Abstract
Background
Little data is available on prognostic biomarkers and effective treatment options for Kidney Renal Papillary Cell Carcinoma (KIRP) patients, to find potential prognostic biomarkers and new targets was an urgent mission for KIRP therapy.
Methods
The differentially expressed autophagy-related genes (DEARGs) were screened out according to the RNA sequencing data in The Cancer Genome Atlas database, then identified survival-related DEARGs to establish a prognostic model for survival predicting of KIRP patients. Then we verified the robustness and validity of the prognostic risk model through clinicopathological data. At last, we evaluate the prognostic value of genes that formed the prognostic risk model individually.
Results
We analyzed the expression of 232 autophagy-related genes (ARGs) in 289 KIRP and 32 non-tumor tissue cases, and 40 mRNAs were screened out as DEARGs. The functional and pathway enrichment analysis was done and protein–protein interaction network was constructed for all DEARGs. To sift candidate DEARGs associated with KIRP patients’ survival and create an autophagy-related risk prognostic model, univariate and multivariate Cox regression analysis were did separately. Eventually 3 desirable independent prognostic DEARGs (P4HB, NRG1, and BIRC5) were picked out and used for construct the autophagy-related risk model. The accuracy of the prognostic risk model for survival prediction was assessed by Kaplan–Meier plotter, receiver-operator characteristic curve, and clinicopathological correlational analyses. The prognostic value of above 3 genes was verified individually by survival analysis and expression analysis on mRNA and protein level.
Conclusions
The autophagy-related prognostic model is accurate and applicable, it can predict OS independently for KIRP patients. Three independent prognostic DEARGs can benefit for facilitate personalized target treatment too.
Similar content being viewed by others
Background
Renal cell carcinoma (RCC) is the sixth/eighth most common tumor in men and women. About 73,750 new cases of RCC and 14,830 RCC-related deaths happened yearly in the United States [1, 2]. Kidney Renal Papillary Cell Carcinoma (KIRP) is the second most commonly diagnosed subtype of RCC which accounting for 15–20% of RCC cases, and the most common subtype is clear cell renal carcinoma (ccRCC) [3, 4]. It has been widely assumed that KIRP has a significantly better prognosis than ccRCC in organ-confined stages [5, 6]. Most KIRP were manifested as localized disease and treated by partial nephrectomy, but substantial numbers of patients will eventually relapse [7]. About 1/3 of KIRP were manifested as metastatic KIRP (m-KIRP) [8]. Most m-KIRP patients needed systemic treatment eventually. In general, m-KIRP had a worse prognosis than metastatic ccRCC [9, 10]. As most KIRP patients had a good prognosis before metastasis, and m-KIRP patients are not so many in contrast to ccRCC, there is little data on the efficacy of available treatment options and few prognostic molecular markers have been discovered. Therefore, there is a need to explore potential prognostic markers and new molecular targets for KIRP therapy.
Traditionally, the TNM stage has been used to evaluate the risk of tumor recurrence for all RCC subtypes. However, it has limited accuracy [11]. Now, some prognostic factors such as grade and pathology stage are also used to evaluate the prognosis. However, these prognostic models were often established for ccRCC only or all RCC subtypes [12, 13]. Therefore, there is a need to refine the prognostic risk model of KIRP and build a more accurate approach for managing this second commonest subtype of RCC.
Autophagy is a non-specific, lysosome-mediated degradation. The process is beneficial for cells internally break down, clearance of damaged or superfluous proteins, and recycle cellular components. Autophagy-Related Genes (ARGs) participates in autophagy, for example, ATG7 involved in energy metabolism is an ARG. Lots of researchers proved that autophagy is associated with the progress of RCC [14,15,16]. For example, inhibiting autophagy in RCC increases the efficacy of many therapies [17, 18]. However, whether the expression level of ARGs has prognostic value is unknown. Hence, this research utilized ARGs to establish the prognostic risk model of KIRP. In this study, the relevance between differentially expressed autophagy-related genes (DEARGs) and clinicopathological parameters in 321 KIRP patients from TCGA database were examined, and an autophagy-related risk prognostic model was constructed as an independent predictor for overall survival of KIRP patients. We verified our risk score model from several aspects and confirmed it’s available, and we hope to provide more helpful guidance for evaluated prognosis and targeted treatment of KIRP with this novel prognostic risk model.
