Abstract
Background
Bladder cancer (BCa) shows its potential immunogenity in current immune-checkpoint inhibitor related immunotherapies. However, its therapeutic effects are improvable and could be affected by tumor immune microenvironment. Hence it is interesting to find some more prognostic indicators for BCa patients concerning immunotherapies.
Methods
In the present study, we retrospect 129 muscle-invasive BCa (MIBC) patients with radical cystectomy in Shanghai General Hospital during 2007 to 2018. Based on the results of proteomics sequencing from 9 pairs of MIBC tissue from Shanghai General Hospital, we focused on 13 immune-related differential expression proteins and their related genes. An immune-related prognostic signature (IRPS) was constructed according to Cancer Genome Atlas (TCGA) dataset. The IRPS was verified in ArrayExpress (E-MTAB-4321) cohort and Shanghai General Hospital (General) cohort, separately. A total of 1010 BCa patients were involved in the study, including 405 BCa patients in TCGA cohort, 476 BCa patients in E-MTAB-4321 cohort and 129 MIBC patients in General cohort.
Result
It can be indicated that high IRPS score was related to poor 5-year overall survival and disease-free survival. The IRPS score was also evaluated its immune infiltration. We found that the IRPS score was adversely associated with GZMB, IFN-γ, PD-1, PD-L1. Additionally, higher IRPS score was significantly associated with more M2 macrophage and resting mast cell infiltration.
Conclusion
The study revealed a novel BCa prognostic signature based on IRPS score, which may be useful for BCa immunotherapies.
Similar content being viewed by others
Introduction
Bladder cancer (BCa) is the fourth most common cancer in men, with estimated 81,400 new incidence and 17,980 deaths in the United States in 2020 [1]. Currently, BCa can be divisible into non-muscle-invasive BCa (NMIBC) and muscle-invasive BCa (MIBC), with diverse clinical outcomes [2,3,4]. The 5-year overall survival of NMIBC patients is over 99% whereas that of MIBC patients drops down dramatically [3, 4]. It is about 50% of MIBC patients that still appear local recurrence or distant metastasis after radical cystectomy and chemotherapy [4, 5]. Classical and traditional prognostic indicators for BCa patients are mainly TNM staging and pathological differentiation [6]. Nevertheless, these parameters may be insufficient for current clinicians’ demands [7, 8]. Hence, it becomes necessary to find some more practical prognostic signatures besides traditional TNM staging and histopathological features [6, 9].
Through the process of malignancy proliferation, cancer cells gradually accelerate the escape from the monitor of normal immune system, resulting in progression and metastasis [10, 11]. Inhibitory immune cells are recruited by cancer cells in the tumor microenvironment to suppress the anti-tumor immune response, which are caused by immune checkpoints and some other cytokines [10, 12, 13]. Currently, some immune-checkpoint inhibitors targeting programmed death-1 (PD-1) and programmed death-ligand 1 (PD-L1) are approved by Food and Drug Administration (FDA) to treat locally advanced or metastatic platinum-ineligible MIBC patients [14]. However, these immune-checkpoint inhibitor targeted drugs can only benefit limited MIBC patients [15,16,17]. Moreover, around 10% MIBC patients treated with immune-checkpoint inhibitors may have hyper-progressive disease [18, 19]. The PD-1/PD-L1 related immunotherapies for MIBC patients are still not satisfactory [20].
In this study, based on the results of proteomics sequencing from 9 pairs of MIBC patient samples, we focused on the immune-related differential proteins with prognostic values. The immune-related prognostic signature (IRPS) was constructed based on Cancer Genome Atlas (TCGA) cohort and verified by ArrayExpress (E-MTAB-4321) cohort and Shanghai General Hospital (General) cohort. A total of 1010 BCa patients were involved in the study. High IRPS score was also correlated with the remodeling of the immune microenvironment.
Materials and methods
Patient samples and ethics statement
In General cohort, totally 129 MIBC patient samples were included in the study. All the involved patients undergone radical cystectomy in Shanghai General Hospital during 2007 to 2018. Patients with concurrent other cancers, autoimmune diseases or HIV infections were excluded in the study. The study was conducted according to the Helsinki Declaration and was approved by the Human Ethics Committee of Shanghai General Hospital. The clinicopathological features of the 129 MIBC patients in General cohort were shown in Table 1.
