Abstract
Background
Cancer-testis antigens (CTAs) have emerged as potential clinical biomarkers targeting immunotherapy. KK-LC-1 is a member of CTAs, which has been demonstrated in a variety of tumors tissues and been found to elicit immune responses in cancer patients. However, the expression level and immune infiltration role of KK-LC-1 in lung adenocarcinoma (LUAD) remains to be elucidated.
Methods
In this study, the mRNA expression and overall survival rate of KK-LC-1 were evaluated by the TIMER and TCGA database in LUAD tissues and KK-LC-1 expression was further validated by clinical serum samples using quantitative RT-PCR. The relationship of KK-LC-1 with clinicopathologic parameters was analyzed. ROC curve result showed that miR-1825 was able to distinguish preoperative breast cancer patients from healthy people and postoperative patients. Then, the ROC curves were used to examine the ability of KK-LC-1 to distinguish preoperative LUAD patients from healthy and postoperative patients. The correlation between KK-LC-1 and infiltrating immune cells and immune marker sets was investigated via TIMER, TISIDB database, and CIBERSORT algorithm. The Kaplan-Meier plotter was used to further evaluate the prognostic value based on the expression levels of KK-LC-1 in related immune cells.
Results
The results showed that KK-LC-1 was significantly over-expressed in LUAD, and high levels of expression of KK-LC-1 were also closely correlated with poor overall survival. We also found that KK-LC-1 associated with TMN stage, NSE and CEA. The ROC curve result showed that KK-LC-1 was able to distinguish preoperative LUAD cancer patients from healthy people and postoperative patients. Moreover, KK-LC-1 had a larger AUC with higher diagnostic sensitivity and specificity than CEA. Based on the TIMER, TISIDB database, and CIBERSORT algorithm, the expression of KK-LC-1 was negatively correlated with CD4+ T cell, Macrophage, and Dendritic Cell in LUAD. Moreover, Based on the TIMER database, KK-LC-1 expression had a remarkable correlation with the type markers of Monocyte, TAM, M1 Macrophage, and M2 Macrophage. Furthermore, KK-LC-1 expression influenced the prognosis of LUAD patients by directly affecting immune cell infiltration by the Kaplan-Meier plotter analysis.
Conclusions
In conclusion, KK-LC-1 may serve as a promising diagnostic and prognostic biomarker in LUAD and correlate with immune infiltration and prognosis.
Similar content being viewed by others
Introduction
Lung cancer, the most common and fatal cancer in the world, causes more than 2.20 million new cases and 1.79 million patients die per year [1, 2]. Among all histological types, non-small cell lung cancer (NSCLC) makes up 80–85% of all lung cancer cases. Lung adenocarcinoma (LUAD) is the main member of NSCLC, which is the most invasive one [3]. Despite the conventional radiotherapy, chemotherapy and surgery have taken a leap forward, the five-year survival rate of NSCLC is still poor [4]. Recently, immunotherapy has emerged as an alternative treatment for lung cancer. For instance, pembrolizumab, an inhibitor of programmed death-1 (PD-1), has already been applied in the clinic [5]. However, the clinical results were still unsatisfactory because of drug resistance and adverse reactions [4]. Therefore, it is urgent to explore a reliable prognostic biomarker that can predict the prognosis of LUAD and improve the immunotherapy of patients.
Cancer-testis antigens (CTAs) may be suitable targets for cancer immunotherapy due to their immune-privileged properties. CTAs expressed restrictively in normal tissue except for testicular germ cells and various tumour types, including epithelial ovarian cancer, lung cancer, and cervical carcinoma [6,7,8]. Many reports found that CTAs were remarkedly correlated with the oncogenesis, metastasis, and unfavorable prognosis of tumors [9]. CTAs have strong immunogenicity and induce humoral immunity in several types of cancers [10, 11]. Kita-kyushu lung cancer antigen 1 (KK-LC-1), called CT83 or CXORF61, is a CTA that has epitope peptides recognised by cytotoxic T lymphocytes (CTLs) [12]. Fukuyama et al. first reported it in LUAD in 2006, which consisted of 556 base pair and located in chromosome Xq22 [12]. In recent years, the researches of KK-LC-1 mainly focused on the change of expression in different cancer tissues, especially gastric cancer, and the different therapeutic strategies such as photodynamic therapy and vaccine [13,14,15,16,17]. However, the expression level of KK-LC-1 in LUAD serum samples and the relationship between the expression level of KK-LC-1 with immune infiltration remain to be elucidated.
