Abstract
Background
Several investigations have reported diverse roles of long non-coding RNA (lncRNA) in biological processes, tumor development, and progression of colorectal cancer (CRC). In this study, we investigated the lncRNA AC087388.1 tumorigenic role in CRC cells.
Methods
The CRC tissues were collected at the Reza Radiotherapy and Oncology Center, Mashhad, Iran. The human SW-48 and HT-29 CRC cell lines were obtained from the national cell bank of Iran. The cells were cultured according to ATCC (the American Type Culture Collection) recommendations. Quantitative real-time PCR was applied to assess the RNA expression. ShRNA transfection was done to downregulate the target gene. MTT and apoptosis assays were conducted to evaluate cell proliferation and viability, respectively. Colony formation assay, wound healing assay, and invasion assay were applied to determine growth, motility, and invasion of the cells, respectively. ENCORI online tool was used as downstream enrichment analysis.
Results
Forty CRC patients were encompassed in this study. The results demonstrated that the lncRNA SLC16A1-AS1, AC087388.1, and ELFN1-AS1 were significantly overexpressed in the CRC tissues in comparison to their normal counterpart margins. All the lncRNAs have shown significant Area Under Curve (AUC) values in the patients. Downregulation of lncRNA AC087388.1 remarkably decreased the cell proliferation and viability of the CRC cells. In addition, the data demonstrated that the downregulation of lncRNA AC087388.1 significantly suppressed cell growth and colony formation capability in the cells. Also, downregulation of lncRNA AC087388.1 attenuated motility and invasion of CRC cells, and significantly decreased the expression of invasion genes. In-silico functional enrichment analysis indicated that the lncRNA AC087388.1 has contributed to crucial signaling pathways in tumorigenesis such as the p53 and Wnt signaling pathways, apoptosis, and cell cycle.
Conclusions
Altogether, we showed that lncRNA AC087388.1 has an oncogenic role in tumorigenesis of CRC, and it can be considered as a novel diagnostic and prognostic biomarker in CRC.
Similar content being viewed by others
Background
Colorectal cancer (CRC) is one of the most frequent malignancies of gastrointestinal (GI) system and according to GLOBOCAN 2020, it accounts for the fifth leading cause of cancer-related death, globally [1, 2]. It has been demonstrated that CRC tumorigenesis is correlated to different kinds of genetic and epigenetic variations, and its development is a complex multi-step biological process [3,4,5]. However, the clear mechanism of CRC tumorigenesis is not completely understood yet.
The gold standard method for screening CRC patients is colonoscopy which combines diagnosis, and treatment, but it is an invasive approach. There are other screening methods such as guaiac fecal occult blood test (gFOBT), and fecal immunochemical test (FIT) which have a lack of sensitivity and specificity [6]. Due to the lack of early and precise diagnosis, and distance metastasis of CRC, the majority of CRC patients are diagnosed in the advanced stages with poor prognoses [7, 8].
In recent years, despite improving the CRC treatment approaches such as surgical resection, radiation, and chemotherapy, unfortunately, the 5-year survival rate of the patients is disappointing (less than 30%) [9, 10]. Therefore, there is an urgent need to discover and develop an efficient diagnostic and prognostic biomarker for CRC.
Recently, a large body of investigations reported diverse roles of non-coding RNA, particularly long non-coding RNA (lncRNA), in biological processes of different sorts of cancer [11,12,13]. LncRNAs are a group of non-coding RNAs with more than 200 nt in length and with no or little capability of coding proteins [14,15,16]. They have been demonstrated to play different canonical roles in diverse biological processes such as cell proliferation, differentiation, and cellular development, carcinogenesis, and metastasis through regulating cornerstone genes expression [4, 17]. Numerous investigations highlighted the crucial role of lncRNAs in cancer development and progression [18]. For instance, it has been demonstrated that lncRNA cCSC1 induced self-renewal capacity and drug resistance (stemness characteristics) in CRC cells through recruiting Hedgehog signaling pathway [19]. In another example, lncRNA SNHG16 has been illustrated to regulate cell proliferation, invasion and metastasis by upregulating MCP1 expression through sponging miR-124-3p in CRC cells [20]. lncRNA MALAT1 has been shown to induce resistance to irradiation in CRC cells via inhibiting miR-101-3p [21]. Altogether, the previous investigations proposed that lncRNAs can be considered as novel therapeutic targets and desired biomarkers in CRC.
According to our previous study [4], we comprehensively demonstrated lncRNA-miRNA-mRNA regulatory networks in patients with CRC by retrieving and analysis of RNA-seq data from The Cancer Genome Atlas (TCGA). Furthermore, we proposed numerous potential diagnostics, and prognostic lncRNA biomarkers such as SLC16A1-AS1, AC087388.1, and ELFN1-AS1 which indicated promising results. In the present study, we investigated these candidate lncRNAs in our patients, and finally, we demonstrated the tumorigenic role of lncRNA AC087388.1 in CRC cells.