In addition, a total of 405 BCa patient samples from TCGA dataset and 476 BCa patient samples from E-MTAB-4321 dataset were used in the study [21]. All the enrolled sample data were with complete gene expression and clinical data.
Proteomics sequencing
A total of 9 pairs of MIBC samples from Shanghai General Hospital were conducted liquid chromatography-mass spectrometry (LC–MS). Each pair included BCa and normal bladder tissue from each individual patient. All the 9 pairs of samples were classified into two groups based on survival. The 3 MIBC patients were survived for 5 years and were considered as good prognosis. The 6 MIBC patients, regarded as poor prognosis, were passed away. All the 9 male MIBC patients were at BCa stage of T2N0M0 and among 62 to 70 years old. The abovementioned MIBC patients were from the institutional cohort (General cohort, n = 129, after excluding the 9 MIBC samples, which were used for LC/MS analysis). Each formalin-fixed and paraffin-embedded sample was used the amount of 10 consecutive section slides with 10 μm thickness. Afterwards, paraffin was removed by xylene, heptane and methanol washes. Samples were treated by 7 M urea, 2 M thiourea, 1 M ammoniumbicarbonate as well as protease and phosphatase inhibitor cocktails. Then samples were heated to 95 °C for 30 min and incubation at 60 °C for 2 h, followed by sonication and centrifugation. Protein concentration was determined by Bradford assay (Bio-Rad). Peptides were desalted and dried on Strata X C18 SPE columns (Phenomenex). Each 100 μg sample was dissolved in 30 μl dissolution buffer and labeled at room temperature with one iTRAQ reagent solution for 2 h, followed by 30 min with 100 μL of Milli-Q water. All the samples were then mixed into one tube and dried. We used the Suveyor high performance liquid chromatography system (Thermo Fisher Scientific) and LTQ-Orbitrap instrument (Thermo Fisher Scientific) to perform the reverse-phase high performance liquid chromatography separation. The flow rate of the reverse-phase high performance liquid chromatography separation was 250 nl/min. We used data-dependent mode to store data. All raw MS files were processed by MaxQuant software and based on the UniProt human database. We set 1% false discovery rate for protein and peptide identification. The minimum peptide length was set as seven amino acids. The fragment ion tolerance was set as 20 ppm. We set carbamidomehtylation of cysteine residues as fixed modification. Variable modifications included oxidation of methionine and N-carbamylation. All the detected proteins were classified by their molecular functions through Gene ontology (GO) enrichment analysis and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis [22].
Construction of the prognosis model for BCa
A total of 13 differentially expressed immune-related genes from proteomics sequencing results were selected as candidate genes. These 13 selected genes were conducted least absolute shrinkage and selection operator (LASSO) cox regression analysis via glmnet package. Afterwards, 7 genes with prognostic values were found and their coefficients were calculated based on TCGA cohort. The number of lambda in LASSO was set up to 1000. The IRPS score was formulated in the following:
Coefi represented the coefficient of each the 7 focused genes in this study and Ri stood for their corresponding mRNA expressions. The detailed calculation of IRPS score was: IRPS score = STAT3 * 0.30218322 + TGFB1 * 0.16910315 + CTSG * 0.05205169—NFKB1 * 0.34611795—SNRPD2 * 0.15818733—PD-1 * 0.09569928—TAP1 * 0.01611763. The IRPS was further validated in the E-MTAB-4321 cohort and the General cohort, covering 605 BCa patients. Both the cut-off values of IRPS score in the two cohorts were set as the median score of each cohort.
Immunohistochemistry (IHC)
MIBC patient samples from Shanghai General Hospital were stained with anti-Cathepsin G (Affinity, 1:100), anti-NF-κB1 (Affinity, 1:100), anti-STAT3 (Affinity, 1:100), anti-TGF-β1 (Affinity, 1:100), anti-SNRPD2 (Affinity, 1:100), anti-TAP1 (Affinity, 1:100) and anti-PD-1 (Affinity, 1:100). All the primary antibodies were incubated overnight at 4℃ and secondary antibodies were incubated for 90 min at room temperature. Slides were visualized with 3, 3′-diaminobenzidine and hematoxylin.
Immune infiltration analysis
We applied the CIBERSORT to estimate the abundances of 22 types of immune cells [23]. Here in this study, the mean expression values of PD1, PD-L1, CTLA-4, LAG-3 and TIM-3 were regarded as immune checkpoint (ICK) score; whereas the mean expression values of granzyme B (GZMB), interferon-γ (IFN-γ) and perforin (PRF1) were set as Effector score [24]. We obtained the lymphocyte infiltration signature score from CRI iAtlas [25].