In this study, we analyzed the KK-LC-1 expression by using The Cancer Genome Atlas (TCGA) database. Quantitative real-time polymerase chain reaction (qRT-PCR) was used to further confirm KK-LC-1 expression in LUAD serum samples. Then, we explored the prognostic value of KK-LC-1 by plotting the survival curve and using the Kaplan-Meier plotter. In addition, we estimated the association between KK-LC-1 expression and tumor-infiltrating immune cells by using the Tumor Immune Estimation Resource (TIMER), CIBERSORT, and TISIDB database. Our results indicated that KK-LC-1 plays a non-redundant role in LUAD and is related to immune response.
Materials and methods
Clinical samples
The serum samples of 74 healthy individuals and 92 LUAD patients were obtained from Fujian Provincial Hospital (Fuzhou, China). All LUAD patients were diagnosed through histopathology without chemotherapy and radiotherapy before surgery. Then, we collected serum from 19 patients before and on the 5th day after surgery so that the serum samples were obtained preoperatively and postoperatively in pair. The clinicopathological data of all LUAD patients were recorded, including age, gender, tumor size, TNM stage etc. This study was approved by the Research Ethics Committee of Fujian Provincial Hospital (K-2021-040-04) and conformed to the ethical standards of the 1964 Helsinki Declaration, and all subjects and voluntarily signed informed consent forms.
Quantitative real-time polymerase chain reaction (qRT-PCR)
We first extracted total RNA from serum samples according to TRIzolTM LS Reagent instructions. Then, the RNA was reversely transcribed into cDNA under 37 °C for 15 min, followed by 85 °C for 5 s. CDNA was finally amplified by TB Green® Premix Ex Taq™ II Kit (RR820A; TAKARA, Tokyo, Japan) manufacturer’s instruction in 40 cycles of denaturation at 95 °C for 10 min, followed by 95 °C for 15 s, with extension at 60 °C for 1 min using LightCycler 480 System. qRT-PCR’s primer sequences were as follows:
-
GAPDH,Forward:GGCCTCCAAGGAGTAAGACC, Reverse:AAGGGGAGATTCAGTGTGGTG;
-
KK-LC-1,Forward:ATGAACTTCTATTTACTCCTAGCGAGC,
-
Reverse: CTACAATATTGAGTGTGGGAAATTATTTAA.
The relative expression levels of KK-LC-1 was calculated by 2-ΔΔCT method.
TIMER database
TIMER database (https://cistrome.shinyapps.io/timer/) is a comprehensive online tool, which can not only systematically evaluate immune infiltrates of different cancers, but also compare the differential expression between cancer and normal tissue [18]. In present study, we used the TIMER database to explore the difference KK-LC-1 expression between various cancers and adjacent normal samples. Then, we analyzed the relationship between KK-LC-1 and immune cell infiltration in LUAD. Moreover, the relationship between immune cell infiltration and corresponding gene markers was analyzed.
TCGA database
The gene expression profiles were downloaded from the TCGA database (http://portal.gdc.cancer.gov/). We analyzed the expression of KK-LC-1 mRNA between LUAD and adjacent para-cancerous lung tissues. In addition, the association between LUAD and matched normal tissues was also further validated by TCGA.
Kaplan-Meier potter analysis
The association between 54,675 genes expression and survival from 21 tumor types can be assessed by Kaplan-Meier potter (http://kmplot.com) [19]. We analyzed the prognosis value of KK-LC-1 expression in LUAD. Besides, prognosis of KK-LC-1 expression based on various immune cell were evaluated by Kaplan–Meier plotter. Log-rank P-values (P < 0.05) and hazard ratio (HR) with 95% confidence intervals were computed.
TISIDB database analysis
The TISIDB (http://cis.hku.hk/TISIDB) database is high-throughput screening techniques, molecular profiling, and para-cancerous multiomic data, as well as various resources for immunological data obtained from seven public databases [20]. TISIDB enables analysis of associations between KK-LC-1 and tumor-infiltrating immune cells.
