Abstract
Background
An increasing number of studies have described the aberrant expression of homeobox (HOX) proteins in gastric cancer (GC), which is critically associated with the prognosis and clinicopathological characteristics of GC. This study was conducted to investigate the clinical value and action mechanisms of HOX proteins in GC.
Methods
A comprehensive search of PubMed, Embase, Web of Science and Cochrane Library was performed in accordance with the Preferred Reporting Items for Systematic Reviews and Meta-Analysis (PRISMA) statement. The pooled hazard ratio (HR) with its 95% confidence interval (95% CI) and the pooled odds ratio (OR) with its 95% CI were used to assess the effect of HOX protein expression on the prognosis and clinicopathological features of GC, respectively.
Results
Nineteen studies containing 3775 patients were selected for this study. Heterogeneity among HRs of overall survival (OS) was markedly high (I2 = 90.5%, p = 0.000). According to the subgroup analysis, increased expression of HOX protein in the downregulated subgroup was associated with a good prognosis for patients with GC (pooled HR: 0.46, 95% CI: 0.36–0.59, I2 = 3.1%, p = 0.377), while overexpression of HOX protein in the upregulated subgroup was correlated with a reduced OS (pooled HR: 2.59, 95% CI: 1.79–3.74, I2 = 73.5%, p = 0.000). The aberrant expression of HOX protein was crucially related to the TNM stage, depth of tumour invasion, tumour size, lymph node metastasis, distant metastasis, vascular invasion, histological differentiation and Lauren classification in patients with GC. In addition, the molecular mechanisms by which HOX proteins regulate tumorigenesis and development of GC were also explored.
Conclusions
HOX proteins play vital roles in GC progression, which might serve as prognostic markers and therapeutic targets for GC.
Similar content being viewed by others
Background
Gastric cancer (GC) is one of the most common cancers. Although the incidence of GC is decreasing, it remains the sixth most common malignancy and accounts for 8.2% of global cancer-related deaths, according to the most recent global cancer statistics reported in 2018 [1]. GC is highly heterogeneous, and patients with GC are usually diagnosed in an advanced stage. Despite significant developments in surgical techniques and adjuvant therapy, the overall prognosis of patients with GC is still poor. Therefore, it is urgent to identify molecular markers to predict the prognosis and evaluate the therapeutic effect of GC.
Homeobox (HOX) genes encode a highly conserved family of 39 transcription factors that are divided into four clusters (HOXA, HOXB, HOXC, and HOXD). HOX proteins play crucial roles in the early development of embryos and organs, including vertebrae, external genitalia, gastrointestinal tract and central nervous system [2]. Because embryogenesis and tumorigenesis share similar characteristics such as growth and differentiation, the deregulation of HOX protein has been observed in abnormal development and malignancy [3]. HOX proteins function as oncogenes or tumour suppressors, depending on the context [4]. An increasing number of HOX proteins have been investigated in various tumours, including acute myeloid leukaemia [5], breast cancer [6], lung cancer [7], and digestive tract neoplasms [8,9,10]. Currently, the implications of HOX proteins in tumorigenesis and development of GC have been reported in many studies. Nevertheless, the roles of HOX proteins in GC remain controversial.
Therefore, considering the conflicting conclusions of current researches, we conducted this systematic review and meta-analysis to explore the prognostic and clinicopathological value of HOX proteins in GC and summarized the molecular mechanisms by which HOX proteins regulate tumorigenesis and development of GC.
Methods
This study was conducted according to the PRISMA guidelines [11].
Search strategies
Two researchers (XJ and LD) independently performed a comprehensive literature search of PubMed, Embase, Web of Science and Cochrane Library through March 6, 2020. The following MeSH terms and text words were used in combination: “genes, homeobox” or “gene, homeobox” or “homeobox gene” or “homeobox genes” or “genes, homeotic” or “gene, homeotic” or “homeotic gene” or “hox genes” or “gene, hox” or “genes, hox” or “hox gene” or “genes, homeo box” or “gene, homeo box” or “homeo box gene” or “homeo box genes” or “homeotic genes” or “homeo box sequence” or “homeo box sequences” or “sequences, homeo box” or “homeo boxes” or “sequence, homeo box” or “homeobox sequence” or “homeobox sequences” or “sequence, homeobox” or “sequences, homeobox” or “homeoboxes” or “homeo box” or “homeobox” or “hoxa1” or “hoxa5” or “hoxa9” or “hoxa10” or “hoxa11” or “hoxa13” or “hoxb5” or “hoxb7” or “hoxb8” or “hoxb9” or “hoxb13” or “hoxc6” or “hoxc9” or “hoxc10” or “hoxd4” or “hoxd9” or “hoxd10” or “hoxd13” or “stomach neoplasms” or “neoplasm, stomach” or “stomach neoplasm” or “neoplasms, stomach” or “gastric neoplasms” or “gastric neoplasm” or “neoplasm, gastric” or “neoplasms, gastric” or “cancer of stomach” or “stomach cancers” or “gastric cancer” or “cancer, gastric” or “cancers, gastric” or “gastric cancers” or “stomach cancer” or “cancer, stomach” or “cancers, stomach” or “cancer of the stomach” or “gastric cancer, familial diffuse”. The references of eligible studies in this field were also searched manually. Two investigators (XJ and LD) reviewed the titles and abstracts of studies and retrieved studies that met the inclusion criteria for full-text evaluation.
Selection criteria
All authors jointly deliberated and set the selection criteria. The following inclusion criteria were established: (1) GC was pathologically and histologically confirmed; (2) HOX protein expression was detected using immunohistochemical (IHC) staining in GC tissues and paired noncancerous mucosae; (3) studies evaluated the correlation between HOX protein expression and the outcome of GC; and (4) studies supplied sufficient information for calculating the HR with its 95% CI for survival and the OR with its 95% CI for clinicopathological parameters. The following exclusion criteria were used: (1) overlapping or duplicate data; (2) reviews, letters, case reports, conference abstracts, retracted articles, editorials, and full texts not published in English; (3) studies of cancer cells or animal models, or irrelevant studies; and (4) studies without adequate information for calculating HRs and 95% CIs.