Methods
Data acquisition
There are 232 genes presently known associated with autophagy were downloaded from HADb (Human Autophagy Database, http://autophagy.lu/). The RNA-seq data and the corresponding clinical data of 289 Kidney Renal Papillary Cell Carcinoma (KIRP) patients and 32 non-tumor samples were obtained from TCGA database (The Cancer Genome Atlas database, https://www.cancer.gov/about-nci/organization/ccg/research/structural-genomics/tcga).
DEARGs screening
The differentially expressed autophagy-related genes (DEARGs) between KIRP tissues and adjacent non-tumor tissues were identified by the Wilcoxon Rank Sum test. The filtering criteria were |log2FoldChange| (|log2FC|) > 1 and false discovery rate (FDR) < 0.05.
Pathway enrichment analysis and Functional annotation for all DEARGs
To reveal the involved pathways and biological function of DEARGs, we performed Kyoto Encyclopedia of Genes and Genomes (KEGG) analysis and Gene Ontology (GO) analysis with the clusterProfiler package of R (version 4.0.1), and p-value < 0.05 was used as a strict cutoff.
Protein–protein interaction (PPI) networks construction
The functional protein–protein interaction (PPI) analysis is performed by STRING database (Search Tool for the Retrieval of Interacting Genes, https://string-db.org/) for all DEARGs. The cut-off criterion of interaction score is 0.4. We make use of the Cytoscape software to search hub genes and achieve two-dimensional (2D) visualization of PPI networks.
Identify the prognostic DEARGs
To identify the prognostic DEARGs whose expression profiles had notable correlation with the overall survival (OS) of patients with KIRP, the univariate Cox regression model was constructed. P < 0.05 is the threshold. These selected DEARGs were regarded as candidate genes had a correlation with patients’ survival.
Prognostic risk model construction and risk score calculation
The prognostic DEARGs identified by make use of the univariate Cox regression analysis were subjected to a multivariate Cox proportional hazards model to remove the genes that might not be an independent indicator in prognosis predicting. After that, several optimal independent survival-related DEARGs were obtained and the risk score composed of expression value of these genes was established. We calculated the risk score for each patient utilizing the regression coefficients of the individual DEARGs obtained from the multivariate Cox hazards model and the expression value of each of the selected DEARGs.
The risk score was calculated based on a linear combination of the relative gene expression level multiplied regression coefficients. The regression coefficients are obtained from the multiple Cox analysis and represents the relative weight of the genes. The risk score is a measure of prognostic risk for KIRP patients. Patients were divided into 2 groups by the median risk score as the critical value. High-risk score group had worse prognosis than low-risk score group.
Assessment of prognostic risk model
To verify the robustness and validity of the prognostic risk model, we plotted the survival curves and assessed the differences in the survival rates between high-risk and low-risk groups using the log-rank test. Then, we evaluated the survival prediction accuracy of the prognostic risk model using receiver-operator characteristic (ROC) curve. The area under the curve (AUC) of ROC curve is a discrimination criterion, it ranges from 0.5 to 1.0, the higher the value, the more accurate the model.
To explore whether the autophagy-related prognostic risk model could be an independent predictor of OS not rely on other clinicopathological parameters, we performed cox proportional hazard regression analysis. The association between risk score and clinical traits were explored. To validate the prognostic value of the risk score model, we took age, sex, pathological stage, tumor grade and T classification (lymphatic metastasis excluded) as candidate risk factors for univariate and multivariate Cox regression analyses.
Evaluation of the prognostic value of 3 prognostic-related DEARGs
As described above, to validate the availability of the prognostic risk score model, we compared the survival differences between high-risk group and low-risk group, which grouping is based on risk scores. Afterwards, we studied the association between the expression level of 3 prognostic-related DEARGs and KIRP patients’ survival individually. KIRP patients’ survival data in TCGA were used for Kaplan–Meier survival analyses.
The expression of 3 independent prognostic-related DEARGs (P4HB, NRG1 and BIRC5) were compared and validated between normal kidney tissues and KIRP tissues in mRNA level and protein level. The mRNA expression of 3 independent prognostic-related DEARGs in kidney and KIRP tissue was analyzed utilizing cancer profiling database called Oncomine (https://www.oncomine.org/resource/main.html). The protein level of 3 independent prognostic-related DEARGs on kidney and KIRP tissue were obtained from The Human Protein Atlas database (https://www.proteinatlas.org/).