Statistical analysis
In this study, the differentially expressed proteins between two different prognosis groups were analyzed by linear models for microarray data (limma). We used either a two-tailed Mann Whitney U test or a one-way analysis of variance to analyze continuous variants. The prognostic values of immune-related genes were analyzed by Cox regression analysis. Kaplan–Meier curves of overall survival (OS) and disease-free survival (DFS) were analyzed by log-rank test. Correction analysis was carried out Spearman method. The statistical analysis and data visualization were achieved by R (3.6.2) and SPSS 24.0. A P value of < 0.05 was considered to be statistically significant. The flow chart diagram were shown in Supplementary Fig. 1.
Results
Investigation of differentially expressed immune-related proteins in MIBC patients
In order to identify differential expression of immune-related proteins among cancer cells and normal bladder cells, we performed a LC–MS analysis based on 9 pairs of samples from MIBC patients resected in Shanghai General Hospital during 2007 to 2018 (Fig. 1A). Between the two different prognosis groups, there were totally 677 differentially expressed proteins, with a P value < 0.05 and |fold change|> 1.5 (Log2(fold change) > 0.584). Considering the GO and KEGG analysis, we found 97 immune-related proteins (Fig. 1B, C). Interestingly, 13 out of 97 immune-related proteins were also detected as differential expression proteins in MIBC patients, including ICAM1, STAT3, TGF-β1, TNFRSF6B, SNRPD3, STAT1, SNRPD2, ICAM3, TAP1, Cathepsin G, PD-1 and NF-κB1 (Fig. 1B, C). Therefore, we decided to further investigate the prognostic values of the 13 proteins related genes.
Identification of immune-related genes with prognostic values. A The heatmap of LC–MS from 9 pairs of MIBC patients with all the differential expression proteins between two different prognosis groups. B A total of 13 out of 677 differentially expressed proteins were classified as immune-related proteins. C The Volcano plot result of all the 677 differential expression protein. D The LASSO analysis of the 13 immune-related genes. E The profile of coefficients of the 7 immune-related genes. F The illustration of expression levels of 7 immune-related proteins in General cohort. LC–MS: liquid chromatography-mass spectrometry. LASSO: least absolute shrinkage and selection operator
Construction of IRPS based on the BCa patients in TCGA cohort
All the 13 immune-related genes were undergone LASSO-Cox regression analysis in the TCGA cohort (Fig. 1D, E). Interestingly, a total of 7 immune-related genes were shown their statistical significance (P < 0.05) in predicting the clinical outcomes of BCa patients, including Cathepsin G, NF-κB1, STAT3, TGF-β1, SNRPD2, TAP1 and PD-1. Subsequently, the IRPS was constructed based on the 7 genes in TCGA dataset. Meanwhile, the IRPS score was established according to the formulation shown in 2 section. In TCGA cohort, 405 BCa patients were classified into IRPS high and low groups based on the cut-off IRPS value. BCa patients with IRPS high score had significantly poorer 5-year OS (P < 0.0001) with hazard ratio of 3.46 (Figs. 2A, 3A). Whereas, BCa patients with IRPS high score were associated with a worse 5-year DFS (P = 0.0069) (Fig. 2B). Additionally, patients with T3/T4 stages and lymph node metastasis were associated with higher IRPS score (P = 0.0013 and P = 0.047, respectively) (Fig. 3B, C).
