Abstract
Background
The clinical significance of GLI1 expression either through canonical Hedgehog signal transduction or through non-canonical mechanisms in rhabdomyosarcoma (RMS) or Ewing sarcoma (EWS) is incompletely understood. We tested a role for Hedgehog (HH) signal transduction and GL11 expression in development of vincristine (VCR) resistance in RMS and EWS.
Methods
We characterized baseline expression and activity of HH pathway components in 5 RMS (RD, Rh18, Ruch-2, Rh30, and Rh41) and 5 EWS (CHLA9, CHLA10, TC32, CHLA258, and TC71) cell lines. We then established VCR-resistant RMS and EWS cell lines by exposing cells to serially increasing concentrations of VCR and determining the IC50. We defined resistance as a ≥ 30-fold increase in IC50 compared with parental cells. We determined changes in gene expression in the VCR-resistant cells compared with parental cells using an 86-gene cancer drug resistance array that included GLI1 and tested the effect of GLI1 inhibition with GANT61 or GLI1 siRNA on VCR resistance.
Results
We found evidence for HH pathway activity and GLI1 expression in RMS and EWS cell lines at baseline, and evidence that GLI1 contributes to survival and proliferation of these sarcoma cells. We were able to establish 4 VCR-resistant cell lines (Ruch-2VR, Rh30VR, Rh41VR, and TC71VR). GLI1 was significantly up-regulated in the Rh30VR, Rh41VR, and TC71VR cells. The only other gene in the drug resistance panel that was significantly up-regulated in each of these VCR-resistant cell lines compared with their corresponding parental cells was the GLI1 direct target and multidrug resistance gene, ATP-binding cassette sub-family B member 1 (MDR1). We established major vault protein (MVP), which was up-regulated in both vincristine-resistant alveolar RMS cell lines (Rh30VR and Rh41VR), as another direct target of GLI1 during development of drug resistance. Treatment of the VCR-resistant cell lines with the small molecule inhibitor GANT61 or GLI1 siRNA together with VCR significantly decreased cell viability at doses that did not reduce viability individually.
Conclusions
These experiments demonstrate that GLI1 up-regulation contributes to VCR resistance in RMS and EWS cell lines and suggest that targeting GLI1 may benefit patients with RMS or EWS by reducing multidrug resistance.
Similar content being viewed by others
Background
Children and young adults with recurrent rhabdomyosarcoma (RMS) or Ewing sarcoma (EWS) fare poorly [1,2,3,4]. Therefore, mechanisms of drug resistance, which contribute to recurrence need to be fully understood so that effective therapeutic approaches can be established to prevent or reverse development of drug resistance.
The Hedgehog (HH) signal transduction pathway functions during normal development and in cancers [5,6,7,8]. The canonical HH signaling pathway is activated when a HH family ligand (Sonic hedgehog [SHH], Indian hedgehog [IHH], or Desert hedgehog [DHH]) interacts with a cell surface receptor in the Patched (PTCH) family (PTCH1 or PTCH2). The interaction between HH and PTCH proteins relieves PTCH-mediated inhibition of the activity of the G protein-coupled seven-span transmembrane protein, Smoothened (SMO). Activation of SMO leads to translocation of GLI family transcription factors (GLI1, GLI2, and GLI3) from cytoplasmic microtubules to the nucleus and transcriptional regulation of target genes. GLI1 and PTCH1 are transcriptional targets of HH signaling and their expression serves as an indicator of pathway activation [9, 10]. Non-canonical GLI1 activation that does not depend on HH, PTCH or SMO, has also been described [11, 12].
In cancer, HH signaling has been implicated in tumorigenicity, cancer stem cell biology, tumor/stromal interactions, and metastasis [13]. In addition, in a wide variety of cancers, including basal cell carcinoma, diffuse large B-cell lymphoma, gliomas, melanoma, myeloid leukemia, and carcinomas of the cervix, colon, esophagus, head/neck, lung, stomach, ovary and prostate, HH signaling has been implicated in the development of resistance to a variety of cytotoxic chemotherapeutic and targeted agents, multidrug resistance, or radiation resistance [14,15,16,17,18,19,20,21,22,23,24,25,26,27].