Single-gene gene set enrichment analysis (GSEA) for 3 prognostic-related DEARGs
To explore the roles of 3 prognostic-related DEARGs in KIRP, GSEA was performed on these genes, respectively. We make use of the KEGG gene sets biological process database (version c2.KEGG.v4.0) to do GSEA. The database was affiliated with the Molecular Signatures Database (Msig DB, http://www.broad.mit.edu/gsea/msigdb/index.jsp). We exhibited 10 signal pathways containing top 5 up-regulated and top 5 downregulated signal pathways respectively, with p < 0.05 as a cutoff criterion. It’s worth noting that, if up-regulated pathways less than 5, we exhibit more downregulated ones instead.
Results
Identification of DEARGs
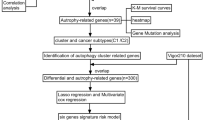
We downloaded the mRNA sequencing data and corresponding clinical data of 289 KIRP tissue samples and 32 normal kidney samples from TCGA database. The gene expression profile and clinical follow-up information of 265 KIRP patients were involved in our subsequent analysis. We extracted expression profile of 232 ARGs. Finally, 40 DEARGs were screened out, involved 31 upregulated ARGs and 9 downregulated ARGs with |log2 (FoldChange)| > 1 and FDR < 0.05 as filter criteria. The flow chart of the overall procedures in this manuscript was showed in Supplementary Fig. 1.
In Fig. 1a, the volcano plot exhibited the distribution of all DEARGs. X-axis of the volcano plot is log2FoldChange and Y-axis represents false discovery rate. The fold change patterns of 40 DEARGs in 32 non-tumor tissues and 289 KIRP tissues were showed in a heat map in Fig. 1b. The scatter plots in Fig. 1c visualized expression of 40 DEARGs between KIRP and normal tissues. In Table 1, we provide a detailed source of information on all DEARGs, including log2FoldChange and statistical significance.
Identification of differentially expressed autophagy-related genes (DEARGs) from KIRP tissues and non-tumor kidney specimens. a Volcano plot of 242 autophagy-related genes (ARGs). Up-regulated genes are marked red, they are DEARGs which |Log2FoldChange| > 1.0 in mRNA level; Down-regulated genes are marked green and they are also DEARGs whose |Log2FoldChange| > 1.0. The genes whose |Log2FoldChange| ≤1.0 are pained black. b Heatmap of the expression levels of 40 DEARGs in KIRP. The color depth represents the intensity of the gene expression level. KIRP, Kidney Renal Papillary Cell Carcinoma. c Box plot of 40 DEARGs’ expression. Red box and green box represent KIRP or non-tumor specimens respectively
PPI network establishment and function annotation of all DEARGs
The interaction of all DEARGs were visualized in Fig. 2a, and there are 12 hub genes arranged in a circle are DEARGs with interaction degree > 15. Biological processes (BP) annotation reminded us that DEARGs had a strong association with enzyme-related process, such as autophagy, regulation of peptidase activity and macroautophagy. In the aspect of the molecular function (MF), DEARGs seems played vital roles in some protein binding related functions, for example, peptidase regulator activity, ubiquitin protein ligase binding and protease binding. Regarding the cellular components (CC), the DEARGs encoded proteins constituted autophagosome membrane, vacuolar membrane, endoplasmic reticulum-Golgi intermediate compartment and so on (Fig. 2b). In Fig. 2c, the mainly pathways that had a positive relation with screened DEARGs was showed, including hepatitis B, pathogenic Escherichia coli infection, human cytomegalovirus infection and Kaposi sarcoma-associated herpesvirus infection. Z-scores of enriched pathways were all > 0, it means that all the pathways were more likely to be enhanced.