CIBERSORT algorithm
CIBERSORT (https://www.biostars.org/p/428905/) is an analytical tool, which aids in evaluating the abundances of member cell types in a mixed cell population through gene expression data [21]. We explored the relationship between high and low expression of KK-LC-1 and 22 types of tumor-infiltrating immune cells using CIBERSORT algorithm.
Statistical analysis
R software package was used to analyze TCGA data after download. Statistical analysis was processed by the Statistical Program for Social Sciences (SPSS) 22.0 software (SPSS, Chicago, IL) and GraphPad Prism 5.0 (GraphPad Software, La Jolla, CA). The qRT-PCR data were analyzed using the unpaired Student’s t-test or paired t-test. The Receiver operating characteristic (ROC) curve, and the area under the curve (AUC) were performed to evaluate the diagnostic value of KK-LC-1. The Kaplan-Meier plotter was employed to generate survival curves. Gene expression corrections were performed in the TIMER databases by Spearman’s correlation analysis with the hazard ratio (HR) and P-values or Cox P-values. P < 0.05 was statistically significant.
Results
The mRNA expression of KK-LC-1 in pan-carcinoma and associated with the prognosis of LUAD
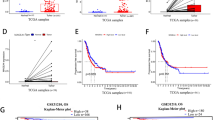
We firstly analyzed KK-LC-1 expression at the mRNA level, and then explored its prognosis value in patients with LUAD. The differential mRNA expression of KK-LC-1 between diverse tumor tissues and matched para-cancerous tissues was analyzed by TIMER database. KK-LC-1 expression was significantly higher in tumor tissue than matched para-cancerous tissues, including LUAD (Fig. 1A). In a dataset from TCGA, the expression level of KK-LC-1 was remarkably increased in 538 LUAD tissues compared with 59 normal tissues (Fig. 1B), and the result was consistent with the 59 matched tissue samples from the LUAD patients (Fig. 1C). In addition, we performed a survival analysis in LUAD, which revealed that the higher expression of KK-LC-1 was correlated with a poorer prognosis (Fig. 1D).
KK-LC-1 expression in various human cancers and related to prognosis in LUAD. A Human KK-LC-1 expression in various tumors according to the TIMER database. B The expression levels of KK-LC-1 in LUAD and para-cancerous lung tissues. C KK-LC-1 expression in LUAD and matched normal tissues by TCGA database. D The overall survival rate of KK-LC-1 expression in LUAD
The mRNA expression of KK-LC-1 in serum and associated with the diagnosis of LUAD
Next, we evaluated the diagnostic significance of KK-LC-1 in the serum of LUAD patients. The expression level of KK-LC-1 in 92 LUAD patient’s serum was higher than 47 healthy control by qRT-PCR (Fig. 2A). Then, the relationship between the clinicopathological features of LUAD samples and the expression level of KK-LC-1 was analyzed. As shown in Table 1, KK-LC-1 expression was correlated with the stage, Carcino-Embryonic Antigen (CEA), and Neuron-Specific Enolase (NSE) (P < 0.05). The receiver operating characteristic (ROC) curves and the area under curve (AUC) analyses were performed for the diagnostic role of KK-LC-1. Compared with CEA, KK-LC-1 had a higher AUC (0.794 VS. 0.565) (Fig. 2B, C). Interestingly, the expression level of KK-LC-1 in 19 LUAD patients’ serum was significantly decreased than that in patients after operation (Fig. 2D), and KK-LC-1 had a good ability to discriminate distinguish preoperative LUAD cancer patients from and postoperative patients with an AUC of 0.720 (Fig. 2E).
