Abstract
Background
Colorectal adenoma (CA) is an important precancerous lesion and early screening target of colorectal cancer (CRC). Lipids with numerous physiological functions are proved to be involved in the development of CRC. However, there is no lipidomic study with large-scale serum samples on diagnostic biomarkers for CA.
Methods
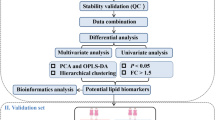
The serum lipidomics of CA patients (n = 50) and normal control (NR) (n = 50) was performed by ultra high performance liquid chromatography-high resolution mass spectrometry with electrospray ionization (UHPLC-ESI-HRMS). Univariate and multivariate statistical analyses were utilized to screen the differential lipids between groups, and combining the constituent ratio analysis and diagnostic efficiency evaluation by receiver operating characteristic (ROC) curve disclosed the potential mechanism and biomarkers for CA.
Results
There were obvious differences in serum lipid profiles between CA and NR groups. Totally, 79 differential lipids were selected by criterion of P < 0.05 and fold change > 1.5 or < 0.67. Triacylglycerols (TAGs) and phosphatidylcholines (PCs) were the major differential lipids with ratio > 60%, indicating these two lipid metabolic pathways showed evident disequilibrium, which could contribute to CA formation. Of them, 12 differential lipids had good diagnostic ability as candidate biomarkers for CA (AUC ≥ 0.900) by ROC analysis.
Conclusions
To our knowledge, this is the first attempt to profile serum lipidomics and explore lipid biomarkers of CA to help early screening of CRC. 12 differential lipids are obtained to act as potential diagnostic markers of CA. PCs and fatty acids were the main dysregulated biomarkers for CA in serum.
Similar content being viewed by others

Background
Colorectal cancer (CRC) is a significant public health problem and lethal disease, causing more than 900,000 deaths annually as a common malignant tumor worldwide [1]. It is reported that the high mortality rate of CRC is primarily due to the diagnosis and discovery of late-stage tumors. The early CRC stage has no specific symptoms, which generally leads to poor diagnostic effects and low detection rate. While the treatment effect in the middle and late stage is less favorable and along with many side effects [2]. The 5-year survival rate of CRC patients diagnosed at the early stage is about 90%, while it dramatically decreases to 14% for patients diagnosed with advanced-stage CRC [3]. In addition, nearly 90% of CRC evolves from colorectal adenoma (CA) [4]. The adenoma-carcinoma sequence is widely regarded as the main pathway for the formation and development of CRC currently [5], and most CA patients are associated with better treatment outcome and prognosis [6]. Thus, CA is an important target for early screening of CRC, and effective CA screening reduces the morbidity and mortality of CRC enormously. At present, the mainstays of CA screening methods including the fecal occult-blood test, stool DNA test, sigmoidoscopy, CT colonography, and optical colonoscopy. However, these methods also have apparent defects, such as poor performance for early diagnosis, high cost and technical requirements, and low patient compliance [6], which seriously limit their application in extensive screening of CA. Accordingly, it is vital to develop a minimally invasive diagnostic strategy with high performance to improve the early screening and prevention of CRC.
Lipids have emerged as important biomolecules involved in the numerous physiological processes of human that plays a diverse role in cell apoptosis, proliferation, signal transduction, and energy metabolism [7,8,9,10]. Growing evidence suggests that metabolic disorder of lipid is closely related to the progression of CRC disease [11,12,13]. As a key branch and advanced technique of metabolomics, lipidomics systematically and comprehensively reflects the changes in lipid profiles and related metabolic pathways within organisms under different physiological or pathological states [14]. Ultra high performance liquid chromatography tandem high resolution mass spectrometry with electrospray ionization (UHPLC-ESI-HRMS) has become the most prevalent analytical tool in lipidomics study due to its advantages of high selectivity, high sensitivity, and high throughput [15]. Currently, lipidomics has developed into a field with widespread application in biology, medicine, and chemistry science, because of its great potential in discovery of molecular mechanisms and biomarkers of diseases, and it has recently become a hotspot in omics research [16]. To date, several lipidomics studies on CRC have been reported. The lipidomics research found that lysophosphatidylcholines (LPCs) and phosphatidylcholines (PCs) are the most strongly related biomarkers of CRC formation [17, 18]. In plasma, ethanolamine plasmalogens and fatty acids (FAs) are considered as early diagnostic biomarkers of CRC [19]. Furthermore, triacylglycerols (TAGs) are found to be the main disturbed lipid markers of CRC progression [20, 21]. However, no study has been reported to explore the biomarkers of CA through serum lipidomics. Plasma untargeted LC-MS-based metabolomics is applied to investigate the potential mechanism of CA, indicating that L-tryptophan, L-proline, and lysoPC (C17:0) could be combined to serve as the biomarker to improve its diagnosis [22]. Studies suggest that total TAGs levels in serum or plasma are elevated may be associated with increased risk of CA [23]. According to some authors, the disruption of polyunsaturated fatty acids (PUFAs) is correlated with CA development [24, 25]. Therefore, the discovery of CA biomarkers based on serum lipidomics still needs to be studied, and which will be expected to solve the shortcomings of current screening methods for CA.