PPI network construction and functional annotation for 40 DEARGs. a PPI network of 40 DEARGs. The color depth of nodes is based on log2FoldChange, red nodes and green nodes denote up-regulated DEARGs and down-regulated DEARGs respectively. The width of links between nodes is positively correlated to combined score of protein interaction. Nodes’ size is inversely related to p-value. Square nodes are hub genes had most interactive protein, the number of interactive protein is > 6. b Gene Ontology enrichment analysis for 40 DEARGs. c KEGG pathway analyses for 40 DEARGs shows the top 10 signaling pathways that 40 DEARGs involved in
Establishment of autophagy-related risk signature
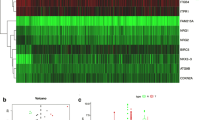
To identify the relationship between the expression of 40 DEARGs and overall survival in KIRP patients, we constructed the univariate Cox proportional hazards model. The results showed that there are 14 DEARGs significantly related to the prognosis of KIRP patients (p < 0.05) (Fig. 3a). In order to raise the robustness, the screened 14 prognostic-related DEARGs were further included in the subsequent multivariate Cox regression analysis. At last, 3 DEARGs (P4HB, NRG1 and BIRC5) were filtered out and used for autophagy-related risk model construction (Fig. 3b). The risk score for each patient was calculated according to the following formula: risk score = (0.8658× expression value of P4HB) + (0.3379× expression value of NRG1) + (1.1201× expression value of BIRC5). Patients were divided into high-risk (n = 132) and low-risk group (n = 133) by the median risk score as the critical value. The risk score was calculated for each patient and list of they belong to low or high-risk group (Supplementary Table 1).
Identify survival related autophagy genes in KIRP patients and development of prognostic model. We make use of univariate and multivariate cox model to filtered out DEARGs whose expression had positive relation with KIRP patients’ survival. a 14 DEARGs are associated with survival of KIRP patients according to univariate cox model. b 3 DEARGs are significantly related to survival of KIRP patients based on multivariate cox model, we exhibited the regression coefficients and p values
Validation of the risk signature
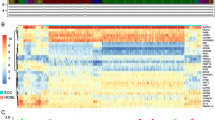
To measure the accuracy of the autophagy-related risk model to predict the prognosis of KIRP patients, we draw Kaplan-Meier plotter to compare the survival time difference between high-risk and low-risk group. Low-risk group patients had more survival probability (p = 4.406E-05) (Fig. 4a). Next, the ROC curves were employed to determine the predictive performance of the prognostic risk model. In Fig. 4b showed, the AUC value of risk score was 0.923, it was larger than AUC values of other indicators except for the pathologic stage, which confirmed that the autophagy-related prognostic model is an excellent and independent prognostic predictor comparing with other clinicopathology indicators. The risk scores of all KIRP patients were visualized from small to large (Fig. 4c). With the increase of the risk score, the death number of KIRP patients is more (Fig. 4d). The expression patterns of 3 prognostic-related DEARGs in different risk groups was exhibited in the heatmap (Fig. 4e).
Validation of the prognostic model. a Kaplan-Meier plotter showed the survival probability in different risk group for KIRP patients. b ROC curves evaluated the accuracy of DEARGs-based risk scores for prognosis predicting. c Scatter plot of KIRP patients distributed as risk scores increase. d Scatter plot exhibited the survival status of KIRP patients as risk scores increase, red plots and green plots represents non-survivors and survivors. e Heatmap of 3 prognostic DEARGs which composed prognostic model as risk scores increase
Clinical verification of prognostic model
To determine the relationship between the autophagy-related prognostic risk model and clinicopathological features in KIPR patients, we put several familiar clinicopathological factors and risk score to do univariate and multivariate cox regression analyses (Fig. 5). Since AUC values are often used to assess the performance of an individual clinicopathological indicator, and the larger of the AUC value, the more accurate of the indicator to predict prognosis. In our study, the AUC values of the clinicopathological features including age and sex to predict OS is less than 0.5 (Fig. 4b), demonstrated that age or sex alone was unable to predict prognosis as an individual indicator. The relationship between risk scores and age/sex was listed in Fig. 5a and Fig. 5b. No difference in risk score was observed between elder patients and younger patients (Fig. 5a). Instead, the AUC values of the clinicopathological features (pathological stage) and risk score is more than 0.9 (Fig. 4b), illustrated that no matter pathological stage or risk score can make a comparatively accurate prediction KIRP patients’ prognosis. Risk scores were lower in pathological stage I than in pathological stage II-IV (p = 6.676e-05) (Fig. 5c), and lower in T classification T1–2 than in T3–4 (p = 5.622e-04) (Fig. 5d). The relationship between the expression of 3 genes composed the risk model and 4 clinicopathological features in KIRP are exhibited in Fig. 5a, b, c, d. The raw TCGA data that containing basic information of all KIRP patients was listed in Supplementary Table 2. In Table 2, pathological stage, T classification, and risk score had obviously positive correlation with prognosis of KIRP patients in univariate Cox analysis, in addition, risk score and pathological stage were independent prognostic predictor of KIRP patients in multivariate Cox analysis. All above results demonstrate that the autophagy-related risk signature can be an excellent prognostic predictor ifor KIRP patients.