Methods
Patients and tissue samples
The CRC tissues were collected by non-random sampling at the Reza radiotherapy, and oncology center, Mashhad, Iran. The age of the patients ranged from 24 to 83 years (mean age 57.25). A total of 40 CRC adenocarcinoma tissue samples were collected and confirmed by the pathological department. Informed consent was completed by participants at the beginning of the project. The study was approved by the Ethical Committee of Mashhad University of Medical Sciences (Code: IR.MUMS.MEDICAL.REC.1399.156).
Cell culture
The human SW-48 and HT-29 CRC cell lines were obtained from the National Cell Bank of Iran (NCI, Tehran, Iran). The cells were cultured, according to ATCC (the American Type Culture Collection) recommendations, in RPMI-1640 medium (for SW48), and DMEM (For HT29) medium, both media from Cegrogen Biothech GmbH, Germany supplemented with 10% fetal bovine serum (FBS, Biosera, France) and 1% penicillin–streptomycin antibiotics (Biosera, France) in a humidified incubator in 5% CO2 at 37 °C. The cells were regularly checked for mycoplasma contamination.
Quantitative Real-time PCR
RNA extraction was conducted by AccuZol™ (Bioneer, Korea) from the tissues and the cell lines. The quality and quantity of RNA extraction were evaluated by the 2% gel electrophoresis and a Nanodrop (Thermo Scientific, USA), respectively. cDNA synthesis was performed by the AccuPower RocketScript™ kit (Bioneer, Korea) according to the manual instruction. The total volume for this reaction was 20 μl that included 1 μg of total RNA. Quantitative Real-time PCR was applied to assess the RNA expression in the cells and tissues by a LightCycler® 96 System (Roche Life Science, Germany) using SYBR green-based kit, RealQ Plus Master Mix Green (Ampliqon, Copenhagen, Denmark). The total volume was 20 μL, including 10 μL of SYBR Green, 1 μL of primer (5 pmol), 2 μL of cDNA, and DEPC water. Thermal cycling conditions were comprised of an activation step at 95 °C for 15 min, followed by 40 cycles, including a denaturation step at 95 °C for 10 s and at 58 °C and 60 °C for 30 s for annealing and extension, respectively. The primer sequences of the target genes are listed in Table S1. GAPDH gene expression was considered as the reference gene. For calculation of relative expression, the 2–ΔΔCT formula was used.
Cell transfection
AC087388.1 small hairpin RNA (shRNA) was synthesized by Metabion (Munich, Germany). The sequence was 5′-GCAAGAATGAGTATATCTATACCTGACCCATATAGATATACTCATTCTTGCTTTTT -3′. A scrambled negative control shRNA was also ordered from Metabion (Munich, Germany). The sequence was 5′- CCGGTACCTCACGTCAGTGGTGATATAGATCAAGAGTCTATATCACCACTGACGTTTTG -3′. The cells lines were incubated with either AC087388.1 shRNA (shRNA) or negative control shRNA (as control) using polyethylenimine (PEI) transfection reagent (Merck KGaA, Darmstadt, Germany) according to the manufacturer protocol.
Cell viability assay
The CRC cells were cultures into 96 well plates (1 × 104 cells/well) for 24 h, 48 h, 72 h, 96 h, and 120 h. Following, the percentage of viable cells was determined by MTT (3-[4,5-dimethylthiazol-2-yl]-2,5 diphenyl tetrazolium bromide) assay, as 10 μl of MTT solution (5 mg/ml; Sigma) was added to each well and incubated with 5% CO2 at 37 °C for 4 h. Then, the supernatant was removed, and 100 μl of DMSO was added to each well as a solvent. Cell viability percentage was assessed by spectrophotometry at 570 and 630 nm using an absorbance microplate reader (BioTek ELx800, USA).
Apoptosis assay
Annexin V and PI staining was carried out using Annexin V/PI-FITC apoptosis detection kit (MabTaq, Germany) according to the manufacturer’s protocol. The results were analyzed using a Partec PAS III flow cytometer (Partec) and WindowsTM FloMax® software (Partec).
Colony formation assay
For colony formation assay, the CRC single-cell suspensions were cultured in 6-well collagen-coated plates (100 cells/well). The plates were further incubated for 7 days, and colonies were stained with 0.5% crystal violet and counted under an inverted microscope.
Wound healing assay
Approximately 1 × 104 the CRC cells were seeded into six-well collagen-coated plates. After overnight incubation, a linear wound was made in the confluent monolayer with a pipette tip. The cultures were washed with 1X phosphate buffer saline (PBS). The migration area was scanned after 5 days by an inverted microscope.
Invasion assay
For evaluating the invasion ability of the cancer cells, transwell culture system were carried out. The CRC cell suspension was seeded (1 × 105 cells/well) with a serum-free medium and cultured in the upper chamber of transwell cell culture chambers (8 mm pore size, Corning Inc., USA) precoated with Matrigel (BD Biosciences, USA). However, the lower chamber was filled with the medium containing 10% FBS. After 48 h incubation, the non-invasive cells remaining in the upper chamber were removed using a cotton swab and cells which passed through the inserts in the lower chamber were fixed with methanol and stained with 5% of crystal violet staining solution at room temperature for 20 min. A camera-equipped light microscope (Olympus, Japan) was applied counting the cells in the lower chamber. The number of invasive tumor cells was counted from five randomly selected 20 × fields per chamber for each assay which was conducted in triplicate.