Quality assessment
Two researchers (YM and JW) assessed the quality of studies using the Newcastle-Ottawa Quality Assessment Scale (NOS) [12]. The NOS consists of three aspects of evaluation: selection, comparability and outcomes between the case group and the control group. Studies that scored ≥6 points were considered high quality. Any disagreement was resolved by discussion.
Data extraction
Two investigators (XJ and LD) independently reviewed all included studies and extracted the available data. The following information was collected: (1) publication details, including the first author’s name, publication year and origin of the studied population; (2) characteristics of the studied population, including HRs with 95% CIs and clinicopathological features. In the studies that reported HRs from both univariate and multivariate models, we extracted the HR from the latter model that had been adjusted for potential confounders. If the HR was not reported, it was extrapolated using Engauge Digitizer v.11.1 software, Tierney’s spreadsheet [13], and (3) a cut-off value.
Statistical analysis
HR and its 95% CI from each study were used to calculate the pooled HR and its 95% CI. The heterogeneity of the combined HR was determined using Cochran’s Q test and Higgins’ I-squared statistics. A p value< 0.05 was considered significant. We used the random effects model if heterogeneity was observed (I2 ≥ 50%). The fixed effects model was used in the absence of heterogeneity (I2<50%) [14]. A subgroup analysis was conducted based on the expression level of HOX proteins in patients with GC. The sensitivity analysis was conducted to evaluate the stability of the results, after excluding each study. Publication bias was assessed using the Begg’s funnel plots and Egger’s tests, and a p value< 0.05 was considered significant. Statistical analyses were performed using Stata statistical software (version 15.0).
Results
Literature search
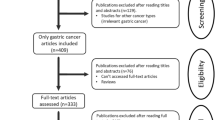
Our search strategy preliminarily identified 329 potential records. One hundred seventy-three articles remained after the removal of duplicated studies. Forty-eight of these studies were removed after perusing the titles and abstracts. Then, reviews, editorials, letters, conference abstracts, retracted articles, full texts not published in English, and studies of cancer cells or animal models were excluded. Subsequently, 18 studies lacking insufficient data were rejected. Finally, 19 studies including 3775 patients with GC were included in this analysis. The selection process is shown in Fig. 1.
Study characteristics
All included studies were conducted in China, Japan and Korea and were published between 2012 and 2019. These studies involve the following HOX proteins: HOXB9 [15], HOXD10 [16], HOXA5 [17, 18], HOXA10 [19,20,21], HOXA13 [22, 23], HOXC6 [24], HOXB7 [25, 26], HOXA1 [27], HOXA9 [28], HOXC9 [29], HOXC10 [30], HOXD4 [31], HOXA11 [32] and HOXD9 [33]. These studies explored the prognostic value of HOX protein expression for determining OS or disease-free survival (DFS) and the correlation between the expression of HOX proteins and clinicopathological characteristics of patients with GC. HOX expression at the protein level was detected using immunohistochemical staining. All included studies divided HOX protein expression into high (positive) and low (negative) groups, but the cut-off value was slightly different among these studies. A detailed description of the characteristics of the included studies is provided in Table 1.
Correlation of HOX protein expression with the prognosis
This meta-analysis included a total of 19 articles containing 14 HOX proteins. HOXB9, HOXD10 and HOXA5 were expressed at low levels in GC and acted as tumour suppressors. In contrast, HOXA13, HOXC6, HOXB7, HOXA1, HOXC9, HOXC10, HOXD4, HOXA11 and HOXD9 were expressed at high levels and functioned as tumour promotors in patients with GC. In addition, HOXA10 expression was increased in GC, but its role in predicting the prognosis of GC was unclear. In a pooled analysis including all studies with data on the prognostic effects of HOX proteins in GC, considerable heterogeneity among pooled HRs for OS was observed. A subgroup analysis stratified by the expression level was performed, and the results revealed different trends between the downregulated subgroup and the upregulated subgroup. High expression of HOX proteins in the downregulated subgroup was associated with a good prognosis for patients with GC (pooled HR: 0.46, 95% CI: 0.36–0.59, I2 = 3.1%, p = 0.377), while the overexpression of HOX proteins in the upregulated subgroup was correlated with a poor OS (pooled HR: 2.59, 95% CI: 1.79–3.74, I2 = 73.5%, p = 0.000) (Fig. 2a). The explanation for the high level of heterogeneity of the upregulated subgroup might be that HOXA10 had different prognostic values in the existing studies. The result of the analysis of the upregulated subgroup after excluding HOXA10 suggested that overexpressed HOX proteins significantly indicated a poor prognosis (pooled HR = 3.03, 95% CI: 2.45–3.74, I2 = 16.5%, p = 0.283) (Fig. 3). DFS was reported in 6 studies involving 5 HOX proteins. HOXA5 expression was associated with an increased DFS in patients with GC (pooled HR = 0.46, 95% CI: 0.23–0.91). In contrast, HOXA13, HOXA10, HOXB7 and HOXA1 expression was associated with a decreased DFS (pooled HR = 3.77, 95% CI: 2.61–5.45) (Fig. 2b).