Kaplan–Meier survival analysis among 3 cohorts. A The OS of MIBC patients in TCGA cohort. B The DFS of MIBC patients in TCGA cohort. C The DFS of BCa patients in E-MTAB-4321 cohort. D The OS of MIBC patients in General cohort. E The DFS of MIBC patients in General cohort. OS: overall survival. DFS: disease-free survival
Correlation of IRPS score and clinicopathological features among 3 cohort. A Cox regression analysis of IRPS score and clinicopathologic features for patients’ OS in TCGA cohort. B The correlation of IRPS score and tumor grade in TCGA cohort. C The correlation of IRPS score and lymph node metastasis in TCGA cohort. D Cox regression analysis of IRPS score and clinicopathologic features for patients’ DFS in E-MTAB-4321 cohort. E The correlation of IRPS score and tumor grade in E-MTAB-4321 cohort. F Cox regression analysis of IRPS score and clinicopathologic features for patients’ OS in General cohort. G The correlation of IRPS score and tumor grade in General cohort. H The correlation of IRPS score and lymph node metastasis in General cohort. OS: overall survival. DFS: disease-free survival
Verification of IRPS among BCa patients in E-MTAB-4321 cohort
We further explored whether the IRPS can be applied as a prognostic model in different dataset. As a result, the IRPS was verified in E-MTAB-4321 cohort. A total of 476 BCa patients were enrolled in the E-MTAB-4321 dataset, including 460 NMIBC and 16 MIBC patients. As expected, high IRPS score BCa patients also showed a statistically significant poorer 5-year DFS outcomes in E-MTAB-4321 cohort (P = 0.0069) with hazard ratio of 1.07 (Figs. 2C, 3D). Moreover, it was also indicated that Cis/Ta-stage BCa patients were correlated with lower IRPS score (P = 0.0235) (Fig. 3E).
Verification of IRPS of MIBC in general cohort
We further verified the effectiveness of IRPS in predicting clinical outcomes of MIBC patients in General cohort. The hazard ratio value of each protein was illustrated in Table 2. The illustration of expression levels of the 7 immune-related proteins were shown in Fig. 1F. MIBC patients in General cohort were subdivided according to the mean value of IRPS score. Consistently, the 5-year OS and DFS of MIBC patients with high IRPS score dropped significantly (P < 0.0001 and P < 0.0001, respectively) (Fig. 2D, E). The hazard ratio of high IRPS score and 5-year OS was 6.00 (Fig. 3F). Also, T3/T4 stages and lymph node metastasis were correlated with higher IRPS score (P = 0.0013 and P = 0.047, respectively) (Fig. 3G, H). These results indicated IRPS an independent prognostic indicator for MIBC.
Correlation of IRPS score and the remodeling of the immune microenvironment
For the purpose of revealing the relationship between the IRPS score and the tumor immune microenvironment, we utilized immune infiltration analysis. The IRPS score was evaluated with the Effector score, ICK score and lymphocyte infiltration signature score (Fig. 4A, D, G). We found that Effector, ICK and lymphocyte infiltration signature score all significantly diminished in IRPS high score groups (P < 0.0001, P < 0.0001 and P = 0.036, respectively) (Fig. 4B, E, H). The IRPS score was adversely associated with GZMB, IFN-γ, PD-1, PD-L1 (Spearman’s R = -0.25, Spearman’s R = -0.33, Spearman’s R = -0.30 and Spearman’s R = -0.18; P < 0.01, P < 0.01, P < 0.01 and P < 0.01, respectively) (Fig. 4C, F). Additionally, higher IRPS score was significantly associated with more M2 macrophage and resting mast cell infiltration (Spearman’s R = 0.13 and Spearman’s R = 0.26; P < 0.01 and P < 0.01, respectively) (Fig. 4I). The complete analysis of IRPS score and immune cell infiltrations were shown in Supplementary Fig. 2.
Correlation of IRPS score and the remodeling of the immune microenvironment. A Heatmap of IRPS score and the expression of effector molecules. B Evaluation of the correlation among IRPS score and Effector score. C Correlation of IRPS score and GZMB or IFN-γ expression. D Heatmap of IRPS score and the expression of ICK molecules. E Evaluation of the correlation among IRPS score and ICK score. F Correlation of IRPS score and PD-1 or PD-L1 expression. G Heatmap of IRPS score and the infiltration of 22 types of immune cells. H Evaluation of the correlation among IRPS score and lymphocyte infiltration signature score. I Correlation of IRPS score and M2 macrophage or resting mast cell infiltration
Discussion
It can be inferred that the potential immunogenity of BCa from the current utilization of PD-1/PD-L1 related immunotherapies [8, 20]. However, considering the immunosuppressive microenvironment in BCa tissue, it would be interesting to investigate the expression of immune-related genes in BCa [26]. Furthermore, the immune-checkpoint inhibitor targeted immunotherapies for MIBC patients need improvements [17]. Therefore, it would be intriguing to find some prognostic biomarkers concerning immune-checkpoint inhibitors for MIBC patients. In the present study, through proteomics sequencing, we established an IRPS for predicting the survival of BCa patients, especially MIBC patients, and analyzed the immune infiltration of IRPS score.