In silico functional enrichment analysis
For more illustration, the functional enrichment analysis of lncRNA AC087388.1 was carried out by applying an online tool, ENCORI: The Encyclopedia of RNA Interactomes (http://starbase.sysu.edu.cn/) to demonstrate considerable CE-RNA networks and KEGG (Kyoto Encyclopedia of Genes and Genomes) signaling pathway analysis [22].
Statistical analysis
All data are presented as mean ± standard deviation (SD) and were evaluated in triplicate against control and collected from three independent experiments. Data were graphed and analyzed by GraphPad Prism Software 7.0 using a two-tailed Student’s t-test for comparing the means between two independent groups, respectively. ROC curve analysis was conducted by SPSS v21. ROC curve was calculated according to the sample of the patients and counterpart control group, and the events was considered as tumor positive participants. P-value < 0.05 was considered as a statistically significant threshold.
Results
In our previous study [4], we retrieved the public RNA-seq, miR-seq, and corresponding clinical data of 459 patients with CRC (primary tumor: 459, and adjacent normal solid tissue: 41) from the TCGA database. The differential gene expression was conducted by the “limma” package in R. Briefly, we demonstrated that 2995 mRNAs, 205 lncRNAs, and 345 miRNAs were differentially expressed in CRC. Gene ontology (GO) and KEGG signaling pathway were conducted and we demonstrated that the main number of the differentially expressed genes were enriched in important pathways in CRC. Furthermore, protein–protein interaction (PPI) was constructed by the STRING database, indicating that the CDKN2A, CCND1, MYC, E2F, CDK4, BRCA2, CDC25B, and CDKN1A proteins were the imperative signaling hubs. In addition, ceRNA network data showed the lncRNA-miRNA-mRNA interaction in the CRC patients (Tables S2 & S3). The diagnostic and prognostic values were evaluated for differentially expressed genes and finally, the data suggested 14 lncRNA as potential novel biomarkers in CRC. The data were sorted according to diagnostic and prognostic values, and the top three genes (lncRNA SLC16A1-AS1 (chr1:112,956,415–112,964,072, intergenic), AC087388.1 (chr17:7,685,260–7,686,371, intronic), ELFN1-AS1 (chr7:1,738,630–1,742,310, intronic) were selected for further investigation in the current study. We investigated the lncRNAs expression in collected CRC samples and determined the role of lncRNA AC087388.1 in CRC tumorigenesis.
Forty patients were enrolled in the study
Forty CRC patients were encompassed in this study. All the patients’ tumors were CRC adenocarcinoma (with different grades). Twenty-three patients were male and 17 of them were female. According to the median age of the patients, 20 ones were more than 58 years and 20 were equal or less than 58 years old. Other features of the patients including tumor size, TNM staging, grading, KRAS, BRAF, and NRAS mutation status were summarized in Table 1.
LncRNA SLC16A1-AS1, AC087388.1, and ELFN1-AS1 showed overexpression in the CRC tissues
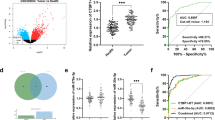
To explore the lncRNA SLC16A1-AS1, AC087388.1, and ELFN1-AS1 expressions in the CRC patients, Quantitative Real-time PCR was applied. The results demonstrated that the lncRNAs were significantly overexpressed in the CRC tissues in comparison to their normal counterpart margins (Fig. 1). Furthermore, we compared the TCGA lncRNA expression data to our patients. They indicated a similar pattern in the same direction (Table S4). Moreover, for determining diagnostic values, ROC curve analyses were conducted. All lncRNAs had significant Area Under Curve (AUC) values. The data are presented in Fig. 1 and Table 2.
Furthermore, we evaluated the gene expression in the different groups of patients according to clinicopathological characteristics. The data showed that the expression of lncRNA AC087388.1 in age 58 < is higher than ≤ 58 years, and expression of lncRNA SLC16A1-AS1 in BRAF negative mutation is significantly higher than BRAF positive group. All the data are presented in Table 3. Moreover, we assessed the association of the lncRNAs expression and clinicopathological characteristics. The lncRNAs expression were divided into low and high expressions according to median expression. The data demonstrated that an increase in age was associated with a significantly high expression of the lncRNA AC087388.1. However, the results did not demonstrate any significant association between the high or low expression and clinicopathological characteristics in the patients. The data are summarized in Table 4.
AC087388.1 small hairpin RNA (shRNA) downregulated lncRNA AC087388.1 in CRC cells
In the next step, by considering the top list lncRNAs and a lack of sufficient studies on the novel candidate lncRNAs, we selected lncRNA AC087388.1 for further investigation. By applying shRNA against AC087388.1 in the CRC cell lines (SW-48 and HT-29), we established stable cell lines producing the shRNA constantly (shRNA). In this study, we used a scrambled shRNA as a negative control (Control). The data illustrated that the shRNA significantly reduced the expression of the lncRNA AC087388.1 in comparison to the control in both cell lines. Figure 2 presents the data.