Correlation of HOX protein expression with clinicopathological characteristics
Seventeen studies with 2899 patients were included to detect the relationship between HOX protein expression and tumour stage. As shown in Fig. 4a, increased expression of HOXB9 and HOXD10 was significantly correlated with an earlier TNM stage (HOXB9: OR = 0.22, 95% CI: 0.12–0.41, HOXD10: OR = 0.21, 95% CI:0.14–0.31), while increased expression of HOXA13, HOXB7, HOXA1, HOXA9, HOXC9, HOXC10, HOXA11 and HOXD9 was notably associated with an advanced TNM stage (I2 = 92.6%, p = 0.000). Due to the high level of heterogeneity, we performed a subgroup analysis based on the expression levels of HOX proteins. The heterogeneity of the upregulated group was decreased but still at a high level (I2 = 75.8%, p = 0.000) (Fig. 4b). A subsequent analysis showed that the studies of HOXA10 contributed a considerable amount of heterogeneity (data not shown). In addition, the difference of scoring systems for assessing expression levels of HOX proteins in the included studies was also one of the main sources of heterogeneity. The pooled analysis of the relationship between HOX proteins and the depth of tumour invasion showed that HOXD10 indicated a low T category (HOXD10: OR = 0.20, 95% CI: 0.09–0.41), while HOXA13, HOXC6, HOXB7 and HOXA1 were related to a high T category (HOXA13 (2013): OR = 4.18, 95% CI: 1.75–10.01; HOXA13 (2018): OR = 1.90, 95% CI: 1.08–3.35; HOXC6: OR = 3.55, 95% CI: 1.11–11.31; HOXB7 (2015): OR = 3.44, 95% CI: 1.32–8.95; HOXB7 (2017): OR = 10.14, 95% CI: 4.36–23.58; and HOXA1: OR = 2.03, 95% CI: 1.18–3.48) (Fig. 5a). We pooled 11 studies including 2087 patients and found that HOXD10, HOXA5 and HOXC10 were associated with a decreased tumour size (HOXD10: OR = 0.37, 95% CI: 0.25–0.54; HOXA5 (2018): OR = 0.20, 95% CI: 0.07–0.55; HOXA5 (2019): OR = 0.23, 95% CI: 0.08–0.67; and HOXC10: OR = 0.38, 95% CI: 0.15–0.98), while the overexpression of HOXA10, HOXB7 and HOXD4 was associated with an increased tumour size (HOXA10 (2015): OR = 2.39, 95% CI: 1.40–4.09; HOXB7 (2017): OR = 2.60, 95% CI: 1.61–4.20; and HOXD4: OR = 2.71, 95% CI: 1.28–5.74) (Fig. 5b). Similarly, the heterogeneity was significantly reduced by conducting a subgroup analysis according to expression levels of HOX proteins (Fig. 5c). Sixteen studies with 3509 patients reported that HOXB9 and HOXD10 were unfavourable factors for lymph node metastasis in patients with GC (HOXB9: OR = 0.35, 95% CI: 0.19–0.63 and HOXD10: OR = 0.24, 95% CI: 0.16–0.37), and overexpression of HOXA13, HOXA1, HOXA9, HOXC10, HOXD4 and HOXD9 was correlated with the presence of lymph node metastasis (HOXA13 (2013): OR = 2.38, 95% CI: 1.02–5.54; HOXA13 (2018): OR = 2.38, 95% CI: 1.39–4.09; HOXA1: OR = 2.45, 95% CI: 1.49–4.04; HOXA9: OR = 2.68, 95% CI: 1.23–5.83; HOXC10: OR = 6.18, 95% CI: 2.22–17.18; HOXD4: OR = 5.53, 95% CI: 2.55–12.02; and HOXD9: OR = 23.11, 95% CI: 6.04–88.49) (Fig. 6a). The results of the pooled analysis revealed that HOXD10 was not conducive to the distant metastasis of GC (HOXD10: OR = 0.34, 95% CI: 0.19–0.60), but that HOXC10 and HOXA11 promoted distant metastasis of GC (HOXC10: OR = 5.55, 95% CI: 1.42–21.61 and HOXA11: OR = 19.02, 95% CI: 1.07–337.91) (Fig. 6b). In addition, the upregulation of HOXB7 promoted vascular invasion in patients with GC (HOXB7 (2017): OR = 5.12, 95% CI: 3.18–8.23) (Fig. 6c). Moreover, HOXB9, HOXD10, HOXA5 and HOXC9 were factors contributing to good or moderate histological differentiation (HOXB9: OR = 0.17, 95% CI: 0.09–0.33, HOXD10: OR = 0.66, 95% CI: 0.44–0.99, HOXA5 (2018): OR = 0.26, 95% CI: 0.10–0.68; and HOXC9: OR = 0.28, 95% CI: 0.11–0.71), and overexpression of HOXA13, HOXA1, HOXA9 and HOXD9 was related to poorly differentiated status of GC (HOXA13 (2013): OR = 2.41, 95% CI: 1.02–5.67; HOXA13 (2018): OR = 1.84, 95% CI: 1.06–3.18; HOXA1: OR = 2.37, 95% CI: 1.41–4.00; HOXA9: OR = 4.98, 95% CI: 2.12 11.70; and HOXD9: OR = 14.63, 95% CI: 4.81–44.43) (Fig. 7a). Additionally, HOXD10 and HOXB7 was correlated with the intestinal phenotype of GC (HOXD10: OR = 5.02, 95% CI: 3.34–7.57 and HOXB7 (2017): OR = 6.27, 95% CI: 3.81–10.31) (Fig. 7b). None of the HOX proteins included in the pooled analysis exhibited significant associations with age (Fig. 8a), sex (Fig. 8b) or tumour location (Fig. 8c). Additionally, the relationships between HOXA5, HOXA10, HOXA13 and HOXB7 expression and clinicopathological characteristics were all explored in several studies. As shown in Fig. 9, HOXA5 expression predicted a smaller tumour size (OR = 0.22, 95% CI: 0.10–0.45) (Fig. 9a), but there is no correlation between HOXA10 expression and clinicopathological features (Fig. 9b). The overexpression of both HOXA13 (Fig. 9c) and HOXB7 (Fig. 9d) was significantly associated with an advanced tumour stage (HOXA13: OR = 2.31, 95% CI: 1.44–3.71 and HOXB7: OR = 3.48, 95% CI: 2.28–5.32) and a high T category (HOXA13: OR = 2.62, 95% CI: 1.23–5.60 and HOXB7: OR = 6.05, 95% CI: 2.08–17.57), and HOXA13 was also related to lymph node metastasis (OR = 2.38, 95% CI: 1.51–3.75) and poor differentiation status (OR = 1.99, 95% CI: 1.25–3.15).
Sensitivity analysis
A sensitivity analysis was performed to verify the robustness of our results. As shown in Fig. 10, the pooled HR was not significantly altered when each study was removed, which confirmed the reliability of overall results for the OS of patients with GC.
Publication bias
Begg’s test and Egger’s test were performed to evaluate publication bias. The results did not reveal substantial publication bias (Fig. 11: Begg’s test: p = 0.576, Egger’s test: p = 0.166).
