faghihloo E, Rezaie F, Salimi V, Naseri M, Mamishi S, Mahmoodi M, et al. Molecular epidemiology of human respiratory syncytial virus in Iran. Acta Virol. 2011;55(1):81–3.
Sigurs N, Gustafsson PM, Bjarnason R, Lundberg F, Schmidt S, Sigurbergsson F, et al. Severe respiratory syncytial virus bronchiolitis in infancy and asthma and allergy at age 13. Am J Respir Crit Care Med. 2005;171(2):137–41.
Zhang L, Peeples ME, Boucher RC, Collins PL, Pickles RJ. Respiratory syncytial virus infection of human airway epithelial cells is polarized, specific to ciliated cells, and without obvious cytopathology. J Virol. 2002;76(11):5654–66.
de Steenhuijsen Piters WA, Heinonen S, Hasrat R, Bunsow E, Smith B, Suarez-Arrabal MC, et al. Nasopharyngeal microbiota, host Transcriptome, and disease severity in children with respiratory syncytial virus infection. Am J Respir Crit Care Med. 2016;194(9):1104–15.
Boyce TG, Mellen BG, Mitchel EF Jr, Wright PF, Griffin MR. Rates of hospitalization for respiratory syncytial virus infection among children in medicaid. J Pediatr. 2000;137(6):865–70.
Garcia CG, Bhore R, Soriano-Fallas A, Trost M, Chason R, Ramilo O, et al. Risk factors in children hospitalized with RSV bronchiolitis versus non-RSV bronchiolitis. Pediatrics. 2010;126(6):e1453–60.
Madi N, Chehadeh W, Asadzadeh M, Al-Turab M, Al-Adwani A. Analysis of genetic variability of respiratory syncytial virus groups a and B in Kuwait. Arch Virol. 2018;163(9):2405–13.
Cheriyath V, Leaman DW, Borden EC. Emerging roles of FAM14 family members (G1P3/ISG 6-16 and ISG12/IFI27) in innate immunity and cancer. J Interferon Cytokine Res. 2011;31(1):173–81.
Martensen PM, Sogaard TM, Gjermandsen IM, Buttenschon HN, Rossing AB, Bonnevie-Nielsen V, et al. The interferon alpha induced protein ISG12 is localized to the nuclear membrane. Eur J Biochem. 2001;268(22):5947–54.
Parker N, Porter AC. Identification of a novel gene family that includes the interferon-inducible human genes 6-16 and ISG12. BMC Genomics. 2004;5(1):8.
Rosebeck S, Leaman DW. Mitochondrial localization and pro-apoptotic effects of the interferon-inducible protein ISG12a. Apoptosis. 2008;13(4):562–72.
Mihalich A, Vigano P, Gentilini D, Borghi MO, Vignali M, Busacca M, et al. Interferon-inducible genes, TNF-related apoptosis-inducing ligand (TRAIL) and interferon inducible protein 27 (IFI27) are negatively regulated in leiomyomas: implications for a role of the interferon pathway in leiomyoma development. Gynecol Endocrinol. 2012;28(3):216–9.
Hsieh WL, Huang YH, Wang TM, Ming YC, Tsai CN, Pang JH. IFI27, a novel epidermal growth factor-stabilized protein, is functionally involved in proliferation and cell cycling of human epidermal keratinocytes. Cell Prolif. 2015;48(2):187–97.
Li S, Xie Y, Zhang W, Gao J, Wang M, Zheng G, et al. Interferon alpha-inducible protein 27 promotes epithelial-mesenchymal transition and induces ovarian tumorigenicity and stemness. J Surg Res. 2015;193(1):255–64.
Labrada L, Liang XH, Zheng W, Johnston C, Levine B. Age-dependent resistance to lethal alphavirus encephalitis in mice: analysis of gene expression in the central nervous system and identification of a novel interferon-inducible protective gene, mouse ISG12. J Virol. 2002;76(22):11688–703.
Fjaerli HO, Bukholm G, Krog A, Skjaeret C, Holden M, Nakstad B. Whole blood gene expression in infants with respiratory syncytial virus bronchiolitis. BMC Infect Dis. 2006;6:175.
Hallen LC, Burki Y, Ebeling M, Broger C, Siegrist F, Oroszlan-Szovik K, et al. Antiproliferative activity of the human IFN-alpha-inducible protein IFI44. J Interferon Cytokine Res. 2007;27(8):675–80.
Hu JG, Fu Y, Xu JJ, Ding XP, Xie HQ, Li-Ling J. Altered gene expression profile in a rat model of gentamicin-induced ototoxicity and nephrotoxicity, and the potential role of upregulated Ifi44 expression. Mol Med Rep. 2017;16(4):4650–8.
McDonald JU, Kaforou M, Clare S, Hale C, Ivanova M, Huntley D, et al. A simple screening approach to prioritize genes for functional analysis identifies a role for interferon regulatory factor 7 in the control of respiratory syncytial virus disease. mSystems. 2016;1(3):e00051.
Bailey CC, Kondur HR, Huang IC, Farzan M. Interferon-induced transmembrane protein 3 is a type II transmembrane protein. J Biol Chem. 2013;288(45):32184–93.
Everitt AR, Clare S, McDonald JU, Kane L, Harcourt K, Ahras M, et al. Defining the range of pathogens susceptible to Ifitm3 restriction using a knockout mouse model. PLoS One. 2013;8(11):e80723.
Pang J, Taylor GR, Munroe DG, Ishaque A, Fung-Leung WP, Lau CY, et al. Characterization of the gene for the human high affinity IgE receptor (Fc epsilon RI) alpha-chain. J Immunol (Baltimore, Md : 1950). 1993;151(11):6166–74.
Knudson CJ, Varga SM. The relationship between respiratory syncytial virus and asthma. Vet Pathol. 2015;52(1):97–106.
Sledz CA, Holko M, de Veer MJ, Silverman RH, Williams BR. Activation of the interferon system by short-interfering RNAs. Nat Cell Biol. 2003;5(9):834–9.
Sadler AJ, Williams BR. Interferon-inducible antiviral effectors. Nat Rev Immunol. 2008;8(7):559–68.
Au WC, Moore PA, Lowther W, Juang YT, Pitha PM. Identification of a member of the interferon regulatory factor family that binds to the interferon-stimulated response element and activates expression of interferon-induced genes. Proc Natl Acad Sci U S A. 1995;92(25):11657–61.
Gonzalez-Sanz R, Mata M, Bermejo-Martin J, Alvarez A, Cortijo J, Melero JA, et al. ISG15 is Upregulated in respiratory syncytial virus infection and reduces virus growth through protein ISGylation. J Virol. 2016;90(7):3428–38.
Acknowledgements
Not applicable.
Funding
Not applicable.
Author information
Authors and Affiliations
Contributions
Literature search: LL, YN and ZS; Data collection: LL and ZY; Study design: YN and ZS; Analysis of data: LL and ZY; Manuscript preparation: LL and FW; Review of manuscript: FW. All authors have read and approved the manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
We obtained the written informed consent from every parent or guardian for participants under 16 years old and the approval from the ethics committee of the Affiliated Hospital of Qingdao University (QYFYW2LL25724).
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Additional file 1: Figure S1.
PCA of three datasets used in this study.
Additional file 2: Table S1.
Clinical features of patients with RSV and controls.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Li, L., Ni, Y.A., Song, Z. et al. Identification of pathogenic genes and transcription factors in respiratory syncytial virus. BMC Pediatr 21, 27 (2021). https://doi.org/10.1186/s12887-020-02480-4
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12887-020-02480-4