Downregulation of lncRNA AC087388.1 suppresses cell proliferation and viability
To evaluate the cell proliferation and viability in downregulation condition of lncRNA AC087388.1, we applied MTT and apoptosis assays. According to the MTT assay results, the proliferation of the shRNA-treated cells was significantly suppressed in comparison to the controls. Also, the apoptosis data showed that downregulation of lncRNA AC087388.1 remarkably decreased cell viability and increased early apoptosis in comparison to the control in the CRC cells. The data are presented in Fig. 3.
Downregulation of lncRNA AC087388.1 remarkably decreased cell viability. A MTT assay for SW-48 cell. B MTT assay for HT-29 cell. C Apoptosis fraction graph of the cells, apoptotic cell death was measured by annexin V staining after 24 h. Annexin V-positive cells are considered early apoptotic, whereas PI uptake indicates necrosis. Cells positive for both stains are considered apoptotic cells. D The percentage of the viable, necrosis, early, and late apoptosis
Downregulation of lncRNA AC087388.1 suppresses cellular growth and colony formation capability
Cell growth and colony formation capability of single-cell suspension were assessed by colony formation assay. The data demonstrated that downregulation of lncRNA AC087388.1 significantly suppressed cell growth and colony formation capability in comparison to the control. The data are presented in Fig. 4.
Downregulation of lncRNA AC087388.1 attenuates cell motility and invasion
We investigated the motility and migration ability of the CRC shRNA-treated cells. Wound healing assay revealed that the downregulation of lncRNA AC087388.1 attenuated motility of the SW-48 cells in comparison with the control group (Fig. 5A). In addition, expression of migration and invasion contributor genes were evaluated by Quantitative Real-time PCR. The data demonstrated that the downregulation of lncRNA AC087388.1 remarkably decreased expression of Vimentin, MMP9, FN1, and N-Cadherin in the SW-48 cell line (Fig. 5B). Furthermore, hereby in transwell cell migration and invasion assay, we showed that the cell invasion and migration of the SW-48 cells decreased (Fig. 5C).
Downregulation of lncRNA AC087388.1 attenuates cell motility and invasion ability. A Downregulation of lncRNA AC087388.1 inhibited the motility of the SW-48 cells as demonstrated by reduced width in would healing assay. B The expression of vimentin, MMP9, FN1, and N-Cadherin were significantly reduced in the downregulation of lncRNA AC087388.1 condition. C As the transwell cell migration and invasion assay represent, the downregulation of lncRNA AC087388.1 inhibits invasion and migration of the CRC cells
LncRNA AC087388.1 has a contribution in canonical signaling pathways in cancer
For further investigation on lncRNA AC087388.1 roles in CRC and to demonstrate the downstream signaling pathways, we conducted In-silico functional studies. In-silico functional enrichment analysis of the lncRNA AC087388.1 by ENCORI online tool, demonstrated that the lncRNA AC087388.1 can regulate varieties of the genes in the human cells (Table 5). Furthermore, the gene set enrichment by KEGG pathway analysis showed that many of the genes were enriched in crucial signaling pathways in cancer such as the p53 signaling pathway, Wnt Signaling pathway, apoptosis, and cell cycle. The data are presented in Fig. 6.
Discussion
CRC is one of the common leading cancer-related deaths with an increasing trend in the world [23]. The early-stage diagnosis of CRC can provide the desired outcome in the patients [24]. Despite huge efforts in developing diagnostic and prognostic methods, a large body of patients is diagnosed in advanced stages, which have shown frustrating outcomes [7]. Thus, an in-depth understanding of CRC’s underlying mechanisms is pivotal. Recently, a number of the investigation highlighted the roles and function of lncRNAs in various cancers particularly in CRC [4, 6, 15, 25, 26]. Recently, we demonstrated overexpression of lncRNA SLC16A1-AS1, AC087388.1, and ELFN1-AS1 in CRC patients on the report of the TCGA public database. Furthermore, due to desired prognostic and diagnostic outcome, we indicated that lncRNA SLC16A1-AS1, AC087388.1, and ELFN1-AS1 could be considered as potential biomarkers in CRC patients [4]. In the current study, we broadly explored the role and the in vitro function of the lncRNA AC087388.1 in CRC cells. The data showed overexpression of lncRNA AC087388.1 in our CRC patients. Furthermore, the data demonstrated that downregulation of lncRNA AC087388.1 inhibits cell proliferation, growth, and invasion in CRC cells.