Among the 7 enrolled immune-related genes, Cathepsin G is a neutrophil serine protease and its interaction with receptor for advanced glycation end products can trigger neutrophil cytotoxicity to kill tumor cells [27, 28]. Studies demonstrate the prognostic value of Cathepsin G in oral squamous cell carcinoma patients [29]. It remains unclear that the detailed function of Cathepsin G in tumor immune response and its prognostic value for BCa patients. NF-κB1 belongs to NF-κB family, involved in regulating cellular proliferation, apoptosis, inflammation and tumor immune response [30]. The dysregulation of NF-κB1 results in hepatocellular carcinoma, acute lymphoblastic leukemia as well as breast cancer [30,31,32]. STAT3 is vital for vertebrate development while its mutations are associated with immunodeficiency, autoimmunity and cancer [33]. As previous studies reported, TGF-β1, a family member of TGF-β, plays a key role in the development and maturation of immune cells [34]. Drugs targeting TGF-β1 pathways for tumor immunotherapies are extremely difficult at present [34]. There are limited studies on SNRPD2, a small nuclear ribonucleoprotein D2 polypeptide [35]. Researchers have evaluated a prognostic signature with 13 key genes including SNRPD2 for hepatocellular carcinoma patients [35]. Detailed molecular mechanisms between SNRPD2 and cancer remain unknown. It is found that TAP1, antigen processing 1, is correlated with good prognosis in colorectal cancer and melanoma patients [36, 37].
Currently, the utility of immune-checkpoint inhibitors in malignancies has increasingly become a hot tendency in cancer treatment [38]. PD-1/PD-L1 is one of the most studied and clinically successful immune-checkpoint inhibitor drug targets [16]. Although it is reported that high levels of PD-L1 and PD-1 expressions in cancers suggest worse prognosis and advanced disease stages, only limited patients could benefit from PD-1/PD-L1 related immune-checkpoint inhibitor treatments [38]. Moreover, the criteria for PD-1/PD-L1 immunochemistry evaluation in BCa are still away from forming the consensus [16, 39]. Different pathologists may even gain a contradictory PD-1/PD-L1 evaluation result from the same pathological image. Therefore, it is necessary to find a convenient and comparatively reliable method for predicting immunotherapy responsiveness. The IRPS score in this study was statistically significant negatively associated with PD-1/PD-L1, which could be a potential clinical utilization of IRPS.
Intriguingly, IRPS score was correlated with distributions of some immune cells, especially positive correlation of macrophage M2 and adverse correlation of CD8 T cells. Currently, PD-1 positive CD8 T cells are the main targets for immune-checkpoint inhibitor therapies [40]. It is demonstrated that BCa patients with heavy CD8 T cell infiltration benefit more immune-checkpoint inhibitor therapies due to the higher accessibility of neoantigens, tumor-specific antigens encoded by mutated genes [38, 40]. Moreover, CD8 T cells can be enhanced by activated memory CD4 T cells [41, 42]. Macrophage M2 can increase immunosuppressive infiltration of Treg cells and decrease the immune-checkpoint inhibitor therapy responsiveness [43]. The correlation of IRPS score and distribution of immune cells implied potential mechanisms behind IRPS and tumor immune microenvironment of BCa.
Moreover, considering the performance of IRPS among TCGA, E-MTAB-4321 and General cohorts, we found that cohorts with higher MIBC patient ratio showed more obvious difference among IRPS high and low score groups. General cohort was consisted of only MIBC patients whereas E-MTAB-4321 cohort mostly was NMIBC patients. It can be suggested that IRPS performed better for MIBC patients. In addition, MIBC patients have much higher mortality and draw more medical attention from clinicians. Via IRPS evaluation, clinicians can briefly judge the prognosis of MIBC patients. At present, immune-checkpoint inhibitor targeted therapies for BCa patients are utilized to treat locally advanced or metastatic platinum-ineligible MIBC patients [14]. IRPS score could be a potential indicator for immunotherapies on MIBC patients.
There were also some limitations in this study. Firstly, the cut-off value was selected as the median value of each cohort for defining high and low IRPS score in this retrospective study. The best cut-off value for IRPS score shall be explored through further prospective studies. Secondly, the outcome and molecular biology of muscle-invasive and non-muscle invasive bladder cancer are considerably different, thus the non-MIBC cases in the E-MTAB-4321 cohort might cause potential bias. But we had also performed external validation in the General cohort with MIBC cases.