In this study, we performed a lipidomics study of serum samples from fifty CA patients and fifty healthy subjects by the UHPLC-ESI-HRMS technique. By compared with serum lipid profiles of CA and normal control (NR) groups, the differential lipids and potential mechanism of lipid metabolism pathways were explored by univariate and multivariate statistical analysis. Then, combined with receiver operating characteristic (ROC) curve analysis and trend change analysis of differential lipids, the potential lipid markers for CA diagnosis were evaluated and selected, which would provide a reference for early screening of CRC.
Methods
Chemicals and reagents
HPLC grade methanol, dichloromethane, isopropanol, acetonitrile, formic acid and ammonium formate were purchased from Merck & Co. (Billerica, MA, USA). Ultrapure water was prepared by a Millipore Milli-Q system (Billerica, MA, USA). Lipid standards including palmitoyl ethanolamide, palmitic acid, methyl palmitate, 2-arachidonoyl glycerol, and 4-dodecylbenzenesulfonic acid were obtained from Sigma-Aldrich (St Louis, MO, USA).
Study cohort and sample collection
Prior to study, the medical ethics approval was obtained from the People’s Hospital of Guangxi Zhuang Autonomous Region (No.KY-DZX202008) and written informed consent was obtained by each subject. The study was carried out in accordance with the Declaration of Helsinki. For serum lipidomics analysis, a total of 100 subjects including 50 NR and 50 CA subjects were enrolled in this study. At the same time, we evaluated for possible sex and age-associated differences by Chi-square test and Student’s test. Detailed characteristics of the study cohort were shown in Table 1. All whole-blood samples were taken after an 8-h fast, left to stand at room temperature for 25 min, and serum was then collected following centrifugation at 5000 rpm/min for 10 min at 4 °C. The serum samples were immediately stored at-80 °C prior to analysis.
Sample preparation for lipidomic analysis
For sample preparation, 50 μL serum sample was added and mixed with 500 μL precooling dichloromethane-methanol (3:1, v/v) solution. After vortexed for 5 min and placed in ice bath for 10 min, the solution was centrifuged at 13,000 rpm/min at 4 °C for 10 min. 300 μL lower dichloromethane solution was dried in vacuum at room temperature. The dried samples were redissolved with 600 μL acetonitrile-isopropanol (1:1, v/v) solution, then vortexed for 2 min and ultrasonicated in ice bath for 5 min. Next vortexed for 1 min, the mixture was centrifuged at 13,000 rpm/min at 4 °C for 15 min, and the supernatant was used for serum lipid analysis. Quality control (QC) samples were prepared by mixing 5 μL of each sample to ensure the stability and reproducibility of data acquisition.
UHPLC–ESI-HRMS-based lipidomic analysis
A Dionex Ultimate 3000 liquid chromatography system (Sunnyvale, CA, USA) (SN: 7254012) coupled to a Thermo Fisher Q Exactive Orbitrap mass spectrometry system (Waltham, MA, USA) (SN: SN02386L) were used for lipidomics analysis. The LC conditions were as follows: column, Waters Acquity UPLC HSS T3 (1.8 μm, 2.1 × 100 mm; Milford, MA, USA); mobile phase A, acetonitrile-water (60:40, v/v) containing 0.1% formic acid and 10 mM ammonium formate; mobile phase B, isopropanol-acetonitrile (90:10, v/v) containing 0.1% formic acid and 10 mM ammonium format; The gradient conditions were set as follows: 0.0–4.0 min, 30 to 60% B; 4.0–9.0 min, 60 to 100% B; 9.0–15.0 min, 100% B; 15.0–18.0 min, 100% B to 30% B. The injection volume was 5 μL, and the column temperature was 50 °C, as well as the flow rate was 0.3 mL/min.
The MS spectrometric parameters were as follows: spray voltage, 3.5 kV; sheath gas flow rate, 50 psi; auxiliary gas flow rate, 13 arb; capillary temperature, 320 °C; auxiliary gas heater temperature, 420 °C; scan modes, full MS (resolution 70,000) and ddMS2 (resolution 17,500 with stepped collision energy (10, 20, and 40 eV); and scan range, m/z 100–1200. All data were acquired using the Thermo Scientific Xcalibur 3.1 software (Waltham, MA, USA).