KK-LC-1 expression in serum and related to diagnosis in LUAD. A The expression level of KK-LC-1in LUAD serum and healthy person’s serum. ROC curve of serum B KK-LC-1 and C CEA. D KK-LC-1 expression in LUAD patient’s preoperative and postoperative serum. E ROC curve of serum KK-LC-1 to validate pre-operative cases from post-operative in LUAD
Association between KK-LC-1 expression and infiltration levels of immune cells in LUAD
We then evaluated the correlation of KK-LC-1 expression with the infiltration levels of immune cells in LUAD based on TIMER database. As shown in Fig. 3A, the expression level of KK-LC-1 was significantly negatively correlated with B cell (r = − 0.131, p = 3.81e–03), CD4+ T cell (r = − 0.122, p = 7.07e–03), macrophage (r = − 0.131, p = 3.85e–03), neutrophil (r = − 0.099, p = 3.04e–02), and dendritic cell (r = − 0.18, p = 6.48e–05). However, there was no significant correlation between KK-LC-1 and tumor purity (r = − 0.043, p = 3.36e–01) and CD8+ T cell (r = − 0.04, p = 3.78e–01) (Fig. 3A). This negative correlation between the expression level of KK-LC-1 and CD4+ T cell (r = − 0.116, p = 8.29e–03), macrophage (r = − 0.1, p = 2.34e–02), Natural killer cell (r = − 0.123, p = 5.13e–03), and dendritic cell (r = − 0.143, p = 1.15e–03) was found in TISIDB database (Fig. 3B). Next, we further explored the difference between infiltrating immune cells and KK-LC-1 expression by CIBERSORT. The results indicated the higher KK-LC-1 expression related to the higher immune cells infiltration in plasma cell, activated CD4+ memory T cell, follicular helper T cell, macrophages M0, while monocyte, macrophages M2, resting dendritic cell, resting mast cell had opposite result (Fig. 3C).
Association of KK-LC-1 expression with immune infiltration in LUAD according to the TIMER database. A The correlation between KKLC and immune cells by TIMER. B The correlation between KKLC and immune cells by TISIDB. C 22 types of tumor-infiltrating immune cells were analyzed in high and low KK-LC-1 expression groups
Association between KK-LC-1 and immune cell type markers
TIMER was used to further investigate the correlation between KK-LC-1 and immune cell markers. After the adjustment for purity, we focused on the immune cell markers of B Cell, T Cell, CD8 + T Cell, Monocyte, M1 Macrophage, M2 Macrophage, TAM, Neutrophil, Natural killer cell, Dendritic cell, and functional T cell markers of Th1, Th2, Tfh, Th17, Treg, T cell exhaustion (Table 2). As shown in Table 2 and Fig. 4, KK-LC-1 expression had a remarkable correlation with CD68 and CSF1R of Monocyte (P < 0.05), CD68 and CCL2 of TAM (P < 0.05), NOS2 of M1 Macrophage (P < 0.05), CD163, VSIG4 and MS4A4A of M2 Macrophage (P < 0.05). Taken together, KK-LC-1 may be participated in the LUAD immune response by regulating the immune cells.
Prognostic value analysis of KK-LC-1 based on immune cells in LUAD
Finally, we performed prognostic analyses based on KK-LC-1 expression in LUAD in different immune cell subgroups. High expression of KK-LC-1 in decreased macrophages cohort in LUAD was associated with better prognosis (Fig. 5H). Meanwhile, high expression of KK-LC-1 in enriched B cells, enriched CD8+ T cells, enriched macrophages, enriched/decreased CD4+ T cells, enriched/decreased natural killer T cells, enriched/decreased regulatory T cells, decreased type 1 T helper cells and enriched/decreased type 2 T helper cells cohorts in LUAD was associated with poor prognosis (Fig. 5A, C-E, G, I-L, N-P). Besides, there was no significant association between low/high KK-LC-1 expression and LUAD patient prognosis in decreased B cells and decreased CD8+ T cells, enriched type 1 T helper cells cohorts (Fig. 5B, F, M). The result suggested that KK-LC-1 expression influenced the prognosis of LUAD patients by directly affecting immune cell infiltration.
Discussion
CTAs referred to as CT antigen is expressed only in testis and embryonic primordial cells, but they can also be abnormally expressed in malignant tumors. Previous researches showed that CTAs were involved in the occurrence and development of tumors [22]. CTA can be used as an immunotherapy target mediated by cytotoxicity T lymphocyte [23]. Some CTAs were identified to have immunogenicity, indicating the possibility of tumor immunotherapy targets [24,25,26]. In previous studies, compared with NY-ESO-1 (10.5%), which has been used in clinical immunotherapy, KK-LC-1 (32.6%) has a higher expression level in NSCLC [27]. KK-LC-1 also has been reported that be highly expressed in lung cancer, gastric cancer, triple-negative breast cancer (TNBC), and hepatocellular carcinoma (HCC) [12, 13, 28, 29]. In this study, we determined the database-derived tissues and clinic-derived serum expression levels of KK-LC-1 in LUAD patients, and analyzed the correlation of KK-LC-1 with immune cell infiltration of LUAD.