HH signal transduction pathway components, including HH ligands, PTCH1, SMO, GLI1, GLI2 or GLI3 are present in RMS and EWS cell lines and patient samples [28,29,30,31,32,33,34,35,36]. The molecular mechanisms that drive HH pathway activation in RMS are incompletely understood [34]. In embryonal RMS (ERMS), there is evidence that HH pathway deregulation sometimes occurs based on loss of heterozygosity at loci for negative regulators of the pathway, including PTCH1 or Suppressor of Fused (SUFU) and based on development of ERMS in Ptc+/− or Sufu+/− knockout mouse models [33, 37,38,39,40]. Gain of 12q13, the GLI1 locus, has been reported more commonly in alveolar RMS (ARMS) [41, 42]. In EWS, GLI1 has been shown to be a direct transcriptional target of the EWSR1-FLI1 fusion-protein, which is found in the majority of EWS cases [35, 36, 43, 44].
The clinical significance of GLI1 activation either through canonical or non-canonical mechanisms is incompletely understood in RMS and EWS. Indeed, debate continues whether markers of HH signaling are present in higher levels in ERMS or ARMS and whether activation of HH signaling correlates with patient outcome [30, 45]. Therefore, we tested the role of HH signal transduction and GLI1 expression in development of a multidrug resistance phenotype in RMS and EWS by establishing vincristine (VCR)-resistant cells.
Methods
RMS and EWS cell lines
We obtained RD cells from ATCC (Manassas, VA). Rh18, Rh30, and Rh41 cells were obtained from Dr. Houghton, Ruch-2 cells from Dr. Schäfer, and UKF-Rhb-1 cells from Dr. Cinatl Jr. We obtained CHLA9, CHLA10, TC32, CHLA258 and TC71 from the Children’s Oncology Group. All cells were cultured in media supplemented with 10–20% fetal bovine serum, 100 U/ml penicillin, and 100 μg/ml streptomycin (Thermo Fisher, MA).
Reverse transcriptase polymerase chain reaction (RT PCR)
We isolated total RNA from the cell lines using the Qiagen RNeasy mini kit (Qiagen, Valencia, CA). We performed RT PCR using the One-Step RT PCR kit (Qiagen, Valencia, CA) or TaqMan Gene Expression Assay reagents (Applied Biosystems, Foster City, CA). We completed 30–35 cycles of PCR, including denaturation for 30 s, annealing for 30 s, and amplification for 1 min. The following primers were used for PCR: DHH sense 5′-GCTCTCCTGACCAATCTACTG-3′ and DHH antisense 5′-TCGTGCCCAACTACAACCC-3′, IHH sense 5′-CAAGCAGTTCAGCCCCAATG-3′ and IHH antisense 5′-CTGGTTCATCACCGAGATAGCC-3′, SHH sense 5′-CAGAGGTGTAAGGACAAGTTGAACG-3′ and SHH antisense 5′-AAAGTGAGGAAGTCGCTGTAGAGC-3′, PTCH1 sense 5′-CCTGGACGACATCCTGAAATCC-3′ and PTCH1 antisense 5′-GCGAGAAATGGCAAAACCTGAG-3′, SMO sense 5′-TGGCTTTGTGCTCATTACCTTCAG-3′ and SMO antisense 5′-ATCCGCTTTGGCTCATCGTC-3′, GLI1 sense 5′-AGTCATACTCACGCCTCGAA-3′ and GLI1 antisense 5′-GACCATGCACTGTCTTGACA-3′, GLI2 sense 5′-AAGGATTGCCACCCAGGACG-3′ and GLI2 antisense 5′-CCGACTCACTGCTCTGCTTGTT-3′, GLI3 sense 5′-CGAACAGATGTGAGCGAGAAAGC-3′ and GLI3 antisense 5′-AAAGATGAGGAGGGTGGTAGTGGG-3′, PAX3-FOX01 sense 5′-CCGACAGCAGCTCTGCCATC-3′ and PAX3-FOX01 antisense 5′-ATGAACTTGCTGTGTAGGGACAG-3′, EWSR1-FLI1 sense 5′- GCACCTCCATCCTACCCTCCT − 3′ and EWSR1-FLI1 antisense 5′- CTTACTGATCGTTTGTGCCCC-3′(long) or EWSR1-FL1I antisense 5′- TGGCAGTGGGTGGGTCTTCAT-3′(short), and GAPDH sense 5′-TGATGACATCAAGAAGGTGGTGAAG-3′ and GAPDH antisense 5′-TCCTTGGAGGCCATGTGGGCCAT-3′.