Statistical analysis
Univariate statistical analysis: Raw data files were imported into the Compound Discoverer™3.1 (Thermo Scientific, Fremont, CA, USA) for data analysis. Lipidomics data (including all ion features with their RT, m/z, and peak intensity) were extracted and normalization was conducted by using QC samples to effectively uncover differential lipids. The feature differences between groups were analyzed with Mann-Whitney U test or Student’ t-test based on distribution characteristics of the data. The value of P < 0.05 was considered to indicate significant differences. A list of potential lipids was identified depending on Thermo mzVault and LipidBlast database. The main parameters were as follows: minimum peak intensity, 500,000, mass error, 10 ppm, RT tolerance, 0.2 min, intensity tolerance, 30%, S/N, 3.
Multivariate statistical analysis: Principal component analysis (PCA) and orthogonal partial least squares discriminant analysis (OPLS-DA) were performed with the software SIMCA-P 14.1 (Umetrics, UMEA, Sweden). To avoid overfitting, 200 times permutation test was carried out on the analytical model. The criteria of fold change > 1.5 or < 0.67 and P < 0.05 were set as the cut-off values for selection of differential lipids between groups. The ROC analysis of the differential lipids was performed by MetaboAnalyst 5.0 (https://www.metaboanalyst.ca/) to evaluate the diagnostic performance and to explore the potential biomarkers for CA patients.
Results
Differential lipid profiles between CA and NR
PCA and OPLS-DA models, the common multivariate statistical methods used in omics study, were utilized to evaluate the differences between groups regarding lipid metabolism of CA and NR groups. Firstly, the PCA model was constructed and its score plots performed on all the samples revealed that the QC samples were clustered closely in both ESI modes, indicating the analysis system with excellent robustness and reproducibility during the batch analysis process (Fig. 1A and B). In addition, most samples contained in the 95% confidence interval apart from a few exceptions. It could be considered individual variations for a few samples outside the confidence interval (Fig. 1A and B). The relatively smaller individual difference of lipid profiles in NR patients was seen compared with the CA patients, which could be due to the pathogenic factors. In addition, a distinct separation between the two groups presented in two modes also reflected their differences in lipid metabolism (Fig. 1A and B). Meanwhile, Relative to the ESI- mode, the ESI+ mode had more obvious separation trend (Fig. 1A and B). To evaluate the effect of gender and age on the difference in lipid profiles between two groups, statistical analysis was conducted and the results showed no significant difference between CA and NR groups for the gender and age in study cohort (Table 1).
Multivariate statistical analysis of differential lipid features between NR and CA groups in both ESI modes. Principal component analysis (PCA) of two groups in ESI+ and ESI- modes, respectively (A, B); orthogonal partial least squares discriminant analysis (OPLS-DA) of the two groups in ESI+ and ESI- modes, respectively (C, D); Overfitting test for OPLS-DA model in ESI+ and ESI- modes, respectively (E, F). Abbreviations: ESI, electrospray ionization; CA, colorectal adenoma; NR, normal control
Furthermore, to maximize the discovery of differential lipid features in the serum between CA and NR groups, the OPLS-DA model was established using all of detected lipid features. Results showed that two groups were clearly discriminated at ESI+ mode (R2X [cum] = 0.386, R2Y [cum] = 0.962, Q2 [cum] = 0.956) and ESI- mode (R2X [cum] = 0.492, R2Y [cum] = 0.945, Q2 [cum] = 0.890), respectively, which indicated remarkable differences between groups in both ESI modes and obvious dysregulation in lipid metabolism of CA relative to NR group (Fig. 1C and D). Meanwhile, a 200 times permutation test was performed to verify the reliability and applicability of OPLS-DA model for data analysis. The intercept of the R2Y and Q2 was 0.668 and − 0.355 in ESI+ mode, and R2Y and Q2 was 0.685 and − 0.358 in ESI- mode, and value of P (CV-ANOVA) was 0.000 in both modes, respectively (Fig. 1E and F). Concurrently, R2Y and Q2 values derived from the permutation test were all lower than corresponding original values, which provided proof that OPLS-DA model was rational and not overfitting for the data analysis in both ESI modes.