When KK-LC-1 was first discovered, it was positively expressed in 50% of 15 lung cancer cell lines and 38% of LUAD tissues [12]. Hsu et al. reported that KK-LC-1 expression level was higher in LUAD than LUSC [30]. The expression of KK-LC-1 was intimately related to tumor stage and lymph node metastasis in lung cancer patients [31]. Our results from TIMER and TCGA databases showed that KK-LC-1 expression was up-regulated in LUAD tissues compared with normal tissues. Meanwhile, high levels of expression of KK-LC-1 were also closely correlated with poor overall survival, suggesting that KK-LC-1 may be a promising biomarker for survival prediction in LUAD. The above results only focus on the expression of KK-LC in tissue and cells but not in serum or plasma. As we all known, compared with tissue samples, serum has the advantages of being less invasive, easy to obtain and repeatable. In our study, it was the first time to analyze KK-LC-1 expression in serum from LUAD patients. As expected, serum KK-LC-1 expression was elevated in serum from LUAD patients, and was significantly correlated with TNM stage, CEA and NSE levels. In addition, serum KK-LC-1 could be used to differentiate LUAD patients from healthy controls. Moreover, serum KK-LC-1 expression was remarkably decreased after surgery, indicating serum KK-LC-1 was closely correlated with tumor occupying. These findings implied that serum KK-LC-1 could serve as a tumor marker for a diagnostic and prognostic predictor in LUAD patients.
Paret et al. demonstrated that KK-LC-1 could induce a strong antigen-specific immune response in TNBC based on specific recognition of TCR epitopes through vitro and vivo experiments [32]. Marcinkowski et al. also reported that KK-LC-1 has great potential in T cell receptor (TCR) gene-engineered T cells therapy for gastric cancer [33]. These evidences suggested that KK-LC-1 might be a progressing immunotherapy target. In our study, we found that KK-LC-1 was negative correlated with immune cell infiltration CD4+ T cell, macrophage, neutrophil and dendritic cell, which was further validated by immune cell surface markers. It is well known that CD4+ T cell, macrophage, neutrophil and dendritic cell play antitumor roles in cancers [34,35,36]. The negative correlation between KK-LC-1 and immune cells further indicated that KK-LC-1 seemed to play a important role in promoting cancer. At the same time, KK-LC-1 directly affected patient outcomes by influencing immune cell infiltration, which was similar with the research by Yoshinobu Ichiki et al. in LUSC [14]. Hsu et al. also reported that KK-LC-1 was related to the abundance of macrophages and CD4 + T cells through QuantiSeq algorithm in lung cancer [30]. Our results showed that KK-LC-1 expression level was strongly correlated with lung cancer-related immune cells infiltration, and KK-LC-1 affected the prognosis of LUAD by modulating the infiltration level of tumor infiltrating immune cells (TIICs).
However, the research on relevant mechanisms is limited in our study. In fact, to date, there are few reports on the mechanism of KK-LC-1 in human malignant tumors. Methylated CpG islands associated with the CT genes in normal somatic cells become demethylated in cancer cells, indicating activation of their expression. Xie et al. reported that PIWIL1 is considered to be a highly expressed CT gene in LUAD, and promoter DNA hypomethylation of PIWIL1 could contribute to its aberrant expression in LUAD [37]. KK-LC-1 expression is activated by treatment with the hypomethylating agent 5-aza-2′-deoxycytidine in KK-LC-1 negative breast cancer cell lines [15, 32].
To sum up, the high expression level of serum KK-LC-1 in patients with LUAD was closely related to TNM stage, CEA and NSE. The expression of serum KK-LC-1 mRNA decreased significantly after surgery. KK-LC-1 expression has a strong correlation with lung cancer-related immune cells infiltration, which can affect the prognosis of LUAD. Our findings suggest that KK-LC-1 played a non-redundant role in tumor immunology and served as a diagnostic biomarker in LUAD. However, it needs a further study to verify the above results with large sample size, and to explore the mechanisms and immunoregulatory functions of KK-LC-1 in LUAD.