Validation of the function of 3 prognostic-related DEARGs in KIRP
According to our results, 3 prognostic-related DEARGs including P4HB, NRG1 and BIRC5 were identified to develop the survival-related risk prognostic model. We analyzed the correlation between 3 prognostic-related DEARGs that composed the prognostic model and survival probability of KIRP patients. The results of the Kaplan-Meier analysis showed that the upregulation of P4HB was obviously associated with the low survival probability of KIRP patients. Also, NRG1 or BIRC5 overexpression leads to worse OS (Fig. 6a). To further compare the expression difference of 3 prognostic-related DEARGs between KIRP and normal tissues, we performed a clinical study using cancer microarray database of Oncomine and human proteome database Human Protein Atlas. The mRNA level of 3 prognostic-related DEARGs was verified and showed in Fig. 6b. The expression trend of 3 prognostic-related DEARGs is in accordance with our previous results obtained from TCGA database which showed in Fig. 1. In Fig. 7a, results of immunohistochemistry (IHC) confirmed the expression of P4HB, NRG1 and BIRC5 protein are stronger in KIRP tissues than normal kidney tissues. Single-gene GSEA of the 3 prognostic-related DEARGs explored the potential roles of 3 prognostic DEARGs in KIRP (Fig. 7b).
Immunohistochemistry analysis of 3 genes (P4HB, NRG1 and BIRC5) which used to develop the risk prognostic model (a). P4HB protein is detected by antibody CAB012463 and HPA018884. BIRC5 protein is detected by antibody HPA002830. Immunohistochemistry of NRG1 in database of The Human Protein Atlas is absence. b Results of single-gene GSEA of 3 prognostic DEARGs which composed the risk signature in KIRP
Discussion
Autophagy is a dynamic and conserved process that can maintain cellular homeostasis [19]. Many researchers had proved autophagy played an important function in cancer [20,21,22]. Some targeted agents aimed at autophagy had been applied in the clinics in RCC patients [23,24,25]. Therefore, we decided to develop an autophagy-related prognostic risk model for prognosis predicting of KIRP patients.
The autophagy-related prognostic model is comprised of 3 genes, including P4HB, BIRC5, and NRG1. The 3 genes are all closely relevant to clinicopathological features of KIRP patients, the expression of them is either correlated with pathological stage or T classification of KIRP patients (p < 0.05). The risk score obtained according to the prognostic model is also significantly associated with clinicopathological features of KIRP patients. Several assessment methods confirmed the autophagy-related signature can be an independently prognosis predictor for KIRP patients. Individual assessment of the function of 3 prognostic-related DEARGs in KIRP further proof P4HB, BIRC5, and NRG1 are up-regulated in KIRP, and overexpression of them is associated with worse survival of KIRP patients. All results proved that risk signature constructed through P4HB, BIRC5, and NRG1 to evaluate the prognosis of KIRP patients is clinically practicable.
P4HB encoded protein disulfide isomerase, it is an autophagy-related gene, Xie et al proved that P4HB was a novel biomarker for ccRCC diagnosis and prognosis predicting [26]. BIRC5 also called survivin, the encoding products of BIRC5 play a significant role in negative regulation of apoptosis or programmed cell death. Philipp et al demonstrated that BIRC5 is of importance for renal pathophysiology and pathology [27]. NRG1 is a growth factor of the epidermal growth factor family, the relationship between NRG1 and RCC had not been explained clearly, but Sushma et al thought that NRG1 fusion is a low frequency event in most tumor types, including RCC [28]. We found that most of prognostic molecular indicators and therapeutic targets were identified and verified in ccRCC only or all RCC subtypes, there is an urgent need to explore possible prognostic molecular indicators and novel targets for KIRP therapy.
Functional enrichment analysis exhibited that 40 DEARGs were mainly involved in infection related pathway. It is generally acknowledged that infection and inflammation are all correlated with carcinogenesis. Kaymakcalan et al reported that RCC patients treated with mTOR inhibitors had a risk of infection [29], Alexander et al reported that urinary tract infection history is positively associated with RCC development [30]. Hence, the affection of infection to KIRP patients should be carefully assessed and managed.