Mechanisms by which HOX proteins regulate GC
In Table 2 and supplementary Fig. 1, we summarize the molecular mechanisms by which HOX proteins included in this study modulate carcinogenesis and development of GC [15,16,17,18,19,20,21,22,23,24,25,26,27,28,29,30,31,32,33,34,35,36,37,38,39,40,41,42,43,44,45,46,47,48,49,50,51,52,53,54,55,56,57]. HOXB9 inhibits GC progression via AKT and NF-κB pathways [34]. HOXD10 suppresses the migration and invasion of GC cells through insulin-like growth factor binding protein-3 (IGFBP3) and RhoC-AKT pathway [36, 39]. HOXA5 suppresses GC progression by inhibiting the G1/S transition during the cell cycle [17]. HOXA13 promotes GC development via TGF-β, ERK1/2, MDM2-p53- MRP1 pathways, and Wnt/β-catenin signalling [23, 44,45,46]. HOXC6 enhances invasive and metastatic abilities of GC cells by upregulating the expression of MMP9 via activating ERK pathway [48]. HOXA1 increases the proliferation of GC cells by upregulating cyclin D1 expression [27]. HOXB7 mediates GC cell malignancy by activating AKT/MAPK signalling, Src-FAK pathway, PIK3R3/AKT pathway, and epithelial mesenchymal transition (EMT) [26, 49, 50]. The miR-182/HOXA9 axis is implicated in RUNX3-mediated GC development [51]. In addition, HOXC9 contributes to GC progression by inducing EMT, MMP2 expression, and stem cell-like properties [29]. HOXC10 activates ATM/NF-kB pathway and MAPK signalling, functioning as an oncogene in GC [30, 54]. HOXD4 increases the proliferation and invasion of GC cells by upregulating c-Myc and cyclin D1 [31]. HOXD9 activates RUFY3, increasing the proliferation, migration and invasion of GC cells [33]. However, the effects of HOXA10 and HOXA11 on tumorigenesis and development of GC are controversial.
Discussion
GC is a main cause of cancer-related mortality. Currently, radical gastrectomy combined with adjuvant chemotherapy is recognized as the most effective treatment for GC. Nevertheless, many patients with GC are usually diagnosed in an advanced stage, missing the opportunity for radical surgical resection. Based on the current situation, it is important to identify factors which is helpful to improving prediction accuracy and promoting curative effect of GC. Most of the HOX genes are generally activated and expressed during embryogenesis, and many of these proteins are aberrantly expressed during tumorigenesis. According to the literatures, HOX proteins are related to the prognosis and clinicopathological features of GC, but the results are controversial. We conducted this study to further clarify the effects of HOX proteins on the prognosis and clinicopathological characteristics of GC and describe the molecular mechanisms by which HOX proteins regulate tumorigenesis and development of GC.
The present systematic review and meta-analysis enrolled 19 eligible studies containing 3775 patients. In the pooled analysis of the effects of HOX proteins on the GC prognosis, HOXB9, HOXD10 and HOXA5 were correlated with a good prognosis in patients with GC, while HOXA13, HOXC6, HOXB7, HOXA1, HOXC9, HOXC10, HOXD4, HOXA1 and HOXD9 were related to a poor prognosis. However, Kato et al. identified positive HOXB9 expression in GC as a marker of a poor prognosis. Unfortunately, the study by Kato was not included in this meta-analysis due to the lack of an analysis of HOXB9 expression in paired noncancerous mucosae [58].
The relationship between HOX proteins and clinicopathological features of GC were also analysed in this study. The results revealed correlations between the expression of HOX proteins and TNM stage, T category, tumour size, lymph node metastasis, distant metastasis, vascular invasion, histological differentiation, and Lauren classification in GC. Based on the results of the meta-analysis described above, we speculated that HOX proteins might predict the prognosis of patients with GC, which was also confirmed in each included original study. Therefore, we inferred that combined detection of the expression of various HOX proteins might provide a novel perspective for predicting the prognosis of patients with GC.
Currently, some clinicopathological parameters such as age, sex, tumour stage, depth of invasion, lymph node metastasis, distant metastasis, and resection margins, have been proven to be prognostic indicators of GC [59, 60]. At the same time, several molecules are under investigation as predictors of survival, such as gene mutations, DNA methylation, RNAs, and proteins [61]. Regrettably, many studies have only explored the individual relationship between clinicopathological characteristics or molecular markers and the prognosis of patients with GC, although a few studies have established prognostic models [62]. Bria et al. combined clinicopathological parameters (sex, age, Lauren classification, stage, margins, grade, site, size, and resected nodes) with molecular markers (HER2, FHIT, and APC) to construct a risk stratification of GC, establishing a scientific model to determine its prognosis. In addition, the authors conducted a large prospective validation with a larger sample size to eliminate all sources of bias in the retrospective study [63]. GC is highly heterogeneous, and even similar clinicopathological features result in different outcomes, suggesting that a more reasonable classification system is needed for predicting the prognosis and therapeutic effect of GC. A novel classification system with four molecular subtypes was developed by The Cancer Genome Atlas (TCGA) [64]. Besides, Sohn et al. developed a scoring system (TCGA risk score) based on TCGA to predict prognosis and adjuvant chemotherapy outcomes in patients with GC, which was validated as an independent prognostic factor for GC in multivariate Cox regression analyses [65]. Analogously, Lin et al. established a novel prognosis scoring system based on TCGA and Gene Expression Omnibus to predict the prognosis of GC, which comprised signatures of tumour protein-coding genes (P), tumour noncoding genes (N) and immune/stroma cells in the tumour microenvironment (M) (PMC score). Furthermore, the combination of PNM scores with American Joint Committeeon Cancer (AJCC) staging significantly increased its predictive value [66]. In addition, Tahara et al. investigated the prognosis and clinicopathological characteristics of GC by combining genetic and epigenetic abnormalities. The CpG island methylator phenotype (CIMP) and TP53 hot spot mutation status (R175, G245, R248, R273, and R282) were sufficient to predict the prognosis and clinicopathological features of GC. Among these features, patients with the CIMP−TP53 hot spot+ subtype presented the worst overall survival [67]. Moreover, Ooi et al. selected three oncogenic pathways (NF-κB, Wnt/β-catenin, and proliferation/stem cells) by analysing a GC pathway heatmap and combined them to predict its prognosis, which was validated in vitro [68].