Screening and identification of diagnostic lipid biomarkers for CA
The difference analysis in serum lipid profiles between NR and CA groups was performed using the univariate and multivariate statistical methods. To minimize false positives, combined with fold change > 1.5 or < 0.67 and P < 0.05, finally, a total of 79 differential lipids were found between the groups, including 4 in ESI- mode and 75 in ESI+ mode (Table 2). These differential lipids mainly included monoacylglycerols (MAGs): 1.27%, diacylglycerols (DAGs): 15.19%, TAGs: 31.65%, FAs: 11.39%, LPCs: 3.80%, PCs: 29.11%, phosphatidylethanolamines (PEs): 1.27%, ceramides (Cers): 2.53% and sphingomyelins (SMs): 3.80% (Fig. 2). TAGs and PCs made up the highest fraction in differential lipid types, followed by DAGs and then FAs. Therefore, the two lipid types accounted for 60.76% of the total proportion, suggesting that dysregulation of PC and TAG metabolism is closely associated with the diagnosis and pathogenesis of CA disease. Furthermore, to learn more about the distribution of relative levels of differential lipids in two groups, the identified lipid data were analyzed using clustering heatmap. Just showed the Table 2 and Fig. 3, the majority of differential lipids features were significantly down-regulated in CA group compared to NR group. Taken together, PCs and TAGs are considered to be the main influencing factor that contributed to the CA formation.
The constituent ratio of differential lipids in serum between CA and NR groups. Abbreviations: CA, colorectal adenoma; NR, normal control; MAGs, monoacylglycerols; DAGs, diacylglycerols; TAGs, triacylglycerols; LPCs, lysophosphatidylcholines; PEs, phosphatidylethanolamines; Cers, ceramides; PCs, phosphatidylcholines; SMs, sphingomyelins; FAs, fatty acids
Level distribution of differential lipids between CA and NR groups. Clustering heatmap was draw using R software by data transforming with log10. The majority of differential lipids in the CA group showed a tendency of significant decrease compared to NR group. Abbreviations: CA, colorectal adenoma; NR, normal control; DAG, diacylglycerol; TAG, triacylglycerol; LPC, lysophosphatidylcholine; PE, phosphatidylethanolamine; Cer, ceramide; PC, phosphatidylcholine; SM, sphingomyelin
Performance evaluation and verification of potential lipid biomarkers of CA
The diagnostic performance of 79 differential lipids between CA and NR was evaluated by ROC analysis using MetaboAnalyst 5.0, which could maximize the area under the curve (AUC) as calculated by the trapezoidal method to select the most suitable cut-off point. Before performing ROC analysis, sum normalization and auto-scaling were carried out for lipidomic data to effectively reduce the influence of individual differences and systematic errors. Generally, the AUC values of the differential lipids ranged from 0.616 to 1.000, and most of them had comparatively low AUC values (Table 2). By combining with AUC ≥ 0.900 as selected criteria, we obtained 12 differential lipids with good diagnostic performance for CA (Fig. 4), mainly including 7 PCs, 4 FAs lipids, and 1 MAG, and which were identified by matching the high resolution MS, MS/MS fragments, and RT from Thermo mzCloud and mzVault with Lipidblast databases (Fig. 5). Among them, PC 30:1, PC 44:5 and 4-dodecylbenzenesulfonic acid had the highest AUC values (1.000 (95% CI: 1.000–1.000)), indicating outstanding diagnostic ability for CA (Fig. 4), while PC 21:4 had the relatively low AUC value (0.900 (95% CI: 0.830–0.969)). Based on the ROC analysis, we further explored the change trend of levels for these 12 potential lipid biomarkers with good distinguish efficacy between two groups. The results showed that five lipids including PC 41:8, PC 36:3, palmitoyl ethanolamide, methyl palmitate, and palmitic acid were significantly up-regulated in the CA group, while the remaining seven lipids including 4-dodecylbenzenesulfonic acid, PC 44:5, PC 30:1, PC 31:2, PC 37:7, PC 21:4, and 2-arachidonoyl glycerol were remarkably down-regulated in the CA group compared with NR group (Fig. 6). Among them, PC 44:5, PC 30:1, palmitic acid and 4-dodecylbenzenesulfonic acid presented the most significant change trend with the fold change more than 10 (Table 2), meanwhile, which was consistent with the clustering heatmap of differential lipids between groups. Additionally, to further confirm the potential lipid biomarkers, we applied the commercial lipid standards to verify the potential lipid biomarkers of CA by matching the exact mass, retention time and fragmentation pattern under the same LC-MS conditions for lipidomic study (Fig. 7). Hence, the above 5 differential lipids identified with lipid standards served as the potential diagnostic biomarkers for CA.