This research constructed a novel prognostic risk model for prognosis predicting of KIRP patients, and proved it is steady and credible by verification with molecular signature combined with clinical features. Several assessment methods confirmed the prognostic risk model is obviously an independently prognosis predictor for KIRP patients. We believe, apart from traditional clinicopathological features (including pathologic stage, T classification and so on), risk score derived from the autophagy-related genes signature could also be incorporated into the clinical evaluation indicators to better predict clinical outcomes. Individual assessment of the function of 3 prognostic-related DEARGs in KIRP further proved P4HB, BIRC5, and NRG1 all play significant roles in KIRP. The 3 prognostic-related DEARGs can benefit personalized target therapy also. All results proved that the risk score calculated according to expression level of P4HB, BIRC5, and NRG1 to evaluate the prognosis of KIRP patients is reliable.
Conclusions
This research analyzed mRNA sequencing data of 289 KIRP tissue specimens and 32 non-tumor specimens and assessed of 232 ARGs’ expression difference in the two groups. We screened out 9 down-regulated DEARGs and 31 up-regulated DEARGs in KIRP with the threshold of |log2FC| > 1.0 and P < 0.05. From 40 DEARGs, 3 prognostic DEARGs (P4HB, NRG1, BIRC5) were determined to establish a prognostic risk model, and the risk score was calculated according to expression of the 3 prognostic DEARGs and fixed regression coefficients. With verification analysis combined using molecular signature and clinical characteristics, the risk score for prognosis predicting of KIRP patients is robustly and accuracy. The genes identified in autophagy-related prognostic model had been verified, and they were all correlated with KIRP patients’ prognosis, and they were all up-regulated in KIRP tissues. What’s more, this research is benefit for illustrating the molecular mechanisms behind KIRP from a new perspective.
Availability of data and materials
There are 232 autophagy-related genes downloaded from HADb (Human Autophagy Database, http://autophagy.lu/). The RNA-seq data and the corresponding clinical data of 289 Kidney Renal Papillary Cell Carcinoma (KIRP) patients and 32 non-tumor samples were obtained from TCGA database (The Cancer Genome Atlas database, https://www.cancer.gov/about-nci/organization/ccg/research/structural-genomics/tcga). The protein level of 3 independent prognostic-related DEARGs on kidney and KIRP tissue were obtained from The Human Protein Atlas database (https://www.proteinatlas.org/).
Abbreviations
- ARGs:
-
Autophagy- related genes
- AUC:
-
Area under the curve
- ccRCC:
-
Clear cell renal carcinoma
- DEARGs:
-
Differentially expressed autophagy- related genes
- DEGs:
-
Differentially expressed genes
- FDR:
-
False discovery rate
- GO:
-
Gene Ontology
- KEGG:
-
Kyoto Encyclopedia of Genes and Genomes
- KIRP:
-
Kidney Renal Papillary Cell Carcinoma
- KM plotter:
-
Kaplan-Meier plotter
- log2FC:
-
Log2Fold Change/Logarithm of Fold Change
- OS:
-
Overall survival
- PPI:
-
Protein-protein interaction
- RCC:
-
Renal cell carcinoma
- ROC:
-
Receiver-operator characteristic
- TCGA:
-
The Cancer Genome Atlas
References
Siegel RL, Miller KD, Jemal A. Cancer statistics, 2020. CA Cancer J Clin. 2020;70(1):7–30. https://doi.org/10.3322/caac.21590.
Capitanio U, Bensalah K, Bex A, Boorjian SA, Bray F, Coleman J, et al. Epidemiology of renal cell carcinoma. Eur Urol. 2019;75(1):74–84. https://doi.org/10.1016/j.eururo.2018.08.036.
Rhoades Smith KE, Bilen MA. A review of papillary renal cell carcinoma and MET inhibitors. Kidney cancer. 2019;3(3):151–61. https://doi.org/10.3233/KCA-190058.
Cancer Genome Atlas Research N, Linehan WM, Spellman PT, Ricketts CJ, Creighton CJ, Fei SS, et al. comprehensive molecular characterization of papillary renal-cell carcinoma. N Engl J Med. 2016;374(2):135–45. https://doi.org/10.1056/NEJMoa1505917.
Steffens S, Janssen M, Roos FC, Becker F, Schumacher S, Seidel C, et al. Incidence and long-term prognosis of papillary compared to clear cell renal cell carcinoma--a multicentre study. Eur J Cancer. 2012;48(15):2347–52. https://doi.org/10.1016/j.ejca.2012.05.002.