The development of GC is determined by both genes and environmental factors, which has been confirmed in mouse models. Microbial infections, particularly Helicobacter pylori (H. pylori) and Epstein-Barr virus, are important environmental factors and have been confirmed as prognostic factors for GC [69, 70]. Although H. pylori infection is the strongest risk factor for GC, very few H. pylori-infected populations develop GC. This outcome is attributed to the duration of infection, strain type and host genetic signatures [71]. The crucial effects of genetic factors on GC development have been revealed using progress in genetic technology, including the construction of genetically engineered mice via recombinant DNA technology to achieve molecular overexpression or deficiency, as well as gene mutations, clarifying the pathogenesis of GC and the interactions between various factors. For example, INS-GAS transgenic mice on the FVB genetic background that overexpress gastrin develop intramucosal carcinomas with submucosal and intravascular invasion in less than 1 year when infected by Helicobacter felis (H. felis) or H. pylori, with males showing a higher prevalence than females, indicating sex differences in GC tumorigenesis [72, 73]. However, INS-GAS mice on a C57BL/6 background infected with H. felis do not progress to GC [74]. Surprisingly, gastrin knockout mice (GAS−/− mice) are also confirmed to be susceptible to GC and exhibit antral GC, in contrast to INS-GAS mice, which develop corpus cancers [75]. Moreover, GAS−/− mice are more susceptible to antral cancer induced by MNU, a gastric carcinogen used in mouse models, compared to WT mice on the same genetic background [76].
Taken together, these studies reveal important roles of genetic signatures in the development of GC, and the external factor such as infection is also indispensable. Thus, the establishment of a comprehensive and detailed scoring system containing the most basic clinicopathological parameters, molecular markers, gene expression profiles, microbial infections, etc., might be more accurate in predicting the prognosis of patients with GC than a single factor. Our manuscript analysing the effects of HOX proteins in GC development aimed to predict the prognosis and provide therapeutic targets for GC. The results of this meta-analysis recommend the inclusion of HOX proteins in the model predicting the prognosis of GC.
Several limitations of this systematic review and meta-analysis should be noted. First, several HRs and their 95% CIs for OS were extracted from the survival curves, which might affect the reliability of the results. Second, the sample size of each study was not large enough, which might affect the accuracy of the results. Third, IHC methodologies including the primary antibody used, antibody dilutions, and the scoring system applied, differed, which might partially contribute to the heterogeneity. Finally, all patients included in our study were Asians, which might restrict the applicability of our results to other races.
Conclusions
This systematic review and meta-analysis firstly generalized and evaluated the significance of HOX proteins in modulating the prognosis and clinicopathological characteristics of GC. We also summarized the molecular mechanisms by which HOX proteins regulate tumorigenesis and development of GC. Based on these findings, HOX proteins might serve as biomarkers and therapeutic targets for GC. Considering the limitations of this study, further large-scale prospective and high-quality studies are required to confirm the potential values of HOX proteins in GC.
Availability of data and materials
All data generated or analysed in this study are included in this published article.
Abbreviations
- AJCC:
-
American Joint Committeeon Cancer
- AKT:
-
Protein kinase B
- ATM:
-
Ataxia telangiectasia mutated
- CI:
-
Confidence interval
- CIMP:
-
CpG island methylator phenotype
- DFS:
-
Disease-free survival
- EMT:
-
Epithelial mesenchymal transition
- ERK:
-
Extracellular regulated protein kinases
- FAK:
-
Focal adhesion kinase
- GC:
-
Gastric cancer
- H. felis :
-
Helicobacter felis
- HOX:
-
Homeobox
- HR:
-
Hazard ratio
- IGFBP3:
-
Insulin-like growth factor binding protein-3
- IHC:
-
Immunohistochemical
- MAPK:
-
Mitogen-activated protein kinase
- MDM2:
-
Murine double minute 2
- MRP1:
-
Multidrug resistance-associated protein 1
- NF-κB:
-
Nuclear factor-kappa B
- NOS:
-
Newcastle-Ottawa Quality Assessment Scale
- OR:
-
Odds ratio
- OS:
-
Overall survival
- PIK3R3:
-
Phosphoinositide-3-kinase, regulatory subunit 3
- RhoC:
-
Ras superfamily of GTP-binding protein
- RUFY3:
-
RUN and FYVE domain containing 3
- TGF-β:
-
Transforming growth factor-β
References
Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394–424.
Mark M, Rijli FM, Chambon P. Homeobox genes in embryogenesis and pathogenesis. Pediatr Res. 1997;42(4):421–9.
Samuel S, Naora H. Homeobox gene expression in cancer: insights from developmental regulation and deregulation. Eur J Cancer (Oxford, England : 1990). 2005;41(16):2428–37.
Shah N, Sukumar S. The Hox genes and their roles in oncogenesis. Nat Rev Cancer. 2010;10(5):361–71.
Nagy A, Osz A, Budczies J, Krizsan S, Szombath G, Demeter J, et al. Elevated HOX gene expression in acute myeloid leukemia is associated with NPM1 mutations and poor survival. J Adv Res. 2019;20:105–16.
de Bessa Garcia SA, Araujo M, Pereira T, Mouta J, Freitas R. HOX genes function in breast Cancer development. Biochimica et biophysica acta Reviews on cancer. 1873;2020(2):188358.
Li L, Zhang X, Liu Q, Yin H, Diao Y, Zhang Z, et al. Emerging role of HOX genes and their related long noncoding RNAs in lung cancer. Crit Rev Oncol Hematol. 2019;139:1–6.
Joo MK, Park JJ, Chun HJ. Impact of homeobox genes in gastrointestinal cancer. World J Gastroenterol. 2016;22(37):8247–56.
Kuo TL, Cheng KH, Chen LT, Hung WC. Deciphering the potential role of Hox genes in pancreatic cancer. Cancers (Basel). 2019;11(5):734.