The change trend of 12 differential lipids with high performance for CA diagnosis between two groups. The levels of differential lipids between CA and NR groups were displayed with mean ± SEM. The “★” represented statistical significance of the variate with P < 0.05 between two groups. Abbreviations: CA, colorectal adenoma; NR, normal control; AUC, area under the curve; PC, phosphatidylcholine; SEM, standard error of mean
Discussion
In this study, serum lipid profiles were found to be distinctly different between the NR and CA groups. After an efficient and robust analysis, a total of 79 differential lipids were found between groups. Among them, TAGs and PCs were the main lipid types, disclosing the metabolic perturbation of TAGs and PCs could be involved in the CA formation. Furthermore, 12 differential lipids showed good diagnostic performance as the potential biomarkers for CA. PCs and FAs are the main dysregulated lipid biomarkers, particularly, three lipids of PC 30:1, PC 44:5, and 4-dodecylbenzenesulfonic acid with outstanding diagnostic ability for CA.
Over the past decade, although some research has overwhelmingly focused on the discovery of potential diagnostic biomarkers of CRC, few studies have committed to exploring the lipid markers for CA diagnosis. Rachieriu C et al. analyzed the serum lipid profiles of CRC patients by HPLC-QTOF-MS, and found 25 potential markers with AUC values > 0.750, principally including PCs, Cers, FAs [26]. Similarly, plasma lipidomics showed that PG 34:0, SM 42:2, Cer 44:5, LPC 18:3, LPC 18:2, O-PE 36:3, O-PE 38:3 and SM 38:8 with good performance (AUC > 0.800) could act as promising diagnostic biomarkers for CRC screening [18]. Moreover, the combination of LPC 17:0, LPC 19:0, LPC 19:1 and LPC 19:2 could better distinguish between NR and CRC patients (AUC = 0.863) through a targeted lipidomic study [27]. As the severe stage of CA, colorectal advanced adenoma (CAA) was also considered as the effective target for CRC prevention. In our recent study, we disclosed that triglyceride (TAG) was the major dysregulated lipids in CAA, and 12 differential lipids served as the potential biomarkers of CAA diagnosis [28]. In addition to lipidomics, metabolomics is also used extensively in the discovery of lipid biomarkers for CA or CRC. Another study based on stool metabolomics showed that combination of ChoE 18:1, ChoE 18:2, ChoE 20:4, PE 16:0/18:1, SM d18:1/23:0, SM 42:3, and TAG 54:1 could effectively discriminate NR and CRC as the diagnostic biomarker, and its integrated performance was good (AUC = 0.821) [29]. In the plasma metabolomics, 48 differential metabolites were uncovered between CA and CRC, mainly including LPCs and PCs, and both of them were down-regulated in CRC [30]. In tissue metabolomics, PC 32:1 was suggested as an invaluable biomarker, which could be used for clinical diagnosis for CRC by imaging mass spectrometry [31]. PCs are the important lipid carrier in plasma, and phospholipids related to choline were considered good biomarkers of CRC [32, 33]. Furthermore, the most abundant metabolic features identified in the CA patients were PCs and PEs, and LPC (P-16:1) could be a putatively novel lipid signature [24]. The disorder of PC metabolism was believed to be strongly linked with the risk of CA [34]. Similarly, our previous study reported that metabolism of linoleic acid and phospholipid exhibited remarkable dysregulation in the CA patients by plasma metabolomics [22]. In this study, the PCs also acted as the main potential lipid markers for CA diagnosis (Figs. 2 and 4).
Apart from PCs, FAs are also potential diagnostic markers for distinguishing CRC or CA from NR according to the previous reports [19, 35]. Altered plasma levels of decanoic acid in CRC could serve as a new diagnostic biomarker [36]. Studies have noted a rise in the level of total TAGs in serum or plasma may be related to the elevated risk of CA [23]. Similar result that TAGs were the main dysregulated lipids in the CA group was observed in our study (Fig. 2). TAGs storage in adipose tissue is the major reservoir for energy metabolism in mammals. During lipolysis, FAs are hydrolyzed from TAGs stores and then transported to other tissues for catabolism [37]. So, the perturbation of TAG metabolism generally dysregulates the FA metabolism. Furthermore, FAs as essential components of biological membranes. It has been found that many cancer cells express higher levels of FAs than corresponding normal cells because cancer cells require substantial lipids for energy synthesis, signal transduction, and more membranes for vigorous metabolism [38, 39]. However, although few FAs including triheptanoin, palmitoyl ethanolamide, palmitic acid, oleoyl ethanolamide, methyl palmitate had increase levels in CA group, most of FAs presented significant down-regulation in CA (Table 2 and Fig. 3). Additionally, the fecal metabolome results found the level of palmitoyl ethanolamide in CA group was visibly higher than NR group, which could serve as putative biomarker of CA [25]. For serum metabolomics [40], the level of palmitic acid in CRC patients showed a noticeable up-regulation trend, meanwhile, which also exhibited significant increase with fold change of 17.64 in CA (Table 2 and Fig. 3), indicating which could be a candidate biomarker of CRC progression. In ROC analysis, the 12 potential lipid biomarkers presented good diagnostic performance for CA screening (AUC ≥ 0.900), containing 7 PCs, 4 FAs, and 1 MAG (Fig. 4), which may contribute to the early discovery and prevention of CRC. In summary, the perturbation of PCs and TAGs metabolism may be closely relevant to CA formation, and the PCs and FAs are the major dysregulated potential biomarkers for CA diagnosis. These discoveries should provide a valuable reference for the early screening and carcinogenesis of CRC.