Deng J, Li L, Xia H, Guo J, Wu X, Yang X, et al. A comparison of the prognosis of papillary and clear cell renal cell carcinoma: evidence from a meta-analysis. Medicine. 2019;98(27):e16309. https://doi.org/10.1097/MD.0000000000016309.
Eggener SE, Yossepowitch O, Pettus JA, Snyder ME, Motzer RJ, Russo P. Renal cell carcinoma recurrence after nephrectomy for localized disease: predicting survival from time of recurrence. J Clin Oncol. 2006;24(19):3101–6. https://doi.org/10.1200/JCO.2005.04.8280.
Buti S, Karakiewicz PI, Bersanelli M, Capitanio U, Tian Z, Cortellini A, et al. Validation of the GRade, age, nodes and tumor (GRANT) score within the surveillance epidemiology and end results (SEER) database: a new tool to predict survival in surgically treated renal cell carcinoma patients. Sci Rep. 2019;9(1):13218. https://doi.org/10.1038/s41598-019-49250-6.
Kroeger N, Xie W, Lee JL, Bjarnason GA, Knox JJ, Mackenzie MJ, et al. Metastatic non-clear cell renal cell carcinoma treated with targeted therapy agents: characterization of survival outcome and application of the international mRCC database consortium criteria. Cancer. 2013;119(16):2999–3006. https://doi.org/10.1002/cncr.28151.
Connor Wells J, Donskov F, Fraccon AP, Pasini F, Bjarnason GA, Beuselinck B, et al. Characterizing the outcomes of metastatic papillary renal cell carcinoma. Cancer Med. 2017;6(5):902–9. https://doi.org/10.1002/cam4.1048.
Sun M, Shariat SF, Cheng C, Ficarra V, Murai M, Oudard S, et al. Prognostic factors and predictive models in renal cell carcinoma: a contemporary review. Eur Urol. 2011;60(4):644–61. https://doi.org/10.1016/j.eururo.2011.06.041.
Leibovich BC, Blute ML, Cheville JC, Lohse CM, Frank I, Kwon ED, et al. Prediction of progression after radical nephrectomy for patients with clear cell renal cell carcinoma: a stratification tool for prospective clinical trials. Cancer. 2003;97(7):1663–71. https://doi.org/10.1002/cncr.11234.
Zisman A, Pantuck AJ, Wieder J, Chao DH, Dorey F, Said JW, et al. Belldegrun AS: risk group assessment and clinical outcome algorithm to predict the natural history of patients with surgically resected renal cell carcinoma. J Clin Oncol. 2002;20(23):4559–66. https://doi.org/10.1200/JCO.2002.05.111.
Wang ZL, Deng Q, Chong T, Wang ZM. Autophagy suppresses the proliferation of renal carcinoma cell. Eur Rev Med Pharmacol Sci. 2018;22(2):343–50. https://doi.org/10.26355/eurrev_201801_14178.
Russell KL, Gorgulho CM, Allen A, Vakaki M, Wang Y, Facciabene A, et al. Inhibiting autophagy in renal cell Cancer and the associated tumor endothelium. Cancer J. 2019;25(3):165–77. https://doi.org/10.1097/PPO.0000000000000374.
Jones TM, Carew JS, Nawrocki ST. Therapeutic targeting of autophagy for renal cell carcinoma therapy. Cancers (Basel). 2020;12(5):1185. https://doi.org/10.3390/cancers12051185.
Zheng B, Zhu H, Gu D, Pan X, Qian L, Xue B, et al. MiRNA-30a-mediated autophagy inhibition sensitizes renal cell carcinoma cells to sorafenib. Biochem Biophys Res Commun. 2015;459(2):234–9. https://doi.org/10.1016/j.bbrc.2015.02.084.
Li H, Jin X, Zhang Z, Xing Y, Kong X. Inhibition of autophagy enhances apoptosis induced by the PI3K/AKT/mTor inhibitor NVP-BEZ235 in renal cell carcinoma cells. Cell Biochem Funct. 2013;31(5):427–33. https://doi.org/10.1002/cbf.2917.
Klionsky DJ, Emr SD. Autophagy as a regulated pathway of cellular degradation. Science. 2000;290(5497):1717–21. https://doi.org/10.1126/science.290.5497.1717.
Chan DA, Giaccia AJ. Targeting cancer cells by synthetic lethality: autophagy and VHL in cancer therapeutics. Cell Cycle. 2008;7(19):2987–90. https://doi.org/10.4161/cc.7.19.6776.