Conclusions
KK-LC-1 may serve as a promising diagnostic and prognostic biomarker in LUAD and is corelated with immune infiltration and prognosis.
Availability of data and materials
The data, which was used in this study can be found at the following websites and thoracic surgery, Fujian Provincial Hospital. All methods were carried out in accordance with relevant guidelines and regulations. The differentially expressed in Pan-cancer was analyzed by TIMER (https://cistrome.shinyapps.io/timer/) and TCGA databases (http://portal.gdc.cancer.gov/). The Kaplan-Meier potter (http://kmplot.com) was utilized to evaluated prognosis. The association between KK-LC-1 and immune infiltration was explored by TIMER and TISIDB database (http://cis.hku.hk/TISIDB), and CIBERSORT (https://www.biostars.org/p/428905/) algorithm. The datasets used during the current study are available from the corresponding author on reasonable request.
Abbreviations
- CTAs:
-
Cancer-testis antigens
- KK-LC-1:
-
Kita-kyushu lung cancer antigen 1
- NY-ESO-1:
-
New York esophageal squamous cell carcinoma 1
- NSCLC:
-
Non-small cell lung cancer
- LUAD:
-
Lung adenocarcinoma
- LUSC:
-
Lung squamous cell carcinoma
- BRCA:
-
Breast invasive carcinoma
- ESCA:
-
Esophageal carcinoma
- PAAD:
-
Pancreatic adenocarcinoma
- STAD:
-
Stomach adenocarcinoma
- TGCT:
-
Testicular germ cell tumors
- TNBC:
-
Triple negative breast cancer
- HCC:
-
Hepatocellular carcinoma
- PD-1:
-
Programmed death-1
- CEA:
-
Carcino-Embryonic Antigen
- NSE:
-
Neuron-Specific Enolase
- CTL:
-
Cytotoxic T lymphocytes
- TCR:
-
T Cell receptor
- qRT-PCR:
-
Quantitative real-time polymerase chain reaction
- TIICs:
-
Tumor infiltrating immune cells
- TCGA:
-
The Cancer Genome Atlas
- TIMER:
-
Tumor Immune Estimation Resource
- TISIDB:
-
An Integrated Repository Portal For Tumor-Immune System Interactions
- ROC:
-
Receiver operating characteristic
- AUC:
-
Area under the curve
- OS:
-
Overall survival
- HR:
-
Hazard Ratio
- SPSS:
-
Statistical Program for Social Sciences
References
Siegel RL, Miller KD, Fuchs HE, Jemal A. Cancer statistics, 2021. CA Cancer J Clin. 2021;71(1):7–33.
Thai AA, Solomon BJ, Sequist LV, Gainor JF, Heist RS. Lung cancer. Lancet. 2021;398(10299):535–54.
Herbst RS, Morgensztern D, Boshoff C. The biology and management of non-small cell lung cancer. Nature. 2018;553(7689):446–54.
Denisenko TV, Budkevich IN, Zhivotovsky B. Cell death-based treatment of lung adenocarcinoma. Cell Death Dis. 2018;9(2):117.
Mok TSK, Wu YL, Kudaba I, Kowalski DM, Cho BC, Turna HZ, et al. Pembrolizumab versus chemotherapy for previously untreated, PD-L1-expressing, locally advanced or metastatic non-small-cell lung cancer (KEYNOTE-042): a randomised, open-label, controlled, phase 3 trial. Lancet. 2019;393(10183):1819–30.
Taherian-Esfahani Z, Dashti S. Cancer-testis antigens: an update on their roles in cancer immunotherapy. Hum Antibodies. 2019;27(3):171–83.
Garg M, Chaurasiya D, Rana R, Jagadish N, Kanojia D, Dudha N, et al. Sperm-associated antigen 9, a novel cancer testis antigen, is a potential target for immunotherapy in epithelial ovarian cancer. Clin Cancer Res. 2007;13(5):1421–8.
Garg M, Kanojia D, Salhan S, Suri S, Gupta A, Lohiya NK, et al. Sperm-associated antigen 9 is a biomarker for early cervical carcinoma. Cancer. 2009;115(12):2671–83.