In conclusion, the present study constructed an IRPS based on LC–MS results and TCGA dataset. The IRPS was verified by E-MTAB-4321 cohort and General cohort. High IRPS score was correlated with worse clinical outcomes in BCa patients. The IRPS score showed its relevance with immune infiltration.
Availability of data and materials
All data generated or analyzed during this study are included in this published article.
References
Siegel RL, Miller KD, Jemal A. Cancer statistics, 2020. CA Cancer J Clin. 2020;70(1):7–30.
Li F, Teng H, Liu M, Liu B, Zhang D, Xu Z, Wang Y, Zhou H. Prognostic value of immune-related genes in the tumor microenvironment of bladder cancer. Front Oncol. 2020;10:1302.
Chen S, Jiang L, Zheng X, Shao J, Wang T, Zhang E, Gao F, Wang X, Zheng J. Clinical use of machine learning-based pathomics signature for diagnosis and survival prediction of bladder cancer. Cancer Sci. 2021;112(7):2905–14.
Flaig TW, Spiess PE, Agarwal N, Bangs R, Boorjian SA, Buyyounouski MK, Downs TM, Efstathiou JA, Friedlander T, Greenberg RE, et al. NCCN Guidelines Insights: bladder cancer Version 5.2018. J Natl Compr Canc Netw. 2018;16(9):1041–53.
Schneider AK, Chevalier MF, Derre L. The multifaceted immune regulation of bladder cancer. Nat Rev Urol. 2019;16(10):613–30.
Chen X, Jin Y, Gong L, He D, Cheng Y, Xiao M, Zhu Y, Wang Z, Cao K. Bioinformatics analysis finds immune gene markers related to the prognosis of bladder cancer. Front Genet. 2020;11:607.
Jordan B, Meeks JJ. T1 bladder cancer: current considerations for diagnosis and management. Nat Rev Urol. 2019;16(1):23–34.
Na L, Bai Y, Sun Y, Wang Z, Wang W, Yuan L, Zhao C. Identification of 9-Core immune-related genes in bladder urothelial carcinoma prognosis. Front Oncol. 2020;10:1142.
Chen S, Jiang L, Zhang E, Hu S, Wang T, Gao F, Zhang N, Wang X, Zheng J. A novel nomogram based on machine learning-pathomics signature and neutrophil to lymphocyte ratio for survival prediction of bladder cancer patients. Front Oncol. 2021;11: 703033.
Wang Y, Chen L, Yu M, Fang Y, Qian K, Wang G, Ju L, Xiao Y, Wang X. Immune-related signature predicts the prognosis and immunotherapy benefit in bladder cancer. Cancer Med. 2020;9(20):7729–41.
Sanmamed MF, Chen L. A Paradigm shift in cancer immunotherapy: from enhancement to normalization. Cell. 2019;176(3):677.
Liu Y, Cao X. Immunosuppressive cells in tumor immune escape and metastasis. J Mol Med. 2016;94(5):509–22.
Kim JW, Tomita Y, Trepel J, Apolo AB. Emerging immunotherapies for bladder cancer. Curr Opin Oncol. 2015;27(3):191–200.
Eckstein M, Cimadamore A, Hartmann A, Lopez-Beltran A, Cheng L, Scarpelli M, Montironi R, Gevaert T. PD-L1 assessment in urothelial carcinoma: a practical approach. Ann Transl Med. 2019;7(22):690.
Lobo N, Mount C, Omar K, Nair R, Thurairaja R, Khan MS. Landmarks in the treatment of muscle-invasive bladder cancer. Nat Rev Urol. 2017;14(9):565–74.
Tan WP, Tan WS, Inman BA. PD-L1/PD-1 Biomarker for metastatic urothelial cancer that progress post-platinum therapy: a systematic review and meta-analysis. Bladder cancer. 2019;5(3):211–23.
Boegemann M, Aydin AM, Bagrodia A, Krabbe LM. Prospects and progress of immunotherapy for bladder cancer. Expert Opin Biol Ther. 2017;17(11):1417–31.
Hwang I, Park I, Yoon SK, Lee JL. Hyperprogressive Disease in Patients With Urothelial Carcinoma or Renal Cell Carcinoma Treated With PD-1/PD-L1 Inhibitors. Clin Genitourin Cancer. 2020;18(2):e122–e133.