Cell proliferation and growth are important in tumorigenesis, being the hallmark of CRC [27, 28]. A variety of signaling pathways such as phosphatidylinositol-3-kinase/protein kinase B (PI3K/AKT) play key roles in cancer cell growth and proliferation [29, 30]. A large body of investigation has shown that lncRNA could regulate cell proliferation and growth in CRC. For instance, it has been shown that the novel lncRNA LINC00460 has been associated with large tumor size, advanced stages of cancer, and poor prognosis in the CRC patients, and has an impact on cell proliferation and apoptosis via sponging EZH2 and miR-149-5p to upregulating KLF2 and CUL4A in CRC, respectively [31]. Furthermore, lncRNA CRNDE (Colorectal Neoplasia Differentially Expressed) has been illustrated overexpression in CRC patients and has been associated with worse clinicopathological outcomes and poor prognosis. lncRNA CRNDE enhances tumorigenesis through epigenetically silencing dual-specificity phosphatase 5 (DUSP5) and CDKN1A by recruiting EZH2 (enhancer of zeste homolog 2) in CRC cells [32]. Constant with previous studies, we presented that lncRNA AC087388.1 overexpressed in our CRC patients. Moreover, it has been shown that the downregulation of lncRNA AC087388.1 remarkably decreased cell proliferation, growth, and cell viability in CRC cells.
Another, crucial hallmark of cancer is invasion and metastasis [27]. Several investigations indicating different sorts of signaling pathways that have a main contribution to invasion and metastasis including epithelial NOTCH, MAPK, STAT3 signaling pathways [33,34,35,36]. Many studies have indicated the lncRNA regulatory effects on invasion and metastasis in CRC cells [37,38,39]. LncRNAs can control cell invasion and metastasis by regulating different signaling pathways such as PI3K/AKT signaling pathway, EGFR/MAPK pathway, and hypoxia-induced signaling pathway in CRC [40,41,42]. For instance, it has been demonstrated that lncRNA SNHG5 enhances cell proliferation and metastasis by increasing CREB5 through downregulating miR-132-3p in CRC cells [43]. According to our results, downregulating of lncRNA AC087388.1 could attenuate cell mobility and invasion in the CRC cells. Furthermore, it reduced the colony formation ability of the cells from single CRC suspension cells. Tumor invasion and migration occur when the basement membranes and extracellular matrix (ECM) are dissolved by matrix metalloproteinases (MMPs) [44, 45]. MMPs are a group of zinc-dependent endopeptidases that work towards ECM turnover [46]. Vimentin, MMP9, FN1, and N-Cadherin are the well-known genes that have the main contribution to metastasis and EMT (Epithelial-to-mesenchymal transition) in cancer [47]. In this study, we showed that downregulation of AC087388.1 remarkably reduced expression of invasion and migration genes including Vimentin, MMP9, FN1, and N-Cadherin in CRC cells which explained the invasive role of this lncRNA.
For more illustration of lncRNA AC087388.1 roles in CRC In-silico functional study was applied to demonstrating the downstream signaling pathways and canonical signaling hubs. The in-silico analysis of our study noticeably demonstrated that the lncRNA AC087388.1 could drive tumorigenesis in various cancers such as prostate cancer, chronic myeloid leukemia, melanoma, and CRC. There are varieties of crucial signaling pathways in CRC which have a major contribution to tumorigenesis such as the Wnt signaling pathway, neurotrophin signaling pathway, p53 signaling pathway [48,49,50,51,52]. In the present study, we reported that the lncRNA AC087388.1 can control a variety of signaling pathways such as the Wnt signaling pathway, neurotrophin signaling pathway, cell cycle and apoptosis, and p53 signaling pathway.
Conclusions
To the best of our knowledge, for the first time, we showed that lncRNA AC087388.1 has an oncogenic role in tumorigenesis of CRC. lncRNA AC087388.1 can be considered as a novel diagnostic and prognostic biomarker in CRC. This study sheds light for further investigation and paves the way for researchers in the field of cancer and lncRNA. Further investigations are needed to illustrate the detailed role of lncRNA AC087388.1 in tumorigenesis particularly in CRC.
Availability of data and materials
The datasets generated and/or analysed during the current study are available in the TCGA database repository, [https://portal.gdc.cancer.gov/].
Abbreviations
- lncRNA:
-
Long non-coding RNA
- CRC:
-
Colorectal cancer
- ATCC:
-
The American Type Culture Collection
- AUC:
-
Area Under Curve
- GI:
-
Gastrointestinal
- gFOBT:
-
Guaiac fecal occult blood test
- FIT:
-
Fecal immunochemical test
- TCGA:
-
The Cancer Genome Atlas
- NCBI:
-
National Cell Bank of Iran
- ATCC:
-
The American Type Culture Collection
- FBS:
-
Fetal bovine serum
- PEI:
-
Polyethylenimine
- MTT:
-
3-[4,5-Dimethylthiazol-2-yl]-2,5 diphenyl tetrazolium bromide
- PBS:
-
Phosphate buffer saline
- ENCORI:
-
The Encyclopedia of RNA Interactomes
- SD:
-
Standard deviation
- GO:
-
Gene ontology
- PPI:
-
Protein–protein interaction
- AUC:
-
Area Under Curve
- KEGG:
-
Kyoto Encyclopedia of Genes and Genomes
- PI3K/AKT :
-
Phosphatidylinositol-3-kinase/protein kinase B
- CRNDE :
-
Colorectal Neoplasia Differentially Expressed
- DUSP5 :
-
dual-specificity phosphatase 5
- EZH2 :
-
Enhancer of zeste homolog 2
- MMPs:
-
Matrix metalloproteinases
- EMT:
-
Epithelial-to-mesenchymal transition
References
Yan Y, Wang Z, Qin B. A novel long noncoding RNA, LINC00483 promotes proliferation and metastasis via modulating of FMNL2 in CRC. Biochem Biophys Res Commun. 2019;509(2):441–7.
Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA cancer J Clin. 2018;68(6):394–424.
Sun J, Jia H, Bao X, Wu Y, Zhu T, Li R, et al. Tumor exosome promotes Th17 cell differentiation by transmitting the lncRNA CRNDE-h in colorectal cancer. Cell Death Dis. 2021;12(1):1–14.
Poursheikhani A, Abbaszadegan MR, Nokhandani N, Kerachian MA. Integration analysis of long non-coding RNA (lncRNA) role in tumorigenesis of colon adenocarcinoma. BMC Med Genomics. 2020;13(1):1–16.
Nokhandani N, Poursheikhani A, NaghaviAlhosseini M, Davoodi H. Bacteria in carcinogenesis and cancer prevention: a review study. Int J Cancer Manag. 2021;14(2):e107956.
Xu W, Zhou G, Wang H, Liu Y, Chen B, Chen W, et al. Circulating lncRNA SNHG11 as a novel biomarker for early diagnosis and prognosis of colorectal cancer. Int J Cancer. 2020;146(10):2901–12.
Mo S, Zhang L, Dai W, Han L, Wang R, Xiang W, et al. Antisense lncRNA LDLRAD4-AS1 promotes metastasis by decreasing the expression of LDLRAD4 and predicts a poor prognosis in colorectal cancer. Cell Death Dis. 2020;11(2):1–16.
Soltani G, Poursheikhani A, Yassi M, Hayatbakhsh A, Kerachian M, Kerachian MA. Obesity, diabetes and the risk of colorectal adenoma and cancer. BMC Endocr Disord. 2019;19(1):1–10.
Sun S, Xia C, Xu Y. HIF-1α induced lncRNA LINC00511 accelerates the colorectal cancer proliferation through positive feedback loop. Biomed Pharmacother. 2020;125:110014.
Liu J, Huang S, Liao X, Chen Z, Li L, Yu L, et al. LncRNA EWSAT1 promotes colorectal cancer progression through sponging miR-326 to modulate FBXL20 expression. Onco Targets Ther. 2021;14:367.
He Q, Long J, Yin Y, Li Y, Lei X, Li Z, et al. Emerging roles of lncRNAs in the formation and progression of colorectal cancer. Front Oncol. 2020;9:1542.
Yousefi H, Poursheikhani A, Bahmanpour Z, Vatanmakanian M, Taheri M, Mashouri L, et al. SARS-CoV infection crosstalk with human host cell noncoding-RNA machinery: an in-silico approach. Biomed Pharmacother. 2020;130:110548.
Rafieenia F, Abbaszadegan MR, Poursheikhani A, Razavi SMS, Jebelli A, Molaei F, et al. In silico evidence of high frequency of miRNA-related SNPs in esophageal squamous cell carcinoma. J Cell Physiol. 2020;235(2):966–78.
Xu L, Yu X, Wei B, Hui H, Sun Y, Dai J, et al. LncRNA SNHG7 promotes the proliferation of esophageal cancer cells and inhibits its apoptosis. Eur Rev Med Pharmacol Sci. 2018;22(9):2653–61.
Poursheikhani A, Bahmanpour Z, Razmara E, Mashouri L, Taheri M, Morshedi Rad D, et al. Non-coding RNAs underlying chemoresistance in gastric cancer. Cell Oncol. 2020;43:1–28.
Yan Y, Fan Q, Wang L, Zhou Y, Li J, Zhou K. LncRNA Snhg1, a non-degradable sponge for miR-338, promotes expression of proto-oncogene CST3 in primary esophageal cancer cells. Oncotarget. 2017;8(22):35750.
Poursheikhani A, Abbaszadegan MR, Kerachian MA. Mechanisms of long non-coding RNA function in colorectal cancer tumorigenesis. Asia Pac J Clin Oncol. 2021;17(1):7–23.
Zhu Y, Hu H, Yuan Z, Zhang Q, Xiong H, Hu Z, et al. LncRNA NEAT1 remodels chromatin to promote the 5-Fu resistance by maintaining colorectal cancer stemness. Cell Death Dis. 2020;11(11):1–11.
Zhou H, Xiong Y, Peng L, Wang R, Zhang H, Fu Z. LncRNA-cCSC1 modulates cancer stem cell properties in colorectal cancer via activation of the Hedgehog signaling pathway. J Cell Biochem. 2020;121(3):2510–24.