Quagliata L, Quintavalle C, Lanzafame M, Matter MS, Novello C, di Tommaso L, et al. High expression of HOXA13 correlates with poorly differentiated hepatocellular carcinomas and modulates sorafenib response in in vitro models. Lab Invest. 2018;98(1):95–105.
Moher D, Liberati A, Tetzlaff J, Altman DG. Preferred reporting items for systematic reviews and meta-analyses: the PRISMA statement. BMJ (Clinical research ed). 2009;339:b2535.
Lo CK, Mertz D, Loeb M. Newcastle-Ottawa scale: comparing reviewers’ to authors’ assessments. BMC Med Res Methodol. 2014;14:45.
Tierney JF, Stewart LA, Ghersi D, Burdett S, Sydes MR. Practical methods for incorporating summary time-to-event data into meta-analysis. Trials. 2007;8:16.
DerSimonian R, Laird N. Meta-analysis in clinical trials revisited. Contemporary Clin Trials. 2015;45(Pt A):139–45.
Sha S, Gu Y, Xu B, Hu H, Yang Y, Kong X, et al. Decreased expression of HOXB9 is related to poor overall survival in patients with gastric carcinoma. Digestive Liver Dis. 2013;45(5):422–9.
Wang YY, Li L, Ye ZY, Zhao ZS, Yan ZL. MicroRNA-10b promotes migration and invasion through Hoxd10 in human gastric cancer. World J Surg Oncol. 2015;13:259.
Peng X, Zha L, Chen A, Wang Z. HOXA5 is a tumor suppressor gene that is decreased in gastric cancer. Oncol Rep. 2018;40(3):1317–29.
Wu Y, Zhou T, Tang Q, Xiao J. HOXA5 inhibits tumor growth of gastric cancer under the regulation of microRNA-196a. Gene. 2019;681:62–8.
Sentani K, Oue N, Naito Y, Sakamoto N, Anami K, Oo HZ, et al. Upregulation of HOXA10 in gastric cancer with the intestinal mucin phenotype: reduction during tumor progression and favorable prognosis. Carcinogenesis. 2012;33(5):1081–8.
Lim JY, Yoon SO, Seol SY, Hong SW, Kim JW, Choi SH, et al. Overexpression of miR-196b and HOXA10 characterize a poor-prognosis gastric cancer subtype. World J Gastroenterol. 2013;19(41):7078–88.
Han Y, Lu S, Wen YG, Yu FD, Zhu XW, Qiu GQ, et al. Overexpression of HOXA10 promotes gastric cancer cells proliferation and HOXA10(+)/CD44(+) is potential prognostic biomarker for gastric cancer. Eur J Cell Biol. 2015;94(12):642–52.
Han Y, Tu WW, Wen YG, Li DP, Qiu GQ, Tang HM, et al. Identification and validation that up-expression of HOXA13 is a novel independent prognostic marker of a worse outcome in gastric cancer based on immunohistochemistry. Med Oncol (Northwood, London, England). 2013;30(2):564.
Han Y, Song C, Wang J, Tang H, Peng Z, Lu S. HOXA13 contributes to gastric carcinogenesis through DHRS2 interacting with MDM2 and confers 5-FU resistance by a p53-dependent pathway. Mol Carcinog. 2018;57(6):722–34.
Zhang Q, Jin XS, Yang ZY, Wei M, Liu BY, Gu QL. Upregulated Hoxc6 expression is associated with poor survival in gastric cancer patients. Neoplasma. 2013;60(4):439–45.
Tu W, Zhu X, Han Y, Wen Y, Qiu G, Zhou C. Overexpression of HOXB7 is associated with a poor prognosis in patients with gastric cancer. Oncol Lett. 2015;10(5):2967–73.
He X, Liu Z, Xia Y, Xu J, Lv G, Wang L, et al. HOXB7 overexpression promotes cell proliferation and correlates with poor prognosis in gastric cancer patients by inducing expression of both AKT and MARKs. Oncotarget. 2017;8(1):1247–61.
Yuan C, Zhu X, Han Y, Song C, Liu C, Lu S, et al. Elevated HOXA1 expression correlates with accelerated tumor cell proliferation and poor prognosis in gastric cancer partly via cyclin D1. J Exp Clin Cancer Res. 2016;35:15.
Ma YY, Zhang Y, Mou XZ, Liu ZC, Ru GQ, Li E. High level of homeobox A9 and PBX homeobox 3 expression in gastric cancer correlates with poor prognosis. Oncol Lett. 2017;14(5):5883–9.
Peng X, Kang Q, Wan R, Wang Z. miR-26a/HOXC9 Dysregulation promotes metastasis and stem cell-like phenotype of gastric Cancer. Cell Physiol Biochem. 2018;49(4):1659–76.
Yao S, He L, Zhang Y, Ye L, Lai Y, Huang L, et al. HOXC10 promotes gastric cancer cell invasion and migration via regulation of the NF-κB pathway. Biochem Biophys Res Commun. 2018;501(3):628–35.
Liu H, Tian H, Zhao J, Jia Y. High HOXD4 protein expression in gastric adenocarcinoma tissues indicates unfavorable clinical outcomes. Saudi J Gastroenterol. 2019;25(1):46–54.
Wang C, Shi M, Ji J, Cai Q, Jiang J, Zhang H, et al. A self-enforcing HOXA11/Stat3 feedback loop promotes stemness properties and peritoneal metastasis in gastric cancer cells. Theranostics. 2019;9(25):7628–47.
Zhu H, Dai W, Li J, Xiang L, Wu X, Tang W, et al. HOXD9 promotes the growth, invasion and metastasis of gastric cancer cells by transcriptional activation of RUFY3. J Exp Clin Cancer Res. 2019;38(1):412.
Zhang L, Wu Q, He C, Liang D, Yi Q, Shi J, et al. HOXB9 inhibits proliferation in gastric carcinoma cells via suppression of phosphorylated-Akt and NF-κB-dependent snail expression. Digestive Liver Dis. 2019;51(1):157–65.
Chang Q, Zhang L, He C, Zhang B, Zhang J, Liu B, et al. HOXB9 induction of mesenchymal-to-epithelial transition in gastric carcinoma is negatively regulated by its hexapeptide motif. Oncotarget. 2015;6(40):42838–53.