Tay C, Qian Y, Sakaguchi S. Hyper-Progressive Disease: The Potential Role and Consequences of T-Regulatory Cells Foiling Anti-PD-1 Cancer Immunotherapy. Cancers (Basel). 2020;13(1):48.
Jiang LR, Zhang N, Chen ST, He J, Liu YH, Han YQ, Shi XQ, Yang JJ, Mu DY, Fu GH, et al. PD-1-Positive tumor-associated macrophages define poor clinical outcomes in patients with muscle invasive bladder cancer through potential CD68/PD-1 complex interactions. Front Oncol. 2021;11: 679928.
Hedegaard J, Lamy P, Nordentoft I, Algaba F, Høyer S, Ulhøi BP, Vang S, Reinert T, Hermann GG, Mogensen K, et al. Comprehensive transcriptional analysis of early-stage urothelial carcinoma. Cancer Cell. 2016;30(1):27–42.
Kanehisa M, Furumichi M, Sato Y, Ishiguro-Watanabe M, Tanabe M. KEGG: integrating viruses and cellular organisms. Nucleic Acids Res. 2021;49(D1):D545–51.
Newman AM, Steen CB, Liu CL, Gentles AJ, Chaudhuri AA, Scherer F, Khodadoust MS, Esfahani MS, Luca BA, Steiner D, et al. Determining cell type abundance and expression from bulk tissues with digital cytometry. Nat Biotechnol. 2019;37(7):773–82.
Zhou Q, Wang Z, Zeng H, Zhang H, Liu Z, Huang Q, Wang J, Chang Y, Bai Q, Liu L, et al. Identification and validation of poor prognosis immunoevasive subtype of muscle-invasive bladder cancer with tumor-infiltrating podoplanin(+) cell abundance. Oncoimmunology. 2020;9(1):1747333.
Thorsson V, Gibbs DL, Brown SD, Wolf D, Bortone DS, Ou Yang TH, Porta-Pardo E, Gao GF, Plaisier CL, Eddy JA, et al. The immune landscape of cancer. Immunity. 2018;48(4):812-830.e814.
Mo Q, Nikolos F, Chen F, Tramel Z, Lee YC, Hayashi K, Xiao J, Shen J, Chan KS. Prognostic power of a tumor differentiation gene signature for bladder urothelial carcinomas. J Natl Cancer Inst. 2018;110(5):448–59.
Sionov RV, Fainsod-Levi T, Zelter T, Polyansky L, Pham CT, Granot Z. Neutrophil Cathepsin G and tumor cell RAGE facilitate neutrophil anti-tumor cytotoxicity. Oncoimmunology. 2019;8(9): e1624129.
Morimoto-Kamata R, Yui S. Insulin-like growth factor-1 signaling is responsible for cathepsin G-induced aggregation of breast cancer MCF-7 cells. Cancer Sci. 2017;108(8):1574–83.
Huang GZ, Wu QQ, Zheng ZN, Shao TR, Li F, Lu XY, Ye HY, Chen GX, Song YX, Zeng WS, et al. Bioinformatics Analyses Indicate That Cathepsin G (CTSG) is a Potential Immune-Related Biomarker in Oral Squamous Cell Carcinoma (OSCC). Onco Targets Ther. 2021;14:1275–89.
Chen X, Zhou Y, Li Z, Wang Z. Mining database for the expression and clinical significance of NF-kappaB family in hepatocellular carcinoma. J Oncol. 2020;2020:2572048.
Grazioli P, Orlando A, Giordano N, Noce C, Peruzzi G, Scafetta G, Screpanti I, Campese AF. NF-kappaB1 regulates immune environment and outcome of notch-dependent t-cell acute lymphoblastic leukemia. Front Immunol. 2020;11:541.
Qadir J, Riaz SK, Sahar NE, Aman D, Khan MJ, Malik MFA. Transcriptional elucidation of tumor necrosis factor-alpha-mediated nuclear factor-kappaB1 activation in breast cancer cohort of Pakistan. J Cancer Res Ther. 2020;16(6):1443–8.
Hillmer EJ, Zhang H, Li HS, Watowich SS. STAT3 signaling in immunity. Cytokine Growth Factor Rev. 2016;31:1–15.
Lodyga M, Hinz B. TGF-beta1 - A truly transforming growth factor in fibrosis and immunity. Semin Cell Dev Biol. 2020;101:123–39.