Chen ZY, Wang XY, Yang YM, et al. LncRNA SNHG16 promotes colorectal cancer cell proliferation, migration, and epithelial–mesenchymal transition through miR-124–3p/MCP-1. Gene Ther. 2020; https://doi.org/10.1038/s41434-020-0176-2.
Guo J, Ding Y, Yang H, Guo H, Zhou X, Chen X. Aberrant expression of lncRNA MALAT1 modulates radioresistance in colorectal cancer in vitro via miR-101–3p sponging. Exp Mol Pathol. 2020;115:104448.
Li J-H, Liu S, Zhou H, Qu L-H, Yang J-H. starBase v2. 0: decoding miRNA-ceRNA, miRNA-ncRNA and protein–RNA interaction networks from large-scale CLIP-Seq data. Nucleic Acids Res. 2014;42(D1):D92–7.
Dacheng W, Songhe L, Weidong J, Shutao Z, Jingjing L, Jiaming Z. LncRNA SNHG3 promotes the growth and metastasis of colorectal cancer by regulating miR-539/RUNX2 axis. Biomed Pharmacother. 2020;125:110039.
Luan Y, Li X, Luan Y, Zhao R, Li Y, Liu L, et al. Circulating lncRNA UCA1 promotes malignancy of colorectal cancer via the miR-143/MYO6 Axis. Mol Ther Nucleic Acids. 2020;19:790–803.
Poursheikhani A, Nokhandani N, Yousefi H, Rad DM, Sahebkar A. Clinicopathological significance of long non-coding RNA GHET1 in human cancers: a meta-analysis. Curr Pharm Biotechnol. 2020;21(14):1422–32.
Zhao K, Ye Z, Li Y, Li C, Yang X, Chen Q, et al. LncRNA FTX contributes to the progression of colorectal cancer through regulating miR-192-5p/EIF5A2 axis. Onco Targets Ther. 2020;13:2677.
Zamarin D, Holmgaard RB, Subudhi SK, Park JS, Mansour M, Palese P, et al. Localized oncolytic virotherapy overcomes systemic tumor resistance to immune checkpoint blockade immunotherapy. Sci Transl Med. 2014;6(226):226ra32-ra32.
Poursheikhani A, Yousefi H, Tavakoli-Bazzaz J, Ghaffari SH. EGFR blockade reverses cisplatin resistance in human epithelial ovarian cancer cells. Iran Biomed J. 2020;24(6):370.
Yousefi H, Momeny M, Ghaffari SH, Parsanejad N, Poursheikhani A, Javadikooshesh S, et al. IL-6/IL-6R pathway is a therapeutic target in chemoresistant ovarian cancer. Tumori. 2019;105(1):84–91.
Hou B, Li W, Li J, Ma J, Xia P, Liu Z, et al. Tumor suppressor LHPP regulates the proliferation of colorectal cancer cells via the PI3K/AKT pathway. Oncol Rep. 2020;43(2):536–48.
Lian Y, Yan C, Xu H, Yang J, Yu Y, Zhou J, et al. A novel lncRNA, LINC00460, affects cell proliferation and apoptosis by regulating KLF2 and CUL4A expression in colorectal cancer. Mol Ther Nucleic Acids. 2018;12:684–97.
Ding J, Li J, Wang H, Tian Y, Xie M, He X, et al. Long noncoding RNA CRNDE promotes colorectal cancer cell proliferation via epigenetically silencing DUSP5/CDKN1A expression. Cell Death Dis. 2017;8(8):e2997-e.
Jackstadt R, van Hooff SR, Leach JD, Cortes-Lavaud X, Lohuis JO, Ridgway RA, et al. Epithelial NOTCH signaling rewires the tumor microenvironment of colorectal cancer to drive poor-prognosis subtypes and metastasis. Cancer Cell. 2019;36(3):319-36.e7.
Peng W, Li J, Chen R, Gu Q, Yang P, Qian W, et al. Upregulated METTL3 promotes metastasis of colorectal cancer via miR-1246/SPRED2/MAPK signaling pathway. J Exp Clin Cancer Res. 2019;38(1):1–16.
Bian Z, Zhang J, Li M, Feng Y, Wang X, Zhang J, et al. LncRNA–FEZF1-AS1 promotes tumor proliferation and metastasis in colorectal cancer by regulating PKM2 signaling. Clin Cancer Res. 2018;24(19):4808–19.
Vojdani S, Ghaderian SMH, Zali A, Rakhshan A, Yazdani SO, Poursheikhani A, et al. Altered expression of EGFR and miR-34a derived from serum and tumoral tissue was associated with glioblastoma multiform. Exp Mol Pathol. 2021;121:104655.
Liang Z-x, Liu H-s, Wang F-w, Xiong L, Zhou C, Hu T, et al. LncRNA RPPH1 promotes colorectal cancer metastasis by interacting with TUBB3 and by promoting exosomes-mediated macrophage M2 polarization. Cell Death Dis. 2019;10(11):1–17.
Shan Y, Ma J, Pan Y, Hu J, Liu B, Jia L. LncRNA SNHG7 sponges miR-216b to promote proliferation and liver metastasis of colorectal cancer through upregulating GALNT1. Cell Death Dis. 2018;9(7):1–13.