Turcotte S, Chan DA, Sutphin PD, Hay MP, Denny WA, Giaccia AJ. A molecule targeting VHL-deficient renal cell carcinoma that induces autophagy. Cancer Cell. 2008;14(1):90–102. https://doi.org/10.1016/j.ccr.2008.06.004.
Carew JS, Espitia CM, Zhao W, Han Y, Visconte V, Phillips J, et al. Disruption of Autophagic degradation with ROC-325 antagonizes renal cell carcinoma pathogenesis. Clin Cancer Res. 2017;23(11):2869–79. https://doi.org/10.1158/1078-0432.CCR-16-1742.
Faivre S, Demetri G, Sargent W, Raymond E. Molecular basis for sunitinib efficacy and future clinical development. Nat Rev Drug Discov. 2007;6(9):734–45. https://doi.org/10.1038/nrd2380.
Grimaldi A, Santini D, Zappavigna S, Lombardi A, Misso G, Boccellino M, et al. Antagonistic effects of chloroquine on autophagy occurrence potentiate the anticancer effects of everolimus on renal cancer cells. Cancer Biol Ther. 2015;16(4):567–79. https://doi.org/10.1080/15384047.2015.1018494.
Haas NB, Appleman LJ, Stein M, Redlinger M, Wilks M, Xu X, et al. Autophagy inhibition to augment mTOR inhibition: a phase I/II trial of Everolimus and Hydroxychloroquine in patients with previously treated renal cell carcinoma. Clin Cancer Res. 2019;25(7):2080–7. https://doi.org/10.1158/1078-0432.CCR-18-2204.
Xie L, Li H, Zhang L, Ma X, Dang Y, Guo J, et al. Autophagy-related gene P4HB: a novel diagnosis and prognosis marker for kidney renal clear cell carcinoma. Aging. 2020;12(2):1828–42. https://doi.org/10.18632/aging.102715.
Lechler P, Wu X, Bernhardt W, Campean V, Gastiger S, Hackenbeck T, et al. The tumor gene survivin is highly expressed in adult renal tubular cells: implications for a pathophysiological role in the kidney. Am J Pathol. 2007;171(5):1483–98. https://doi.org/10.2353/ajpath.2007.070132.
Jonna S, Feldman RA, Swensen J, Gatalica Z, Korn WM, Borghaei H, et al. Detection of NRG1 gene fusions in solid tumors. Clin Cancer Res. 2019;25(16):4966–72. https://doi.org/10.1158/1078-0432.CCR-19-0160.
Kaymakcalan MD, Je Y, Sonpavde G, Galsky M, Nguyen PL, Heng DY, et al. Risk of infections in renal cell carcinoma (RCC) and non-RCC patients treated with mammalian target of rapamycin inhibitors. Br J Cancer. 2013;108(12):2478–84. https://doi.org/10.1038/bjc.2013.278.
Parker AS, Cerhan JR, Lynch CF, Leibovich BC, Cantor KP. History of urinary tract infection and risk of renal cell carcinoma. Am J Epidemiol. 2004;159(1):42–8. https://doi.org/10.1093/aje/kwh014.
Acknowledgements
Not applicable.
Funding
The study was supported by the Nosocomial Scientific Research Fund Projects from the International Peace Maternity and Child Health Hospital of Shanghai Jiao Tong University School of Medicine (No.GFY5801), clinical research special projects from Shanghai Municipal Health Commission (No.20204Y0230) and Shanghai Sailing Program from Shanghai Science and Technology Committee (No.19YF1452200, No. 18YF1424800), National Natural Science Foundation of China (No.81901495), the National Key R&D Program of China (No.2018YFC1004901) and Shanghai “Rising Stars of Medical Talent” Youth Development Program Clinical Laboratory Practitioners Program (No.201972).
Author information
Authors and Affiliations
Contributions
HF designed this study and drafted this paper. SC and HF conducted experiments and did statistical analysis. HF and SC searched reference and participate in making Fig. CX revised the manuscript and supervised all the work. The author(s) read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors have no conflicts of interest to declare.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Additional file 1: Figure S1.
The flow chart of the overall process in our manuscript.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Fei, H., Chen, S. & Xu, C. Construction autophagy-related prognostic risk signature combined with clinicopathological validation analysis for survival prediction of kidney renal papillary cell carcinoma patients. BMC Cancer 21, 411 (2021). https://doi.org/10.1186/s12885-021-08139-2
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12885-021-08139-2