Gu Y, Li J, Guo D, Chen B, Liu P, Xiao Y, Yang K, Liu Z, Liu Q. Identification of 13 Key genes correlated with progression and prognosis in hepatocellular carcinoma by weighted gene co-expression network analysis. Front Genet. 2020;11:153.
Ling A, Lofgren-Burstrom A, Larsson P, Li X, Wikberg ML, Oberg A, Stenling R, Edin S, Palmqvist R. TAP1 down-regulation elicits immune escape and poor prognosis in colorectal cancer. Oncoimmunology. 2017;6(11): e1356143.
Lazaridou MF, Gonschorek E, Massa C, Friedrich M, Handke D, Mueller A, Jasinski-Bergner S, Dummer R, Koelblinger P, Seliger B. Identification of miR-200a-5p targeting the peptide transporter TAP1 and its association with the clinical outcome of melanoma patients. Oncoimmunology. 2020;9(1):1774323.
Lopez-Beltran A, Cimadamore A, Blanca A, Massari F, Vau N, Scarpelli M, Cheng L, Montironi R. Immune Checkpoint Inhibitors for the Treatment of Bladder Cancer. Cancers (Basel). 2021;13(1):131.
Tu MM, Ng TL, De Jong FC, Zuiverloon TCM, Fazzari FGT, Theodorescu D. Molecular biomarkers of response to PD-1/PD-L1 immune checkpoint blockade in advanced bladder cancer (vol 5, pg 131, 2019). Bladder Cancer. 2020;6(4):559–559.
Durgeau A, Virk Y, Corgnac S, Mami-Chouaib F. Recent Advances in Targeting CD8 T-Cell immunity for more effective cancer immunotherapy. Front Immunol. 2018;9:14.
MacLeod MK, Clambey ET, Kappler JW, Marrack P. CD4 memory T cells: what are they and what can they do? Semin Immunol. 2009;21(2):53–61.
Imai N, Tawara I, Yamane M, Muraoka D, Shiku H, Ikeda H. CD4(+) T cells support polyfunctionality of cytotoxic CD8(+) T cells with memory potential in immunological control of tumor. Cancer Sci. 2020;111(6):1958–68.
Chen D, Xie J, Fiskesund R, Dong W, Liang X, Lv J, Jin X, Liu J, Mo S, Zhang T, et al. Chloroquine modulates antitumor immune response by resetting tumor-associated macrophages toward M1 phenotype. Nat Commun. 2018;9(1):873.
Acknowledgements
Not applicable.
Funding
This work was supported by the Shanghai Municipal Key Clinical Specialty (shslczdzk01303), the National Natural Science Foundation of China (No.81902569) and the National Natural Science Foundation of China (No.82002665).
Author information
Authors and Affiliations
Contributions
LJ conceived the idea for the study and was a major contributor in writing the manuscript. SC conducted the bioinformatics analysis and was a major contributor for revising the manuscript. QP collected samples with clinicopathological data, funded the research and was contributed for manuscript writing. JZ made the tissue microarrays and contributed for manuscript revising. JH performed the IHC staining for tissue microarrays. JS assisted to collect patients’ clinicopathological data. YH prepared HE stained slides and helped to prepare tissue microarrays. JY gathered all the patient consent forms as well as the ethics statements. NZ funded the research, and was the second leading contributor for manuscript preparation. GHF provided suggestions on the project throughout the study, joined the interpretation of the results and was the major contributor for manuscript revision. FG developed the idea for the study, contributed the central idea, funded the research, and was the leading contributor for manuscript revision. All authors contributed to the revision and approved the final manuscript.
Corresponding authors
Ethics declarations
Ethics approval consent to participate
The study was conducted according to the Helsinki Declaration and was approved by the Human Ethics Committee of Shanghai General Hospital.
All the participated patients were well informed and informed consents were obtained from all patients.
Consent for publication
Not applicable.
Competing interests
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
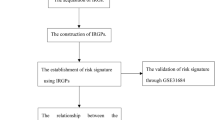
Additional file 1.
The workflow of the study.
Additional file 2.
Correlation analysis of IRPS score and immune infiltration cells.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Jiang, L., Chen, S., Pan, Q. et al. The feasibility of proteomics sequencing based immune-related prognostic signature for predicting clinical outcomes of bladder cancer patients. BMC Cancer 22, 676 (2022). https://doi.org/10.1186/s12885-022-09783-y
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12885-022-09783-y