Conclusions
To our knowledge, the present study is the first to explore lipid biomarkers for CA based on serum lipidomics with large-scale samples. In this research, obvious differences in the serum lipid profiles between CA and NR were observed, and 79 differential lipids were selected by UHPLC-ESI-HRMS-based serum lipidomics. TAGs and PCs were predominant components of differential lipids, indicating the abnormal metabolism of them should contribute to the formation of CA. In addition, 12 serum differential lipids were verified as the potential biomarkers for CA screening owing to their excellent diagnostic performance. Generally, this study provides a novel light into the lipid metabolism pathways associated with CA formation. Simultaneously, the discovery of lipid biomarkers for CA may also offer new insights for its clinical diagnosis. Due to limitations in sample sizes and study models, undoubtedly, further validation is needed for our discovery.
Availability of data and materials
The datasets supporting the conclusions of this article are included within the article.
Abbreviations
- UHPLC:
-
Ultra high performance liquid chromatography
- ESI:
-
Electrospray ionization
- HRMS:
-
High resolution mass spectrometry
- CA:
-
Colorectal adenoma
- NR:
-
Normal control
- CRC:
-
Colorectal cancer
- AUC:
-
Area under the curve
- RT:
-
Retention time
- m/z :
-
Mass to charge ratio
- MAGs:
-
Monoacylglycerols
- DAGs:
-
Diacylglycerols
- TAGs:
-
Triacylglycerols
- LPCs:
-
Lysophosphatidylcholines
- PEs:
-
Phosphatidylethanolamines
- Cers:
-
Ceramides
- PCs:
-
Phosphatidylcholines
- SMs:
-
Sphingomyelins
- FAs:
-
Fatty acids
References
Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, et al. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J Clin. 2021;71(3):209–49.
Lee PY, Chin S-F, Low TY, Jamal R. Probing the colorectal cancer proteome for biomarkers: current status and perspectives. J Proteome. 2018;187:93–105.
Sun M, Xu J, Shamul JG, Lu X, Husain S, He X. Creating a capture zone in microfluidic flow greatly enhances the throughput and efficiency of cancer detection. Biomaterials. 2019;197:161–70.
Vacante M, Ciuni R, Basile F, Biondi A. Gut microbiota and colorectal Cancer development: a closer look to the adenoma-carcinoma sequence. Biomedicines. 2020;8(11):489.
Song M, Lee IM, Manson JE, Buring JE, Dushkes R, Gordon D, et al. Effect of Supplementation With Marine ω-3 Fatty Acid on Risk of Colorectal Adenomas and Serrated Polyps in the US General Population: A Prespecified Ancillary Study of a Randomized Clinical Trial. JAMA Oncol. 2020;6(1):108–15.
Strum WB. Colorectal Adenomas. N Engl J Med. 2016;374(11):1065–75.
Poulose N, Amoroso F, Steele RE, Singh R, Ong CW, Mills IG. Genetics of lipid metabolism in prostate cancer. Nat Genet. 2018;50(2):169–71.
Silvente-Poirot S, Poirot M. Cancer. Cholesterol and cancer, in the balance. Science. 2014;343(6178):1445–6.
Atilla-Gokcumen GE, Muro E, Relat-Goberna J, Sasse S, Bedigian A, Coughlin ML, et al. Dividing cells regulate their lipid composition and localization. Cell. 2014;156(3):428–39.
Fernández R, Garate J, Lage S, Terés S, Higuera M, Bestard-Escalas J, et al. Identification of biomarkers of necrosis in xenografts using imaging mass spectrometry. J Am Soc Mass Spectrom. 2016;27(2):244–54.
Cheng C, Geng F, Cheng X, Guo D. Lipid metabolism reprogramming and its potential targets in cancer. Cancer Commun (Lond). 2018;38(1):27.