Liu Z, Zhu J, Cao H, Ren H, Fang X. miR-10b promotes cell invasion through RhoC-AKT signaling pathway by targeting HOXD10 in gastric cancer. Int J Oncol. 2012;40(5):1553–60.
Li C, Huo B, Wang Y, Cheng C. Downregulation of microRNA-92b-3p suppresses proliferation, migration, and invasion of gastric cancer SGC-7901 cells by targeting Homeobox D10. J Cell Biochem. 2019;120(10):17405–12.
Wang L, Chen S, Xue M, Zhong J, Wang X, Gan L, et al. Homeobox D10 gene, a candidate tumor suppressor, is downregulated through promoter hypermethylation and associated with gastric carcinogenesis. Molecular Med (Cambridge, Mass). 2012;18(1):389–400.
Xue M, Fang Y, Sun G, Zhuo W, Zhong J, Qian C, et al. IGFBP3, a transcriptional target of homeobox D10, is correlated with the prognosis of gastric cancer. PLoS One. 2013;8(12):e81423.
Shao L, Chen Z, Peng D, Soutto M, Zhu S, Bates A, et al. Methylation of the HOXA10 promoter directs miR-196b-5p-dependent cell proliferation and invasion of gastric Cancer cells. Mol Cancer Res. 2018;16(4):696–706.
Chen W, Wu G, Zhu Y, Zhang W, Zhang H, Zhou Y, et al. HOXA10 deteriorates gastric cancer through activating JAK1/STAT3 signaling pathway. Cancer Manag Res. 2019;11:6625–35.
Song C, Han Y, Luo H, Qin Z, Chen Z, Liu Y, et al. HOXA10 induces BCL2 expression, inhibits apoptosis, and promotes cell proliferation in gastric cancer. Cancer Med. 2019;8(12):5651–61.
Chang S, Liu J, Guo S, He S, Qiu G, Lu J, et al. HOTTIP and HOXA13 are oncogenes associated with gastric cancer progression. Oncol Rep. 2016;35(6):3577–85.
He YX, Song XH, Zhao ZY, Zhao H. HOXA13 upregulation in gastric cancer is associated with enhanced cancer cell invasion and epithelial-to-mesenchymal transition. Eur Rev Med Pharmacol Sci. 2017;21(2):258–65.
Qin Z, Chen Z, Weng J, Li S, Rong Z, Zhou C. Elevated HOXA13 expression promotes the proliferation and metastasis of gastric cancer partly via activating Erk1/2. Onco Targets Ther. 2019;12:1803–13.
Qu LP, Zhong YM, Zheng Z, Zhao RX. CDH17 is a downstream effector of HOXA13 in modulating the Wnt/β-catenin signaling pathway in gastric cancer. Eur Rev Med Pharmacol Sci. 2017;21(6):1234–41.
Lin J, He J, He X, Wang L, Xue M, Zhuo W, et al. HoxC6 functions as an oncogene and isoform HoxC6-2 may play the primary role in gastric carcinogenesis. Dig Dis Sci. 2020.
Chen SW, Zhang Q, Xu ZF, Wang HP, Shi Y, Xu F, et al. HOXC6 promotes gastric cancer cell invasion by upregulating the expression of MMP9. Mol Med Rep. 2016;14(4):3261–8.
Wu J, Long Z, Cai H, Yu S, Liu X. Homeobox B7 accelerates the cancer progression of gastric carcinoma cells by promoting epithelial-mesenchymal transition (EMT) and activating Src-FAK pathway. Onco Targets Ther. 2019;12:3743–51.
Cai JQ, Xu XW, Mou YP, Chen K, Pan Y, Wu D. Upregulation of HOXB7 promotes the tumorigenesis and progression of gastric cancer and correlates with clinical characteristics. Tumour Biol. 2016;37(2):1641–50.
Yu J, Tian X, Chang J, Liu P, Zhang R. RUNX3 inhibits the proliferation and metastasis of gastric cancer through regulating miR-182/HOXA9. Biomedicine & pharmacotherapy = Biomedecine & pharmacotherapie. 2017;96:782–91.
Kim J, Bae DH, Kim JH, Song KS, Kim YS, Kim SY. HOXC10 overexpression promotes cell proliferation and migration in gastric cancer. Oncol Rep. 2019;42(1):202–12.
Zheng J, Ge P, Liu X, Wei J, Wu G, Li X. MiR-136 inhibits gastric cancer-specific peritoneal metastasis by targeting HOXC10. Tumour Biol. 2017;39(6):1010428317706207.
Guo C, Hou J, Ao S, Deng X, Lyu G. HOXC10 up-regulation promotes gastric cancer cell proliferation and metastasis through MAPK pathway. Chinese J Cancer Res = Chung-kuo yen cheng yen chiu. 2017;29(6):572–80.
Li J, Tong G, Huang C, Luo Y, Wang S, Zhang Y, et al. HOXC10 promotes cell migration, invasion, and tumor growth in gastric carcinoma cells through upregulating proinflammatory cytokines. J Cell Physiol. 2020;235(4):3579–91.
Bai Y, Fang N, Gu T, Kang Y, Wu J, Yang D, et al. HOXA11 gene is hypermethylation and aberrant expression in gastric cancer. Cancer Cell Int. 2014;14:79.
Cui Y, Gao D, Linghu E, Zhan Q, Chen R, Brock MV, et al. Epigenetic changes and functional study of HOXA11 in human gastric cancer. Epigenomics. 2015;7(2):201–13.
Kato F, Wada N, Hayashida T, Fukuda K, Nakamura R, Takahashi T, et al. Experimental and clinicopathological analysis of HOXB9 in gastric cancer. Oncol Lett. 2019;17(3):3097–102.
Park SR, Kim MJ, Ryu KW, Lee JH, Lee JS, Nam BH, et al. Prognostic value of preoperative clinical staging assessed by computed tomography in resectable gastric cancer patients: a viewpoint in the era of preoperative treatment. Ann Surg. 2010;251(3):428–35.