Zhang Y, Liu X, Li Q, Zhang Y. lncRNA LINC00460 promoted colorectal cancer cells metastasis via miR-939-5p sponging. Cancer Manag Res. 2019;11:1779.
Wang Y, Kuang H, Xue J, Liao L, Yin F, Zhou X. LncRNA AB073614 regulates proliferation and metastasis of colorectal cancer cells via the PI3K/AKT signaling pathway. Biomed Pharmacother. 2017;93:1230–7.
Tang R, Chen J, Tang M, Liao Z, Zhou L, Jiang J, et al. LncRNA SLCO4A1-AS1 predicts poor prognosis and promotes proliferation and metastasis via the EGFR/MAPK pathway in colorectal cancer. Int J Biol Sci. 2019;15(13):2885.
Zhang W, Yuan W, Song J, Wang S, Gu X. LncRNA CPS1-IT1 suppresses EMT and metastasis of colorectal cancer by inhibiting hypoxia-induced autophagy through inactivation of HIF-1α. Biochimie. 2018;144:21–7.
Zhang M, Li Y, Wang H, Yu W, Lin S, Guo J. LncRNA SNHG5 affects cell proliferation, metastasis and migration of colorectal cancer through regulating miR-132-3p/CREB5. Cancer Biol Ther. 2019;20(4):524–36.
Castro MG, Campos LE, Rodriguez YI, Alvarez SE. In vitro methods to study the modulation of migration and invasion by sphingosine-1-phosphate. Sphingosine-1-Phosphate. Methods Mol Biol. 2018;1697:117–31.
Edatt L, Maurya AK, Raji G, Kunhiraman H, Kumar SV. MicroRNA106a regulates matrix metalloprotease 9 in a sirtuin-1 dependent mechanism. J Cell Physiol. 2018;233(1):238–48.
Fares J, Fares MY, Khachfe HH, Salhab HA, Fares Y. Molecular principles of metastasis: a hallmark of cancer revisited. Signal Transduct Target Ther. 2020;5(1):1–17.
Chen Z, Tao Q, Qiao B, Zhang L. Silencing of LINC01116 suppresses the development of oral squamous cell carcinoma by up-regulating microRNA-136 to inhibit FN1. Cancer management and research. 2019;11:6043.
Spengler G, Molnar J, Viveiros M, Amaral L. Thioridazine induces apoptosis of multidrug-resistant mouse lymphoma cells transfected with the human ABCB1 and inhibits the expression of P-glycoprotein. Anticancer Res. 2011;31(12):4201–5.
Griffin N, Faulkner S, Jobling P, Hondermarck H. Targeting neurotrophin signaling in cancer: the renaissance. Pharmacol Res. 2018;135:12–7.
Feng Q, Song D, Wang X. Pan-cancer analysis reveals that neurotrophin signaling correlates positively with anti-tumor immunity, clinical outcomes, and response to targeted therapies and immunotherapies in cancer. Life Sci. 2021;282:119848.
Slattery ML, Mullany LE, Wolff RK, Sakoda LC, Samowitz WS, Herrick JS. The p53-signaling pathway and colorectal cancer: interactions between downstream p53 target genes and miRNAs. Genomics. 2019;111(4):762–71.
Li XL, Subramanian M, Jones MF, Chaudhary R, Singh DK, Zong X, et al. Long noncoding RNA PURPL suppresses basal p53 levels and promotes tumorigenicity in colorectal cancer. Cell Rep. 2017;20(10):2408–23.
Acknowledgements
Our special thanks go to Ms. Marjan Azghandi for her technical assistance. This study was part of a Ph.D. dissertation (AP).
Funding
This study was supported financially by Mashhad University of Medical Sciences (grant# 981287), which played no role in the design of the study and collection, analysis, and interpretation of data and in writing the manuscript.
Author information
Authors and Affiliations
Contributions
AP, MRA, and MAK were all participated in study design, data analysis, and preparation of the drafted manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Informed written consent was obtained from all participants. The current study was approved by Mashhad University of Medical Sciences (MUMS) ethics committee (Ethical Code# IR.MUMS.MEDICAL.REC.1399.156).
Consent for publication
Not Applicable
Competing interests
The authors have no relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript. This includes employment, consultancies, honoraria, stock ownership or options, expert testimony, grants, or patents received or pending, or royalties.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Additional file 1:
Table S1. The primer sequence sets of the genes.
Additional file 2: Table S2.
The number of miRNA interactions to lncRNAs and mRNAs.
Additional file 3: Table S3.
The miRNA targets to lncRNAs and mRNAs.
Additional file 4: Table S4.
Comparisonof the TCGA gene expression with our patients.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Poursheikhani, A., Abbaszadegan, M.R. & Kerachian, M.A. Long non-coding RNA AC087388.1 as a novel biomarker in colorectal cancer. BMC Cancer 22, 196 (2022). https://doi.org/10.1186/s12885-022-09282-0
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12885-022-09282-0