Wang Y, Hinz S, Uckermann O, Hönscheid P, von Schönfels W, Burmeister G, et al. Shotgun lipidomics-based characterization of the landscape of lipid metabolism in colorectal cancer. Biochim Biophys Acta Mol Cell Biol Lipids. 1865;2020(3):158579.
Cotte AK, Aires V, Fredon M, Limagne E, Derangère V, Thibaudin M, et al. Lysophosphatidylcholine acyltransferase 2-mediated lipid droplet production supports colorectal cancer chemoresistance. Nat Commun. 2018;9(1):322.
Wenk MR. Lipidomics: new tools and applications. Cell. 2010;143(6):888–95.
Cui L, Lu H, Lee YH. Challenges and emergent solutions for LC-MS/MS based untargeted metabolomics in diseases. Mass Spectrom Rev. 2018;37(6):772–92.
Yang K, Han X. Lipidomics: techniques, applications, and outcomes related to biomedical sciences. Trends Biochem Sci. 2016;41(11):954–69.
Mirnezami R, Spagou K, Vorkas PA, Lewis MR, Kinross J, Want E, et al. Chemical mapping of the colorectal cancer microenvironment via MALDI imaging mass spectrometry (MALDI-MSI) reveals novel cancer-associated field effects. Mol Oncol. 2014;8(1):39–49.
Shen S, Yang L, Li L, Bai Y, Cai C, Liu H. A plasma lipidomics strategy reveals perturbed lipid metabolic pathways and potential lipid biomarkers of human colorectal cancer. J Chromatogr B Analyt Technol Biomed Life Sci. 2017;1068–1069(8):41.
Liu T, Tan Z, Yu J, Peng F, Guo J, Meng W, et al. A conjunctive lipidomic approach reveals plasma ethanolamine plasmalogens and fatty acids as early diagnostic biomarkers for colorectal cancer patients. Expert Rev Proteomics. 2020;17(3):233–42.
Liu T, Peng F, Yu J, Tan Z, Rao T, Chen Y, et al. LC-MS-based lipid profile in colorectal cancer patients: TAGs are the main disturbed lipid markers of colorectal cancer progression. Anal Bioanal Chem. 2019;411(20):5079–88.
Mika A, Pakiet A, Czumaj A, Kaczynski Z, Liakh I, Kobiela J, et al. Decreased triacylglycerol content and elevated contents of cell membrane lipids in colorectal cancer tissue: a lipidomic study. J Clin Med. 2020;9(4):1095.
Zhang Q, Nong Y, Liu Z, Gong L. Proteinase K combining two-step liquid-liquid extraction for plasma untargeted liquid chromatography-mass spectrometry-based metabolomics to discover the potential mechanism of colorectal adenoma. Anal Chem. 2019;91(22):14458–66.
Pakiet A, Kobiela J, Stepnowski P, Sledzinski T, Mika A. Changes in lipids composition and metabolism in colorectal cancer: a review. Lipids Health Dis. 2019;18(1):29.
Fernandes Messias MC, Mecatti GC, Figueiredo Angolini CF, Eberlin MN, Credidio L, Real Martinez CA, et al. Plasma Lipidomic signature of rectal adenocarcinoma reveals potential biomarkers. Front Oncol. 2018;7:325.
Kim M, Vogtmann E, Ahlquist DA, Devens ME, Kisiel JB, Taylor WR, et al. Fecal metabolomic signatures in colorectal adenoma patients are associated with gut microbiota and early events of colorectal cancer pathogenesis. mBio. 2020;11(1):e03186–19.
Rachieriu C, Eniu DT, Mois E, Graur F, Socaciu C, Socaciu MA, et al. Lipidomic signatures for colorectal Cancer diagnosis and progression using UPLC-QTOF-ESI(+)MS. Biomolecules. 2021;11(3):417.
Zhang Q, Xu H, Liu R, Gao P, Yang X, Jin W, et al. A novel strategy for targeted Lipidomics based on LC-tandem-MS parameters prediction, quantification, and multiple statistical data mining: evaluation of Lysophosphatidylcholines as potential Cancer biomarkers. Anal Chem. 2019;91(5):3389–96.
Zhu Y, Wang L, Nong Y, Liang Y, Huang Z, Zhu P, et al. Serum untargeted UHPLC-HRMS-based Lipidomics to discover the potential biomarker of colorectal advanced adenoma. Cancer Manag Res. 2021;13:8865–78.