Nagata T, Ichikawa D, Komatsu S, Inoue K, Shiozaki A, Fujiwara H, et al. Prognostic impact of microscopic positive margin in gastric cancer patients. J Surg Oncol. 2011;104(6):592–7.
Machlowska J, Maciejewski R, Sitarz R. The Pattern of Signatures in Gastric Cancer Prognosis. Int J Mol Sci. 2018;19(6):1658.
Mohri Y, Tanaka K, Ohi M, Yokoe T, Miki C, Kusunoki M. Prognostic significance of host- and tumor-related factors in patients with gastric cancer. World J Surg. 2010;34(2):285–90.
Bria E, De Manzoni G, Beghelli S, Tomezzoli A, Barbi S, Di Gregorio C, et al. A clinical-biological risk stratification model for resected gastric cancer: prognostic impact of Her2, Fhit, and APC expression status. Ann Oncol. 2013;24(3):693–701.
Bass JA, Thorsson V, Shmulevich L, Reynolds MS, Miller M, Bernard B, et al. Comprehensive molecular characterization of gastricadenocarcinoma. Nature. 2014;513(7517):202–9.
Sohn BH, Hwang JE, Jang HJ, Lee HS, Oh SC, Shim JJ, et al. Clinical significance of four molecular subtypes of gastric Cancer identified by the Cancer genome atlas project. Clin Cancer Res. 2017.
Lin S, Zhou R, Zeng D, Wu J, Wu J, Zhang J, et al. A novel assessing system for predicting the prognosis of gastric cancer. Epigenomics. 2019;11(11):1251–66.
Tahara T, Tahara S, Horiguchi N, Okubo M, Terada T, Yamada H, et al. Molecular subtyping of gastric cancer combining genetic and epigenetic anomalies provides distinct clinicopathological features and prognostic impacts. Hum Mutat. 2019;40(3):347–54.
Ooi CH, Ivanova T, Wu J, Lee M, Tan IB, Tao J, et al. Oncogenic pathway combinations predict clinical prognosis in gastric cancer. PLoS Genet. 2009;5(10):e1000676.
Wang J, Liu X. Correlation analysis between helicobacter pylori infection status and tumor clinical pathology as well as prognosis of gastric Cancer patients. Iran J Public Health. 2018;47(10):1529–36.
Shinozaki-Ushiku A, Kunita A, Fukayama M. Update on Epstein-Barr virus and gastric cancer (review). Int J Oncol. 2015;46(4):1421–34.
Cover TL. Helicobacter pylori Diversity and Gastric Cancer Risk. mBio. 2016;7(1):e01869–15.
Wang TC, Dangler CA, Chen D, Goldenring JR, Koh T, Raychowdhury R, et al. Synergistic interaction between hypergastrinemia and helicobacter infection in a mouse model of gastric cancer. Gastroenterology. 2000;118(1):36–47.
Fox JG, Rogers AB, Ihrig M, Taylor NS, Whary MT, Dockray G, et al. Helicobacter pylori-associated gastric cancer in INS-GAS mice is gender specific. Cancer Res. 2003;63(5):942–50.
Takaishi S, Tu S, Dubeykovskaya ZA, Whary MT, Muthupalani S, Rickman BH, et al. Gastrin is an essential cofactor for helicobacter-associated gastric corpus carcinogenesis in C57BL/6 mice. Am J Pathol. 2009;175(1):365–75.
Zavros Y, Eaton KA, Kang W, Rathinavelu S, Katukuri V, Kao JY, et al. Chronic gastritis in the hypochlorhydric gastrin-deficient mouse progresses to adenocarcinoma. Oncogene. 2005;24(14):2354–66.
Tomita H, Takaishi S, Menheniott TR, Yang X, Shibata W, Jin G, et al. Inhibition of gastric carcinogenesis by the hormone gastrin is mediated by suppression of TFF1 epigenetic silencing. Gastroenterology. 2011;140(3):879–91.
Acknowledgements
We would like to thank all researchers for their contributions to this study.
Funding
Not applicable.
Author information
Authors and Affiliations
Contributions
XJ designed the research, searched the literatures, extracted and analysed the data, and wrote the manuscript. LD searched the literatures, extracted and analysed the data. YM and JW conducted literatures quality assessment. XJ, LD, YM, JW, HY, YJ, XZ and ZL established selection criteria. All authors read, reviewed and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare no conflict of interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Additional file 1: Figure S1.
Molecular mechanisms how HOX proteins regulate tumorigenesis and development of GC.↑: promote; ⊥: inhibit; AKT: protein kinase B; ATM: ataxia telangiectasia mutated; BCL2: B cell lymphoma-2; CDH17: cadherin 17; CST1: cystatin SN; DHRS2: dehydrogenase/reductase 2; EGF: epidermal growth factor; ERK: extracellular regulated protein kinases; FAK: focal adhesion kinase; IGFBP3: insulin-like growth factor binding protein-3; JAK1: janus kinase 1; MAPK: mitogen-activated protein kinase; MDM2: murine double minute 2; MET: mesenchymal epithelial transition; MMP2: matrix metalloproteinase 2; MMP9: matrix metalloproteinase 9; MMP14: matrix metalloproteinase 14; MRP1: multidrug resistance-associated protein 1; NF-κB: nuclear factor-kappa B; NKD1: naked cuticle homolog 1; PIK3R3: phosphoinositide-3-kinase, regulatory subunit 3; RhoC: ras superfamily of GTP-binding protein; RUFY3: RUN and FYVE domain containing 3; RUNX3: runt-related transcription factor 3; Src: steroid receptor coactivator; STAT3: signal transducers and activators of transcription 3; TFF1: trefoil factor 1; TGF-β: transforming growth factor-β; TNF-α: tumour necrosis factor-α; uPA: urokinase-type plasminogen activator; uPAR: urokinase-type plasminogen activator receptor.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Jin, X., Dai, L., Ma, Y. et al. Homeobox proteins are potential biomarkers and therapeutic targets in gastric cancer: a systematic review and meta-analysis. BMC Cancer 20, 866 (2020). https://doi.org/10.1186/s12885-020-07346-7
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12885-020-07346-7