Whitehurst AW. Cause and consequence of cancer/testis antigen activation in cancer. Annu Rev Pharmacol Toxicol. 2014;54:251–72.
Chen Z, Li M, Yuan Y, Wang Q, Yan L, Gu J. Cancer/testis antigens and clinical risk factors for liver metastasis of colorectal cancer: a predictive panel. Dis Colon Rectum. 2010;53(1):31–8.
Astaneh M, Dashti S, Esfahani ZT. Humoral immune responses against cancer-testis antigens in human malignancies. Hum Antibodies. 2019;27(4):237–40.
Fukuyama T, Hanagiri T, Takenoyama M, Ichiki Y, Mizukami M, So T, et al. Identification of a new cancer/germline gene, KK-LC-1, encoding an antigen recognized by autologous CTL induced on human lung adenocarcinoma. Cancer Res. 2006;66(9):4922–8.
Ji J, Chen J, Wang A, Zhang W, Ju H, Liu Y, et al. KK-LC-1 may be an effective prognostic biomarker for gastric cancer. BMC Cancer. 2021;21(1):267.
Ichiki Y, Fukuyama T, Ohmiya H, Ueno M, Yanagi S, Kanasaki Y, et al. Relationship between Kita-Kyushu lung Cancer antigen-1 expression and prognosis of cases with lung squamous cell carcinoma. Transl Cancer Res. 2021;10(12):5212–21.
Chen C, Gao D, Huo J, Qu R, Guo Y, Hu X, et al. Multiomics analysis reveals CT83 is the most specific gene for triple negative breast cancer and its hypomethylation is oncogenic in breast cancer. Sci Rep. 2021;11(1):12172.
Herrera LRM. Reverse Vaccinology approach in constructing a multi-epitope vaccine against Cancer-testis antigens expressed in non-small cell lung Cancer. Asian Pac J Cancer Prev. 2021;22(5):1495–506.
Ye Z, Liang Y, Ma Y, Lin B, Cao L, Wang B, et al. Targeted photodynamic therapy of cancer using a novel gallium (III) tris (ethoxycarbonyl) corrole conjugated-mAb directed against cancer/testis antigens 83. Cancer Med. 2018.
Li T, Fan J, Wang B, Traugh N, Chen Q, Liu JS, et al. TIMER: a web server for comprehensive analysis of tumor-infiltrating immune cells. Cancer Res. 2017;77(21):e108–10.
Lanczky A, Nagy A, Bottai G, Munkacsy G, Szabo A, Santarpia L, et al. miRpower: a web-tool to validate survival-associated miRNAs utilizing expression data from 2178 breast cancer patients. Breast Cancer Res Treat. 2016;160(3):439–46.
Edgar R, Domrachev M, Lash AE. Gene expression omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002;30(1):207–10.
Le T, Aronow RA, Kirshtein A, Shahriyari L. A review of digital cytometry methods: estimating the relative abundance of cell types in a bulk of cells. Brief Bioinform. 2021;22(4).
Chen L, Wu Q, Xu X, Yang C, You J, Chen F, et al. Cancer/testis antigen LDHC promotes proliferation and metastasis by activating the PI3K/Akt/GSK-3beta-signaling pathway and the in lung adenocarcinoma. Exp Cell Res. 2021;398(2):112414.
Grizzi F, Mirandola L, Qehajaj D, Cobos E, Figueroa JA, Chiriva-Internati M. Cancer-testis antigens and immunotherapy in the light of cancer complexity. Int Rev Immunol. 2015;34(2):143–53.
Liu X, Xu Y, Xiong W, Yin B, Huang Y, Chu J, et al. Development of a TCR-like antibody and chimeric antigen receptor against NY-ESO-1/HLA-A2 for cancer immunotherapy. J Immunother Cancer. 2022;10(3).
Kim YR, Kim KU, Lee JH, Kim DW, Chung JH, Kim YD, et al. Cancer testis antigen, NOL4, is an immunogenic antigen specifically expressed in small-cell lung Cancer. Curr Oncol. 2021;28(3):1927–37.
Zhang Y, Yu X, Liu Q, Gong H, Chen AA, Zheng H, et al. SAGE1: a potential target antigen for lung Cancer T-cell immunotherapy. Mol Cancer Ther. 2021;20(11):2302–13.