Cubiella J, Clos-Garcia M, Alonso C, Martinez-Arranz I, Perez-Cormenzana M, Barrenetxea Z, et al. Targeted UPLC-MS metabolic analysis of human Faeces reveals novel Low-invasive candidate markers for colorectal Cancer. Cancers (Basel). 2018;10(9):300.
Gumpenberger T, Brezina S, Keski-Rahkonen P, Baierl A, Robinot N, Leeb G, et al. Untargeted metabolomics reveals major differences in the plasma metabolome between colorectal Cancer and colorectal adenomas. Metabolites. 2021;11(2):119.
Kurabe N, Hayasaka T, Ogawa M, Masaki N, Ide Y, Waki M, et al. Accumulated phosphatidylcholine (16:0/16:1) in human colorectal cancer; possible involvement of LPCAT4. Cancer Sci. 2013;104(10):1295–302.
Zhao Z, Xiao Y, Elson P, Tan H, Plummer SJ, Berk M, et al. Plasma lysophosphatidylcholine levels: potential biomarkers for colorectal cancer. J Clin Oncol. 2007;25(19):2696–701.
Li S, Guo B, Song J, Deng X, Cong Y, Li P, et al. Plasma choline-containing phospholipids: potential biomarkers for colorectal cancer progression. Metabolomics. 2013;9(1):202–12.
Cho E, Willett WC, Colditz GA, Fuchs CS, Wu K, Chan AT, et al. Dietary choline and betaine and the risk of distal colorectal adenoma in women. J Natl Cancer Inst. 2007;99(16):1224–31.
Cottet V, Collin M, Gross AS, Boutron-Ruault MC, Morois S, Clavel-Chapelon F, et al. Erythrocyte membrane phospholipid fatty acid concentrations and risk of colorectal adenomas: a case-control nested in the French E3N-EPIC cohort study. Cancer Epidemiol Biomark Prev. 2013;22(8):1417–27.
Crotti S, Agnoletto E, Cancemi G, Di Marco V, Traldi P, Pucciarelli S, et al. Altered plasma levels of decanoic acid in colorectal cancer as a new diagnostic biomarker. Anal Bioanal Chem. 2016;408(23):6321–8.
Chitraju C, Mejhert N, Haas JT, Diaz-Ramirez LG, Grueter CA, Imbriglio JE, et al. Triglyceride Synthesis by DGAT1 protects adipocytes from lipid-induced ER stress during lipolysis. Cell Metab. 2017;26(2):407–18.e3.
Yi M, Li J, Chen S, Cai J, Ban Y, Peng Q, et al. Emerging role of lipid metabolism alterations in Cancer stem cells. J Exp Clin Cancer Res. 2018;37(1):118.
Stepanova DS, Semenova G, Kuo Y-M, Andrews AJ, Ammoun S, Hanemann CO, et al. An essential role for the tumor-suppressor Merlin in regulating fatty acid synthesis. Cancer Res. 2017;77(18):5026–38.
Farshidfar F, Weljie AM, Kopciuk KA, Hilsden R, McGregor SE, Buie WD, et al. A validated metabolomic signature for colorectal cancer: exploration of the clinical value of metabolomics. Br J Cancer. 2016;115(7):848–57.
Acknowledgments
The authors sincerely wish to thank the support and help of workers in State Key Laboratory for Conservation and Utilization of Subtropical Agro-Bioresources of Guangxi University.
Funding
This research was funded by the Fund Project of Guangxi University, Guangxi, China (No. A3370051006), the Natural Science Foundation of Guangxi, Guangxi, China (No. 2017GXNSFAA198310), and Bama County Program for Talents in Science and Technology, Guangxi, China (No. AE33700024).
Author information
Authors and Affiliations
Contributions
Conceptualization and investigation, Hailin Zhou and Yanying Nong.; writing—original draft preparation, Hailin Zhou.; methodology, Pingchuang Zhu and Jiahao Zhang.; formal analysis, Yunxiao Liang.; data curation, Yifan Zhu.; visualization, Yifan Zhu and Hongwei Chen.; writing—review and editing, project administration, and funding acquisition, Qisong Zhang. All authors have read and agreed to the published version of the manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
This study was approved by the Ethics Committee of the People’s Hospital of Guangxi Zhuang Autonomous Region (No.KY-DZX202008), and the written informed consent was obtained from each subject. The study was carried out in accordance with the Declaration of Helsinki.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Zhou, H., Nong, Y., Zhu, Y. et al. Serum untargeted lipidomics by UHPLC-ESI-HRMS aids the biomarker discovery of colorectal adenoma. BMC Cancer 22, 314 (2022). https://doi.org/10.1186/s12885-022-09427-1
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12885-022-09427-1