Thomas R, Al-Khadairi G, Roelands J, Hendrickx W, Dermime S, Bedognetti D, et al. NY-ESO-1 based immunotherapy of Cancer: current perspectives. Front Immunol. 2018;9:947.
Kondo Y, Fukuyama T, Yamamura R, Futawatari N, Ichiki Y, Tanaka Y, et al. Detection of KK-LC-1 protein, a Cancer/testis antigen, in Patients with Breast Cancer. Anticancer Res. 2018;38(10):5923–8.
Chen Z, Zuo X, Pu L, Zhang Y, Han G, Zhang L, et al. Hypomethylation-mediated activation of cancer/testis antigen KK-LC-1 facilitates hepatocellular carcinoma progression through activating the Notch1/Hes1 signalling. Cell Prolif. 2019;52(3):e12581.
Hsu R, Baca Y, Xiu J, Wang R, Bodor JN, Kim C, et al. Molecular characterization of Kita-Kyushu lung cancer antigen (KK-LC-1) expressing carcinomas. Oncotarget. 2021;12(25):2449–58.
Jin S, Cao S, Li J, Meng Q, Wang C, Yao L, et al. Cancer/testis antigens (CTAs) expression in resected lung cancer. Onco Targets Ther. 2018;11:4491–9.
Paret C, Simon P, Vormbrock K, Bender C, Kolsch A, Breitkreuz A, et al. CXorf61 is a target for T cell based immunotherapy of triple-negative breast cancer. Oncotarget. 2015;6(28):25356–67.
Marcinkowski B, Stevanovic S, Helman SR, Norberg SM, Serna C, Jin B, et al. Cancer targeting by TCR gene-engineered T cells directed against Kita-Kyushu lung Cancer Antigen-1. J Immunother Cancer. 2019;7(1):229.
Waniczek D, Lorenc Z, Snietura M, Wesecki M, Kopec A, Muc-Wierzgon M. Tumor-associated macrophages and regulatory T cells infiltration and the clinical outcome in colorectal Cancer. Arch Immunol Ther Exp. 2017;65(5):445–54.
Farhood B, Najafi M, Mortezaee K. CD8(+) cytotoxic T lymphocytes in cancer immunotherapy: a review. J Cell Physiol. 2019;234(6):8509–21.
Zhang H, Liu H, Shen Z, Lin C, Wang X, Qin J, et al. Tumor-infiltrating neutrophils is prognostic and predictive for postoperative adjuvant chemotherapy benefit in patients with gastric Cancer. Ann Surg. 2018;267(2):311–8.
Xie K, Zhang K, Kong J, Wang C, Gu Y, Liang C, et al. Cancer-testis gene PIWIL1 promotes cell proliferation, migration, and invasion in lung adenocarcinoma. Cancer Med. 2018;7(1):157–66.
Acknowledgements
We thank the Fujian Provincial hospital for providing experimental support for this study.
Funding
This work was supported by the Medical Innovation Grant of Fujian Province (No.2019-CXB-1); Fujian Natural Science Foundation of China (No. 2021 J1375).
Author information
Authors and Affiliations
Contributions
Yanli Kang and Yuhan Gan performed the bioinformatics analysis, drafted the manuscript and prepared the figures. Yingfeng Jiang, Jianbin You, Chen Huang, Qianshun Chen and Xunyu Xu collected the related references and participated in discussion. Liangyuan Chen and Falin Chen made substantial contributions to conception and design of the research. All authors contributed to this manuscript. The author(s) read and approved the final manuscript.
Corresponding authors
Ethics declarations
Ethics approval and consent to participate
This study was approved by the Research Ethics Committee of Fujian Provincial Hospital (K-2021-040-04) and conformed to the ethical standards of the 1964 Helsinki Declaration, and all subjects and voluntarily signed informed consent forms.
Consent for publication
Not applicable.
Competing interests
The authors declare that have no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Kang, Y., Gan, Y., Jiang, Y. et al. Cancer-testis antigen KK-LC-1 is a potential biomarker associated with immune cell infiltration in lung adenocarcinoma. BMC Cancer 22, 834 (2022). https://doi.org/10.1186/s12885-022-09930-5
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12885-022-09930-5