Western blot analysis
We prepared cell lysates using Tris.HCl buffer (pH 7.4), containing 150 mM NaCl, protease inhibitor cocktail (Thermo Fisher, MA), 0.5 mM DTT, and 1% TritonX-100. We loaded 50–100 μg of protein onto 4–15% SDS-PAGE gels (BioRad, Hercules, CA). After electrophoresis, we blotted the proteins onto nitrocellulose membranes (BioRad, Hercules, CA) and probed with polyclonal antibodies against human GLI1 protein (Cell Signaling, Danvers, MA) or against GAPDH (Santa Cruz Biotech, Santa Cruz, CA). We visualized the protein using a chemiluminescence kit (Pierce Inc., Rockford, IL).
Quantitative RT PCR (qRT PCR)
cDNA was synthesized using a high capacity cDNA reverse transcription kit (Applied Biosystems, Foster City, CA). PCR was performed using TaqMan universal PCR master mix and the following conditions; 50 °C for 2 min, 95 °C for 10 min and 40 cycles of 95 °C for 15 s and 60 °C for 1 min (Applied Biosystems, Foster City, CA). Primers and probes for GLI1 (Hs00171790_ml), PTCH1 (Hs00970980_ml), and GAPDH (Hs99999905_ml) were purchased from Applied Biosystems (Foster City, CA). The experiments were completed in triplicate and an average and standard deviation were calculated.
Immunofluorescence for primary cilia
We grew RMS and EWS cells in 8-well chamber slides (Nunc, Rochester, NY). Cells were fixed with 4% paraformaldehyde/0.5% TritonX-100 in PBS for 30 min at room temperature with rocking. Cells were washed again with PBS at room temperature and were then blocked for 1 h with 10% donkey serum. We incubated the cells with anti-acetylated alpha-tubulin (Sigma, St. Louis, MO, 1:5000 dilution), and pericentrin (Abcam, Cambridge, MA, 1:500 dilution) or pericentrin2 (Santa Cruz Biotech, Santa Cruz, CA, 1:500 dilution) antibodies overnight at 4 °C. Cells were washed with PBS and incubated with the secondary antibodies at 1:300 dilution (donkey anti-goat IgG Alexa488, [Alexa, Eugene, OR], or donkey anti-mouse IgG Alexa568, [Alexa, Eugene, OR]) for 1 h at room temperature. The nuclei were stained with 4′,6-diamidino-2-phenyindole (DAPI) at 1:2000 dilution (Biotium, Hayward, CA) for 20 min at room temperature with rocking. Immunofluorescence was observed using a Zeiss LSM510 or LSM880 META confocal laser scanning microscope.
Treatment of cells with hedgehog ligands
We exposed RMS and EWS cells to 1 μg/ml of DHH, IHH, or SHH peptide (R&D Systems, Minneapolis, MN) for 24 h at 37 °C in 5% CO2 with serum-free culture media.
Treatment of cells with SMO or GLI1 inhibitors and/or VCR
We added 2500–5000 cells in 0.1 ml of culture media supplemented with 10–20% (v/v) fetal bovine serum to Falcon 96-well cell culture plates (Becton Dickinson, Franklin Lakes, NJ) and cultured overnight. We then added 0–10 μM vismodegib (LC Lab, Woburn, MA), 0–40 μM GANT61 (Sigma, St. Louis, MO), 0–8 nM GLI1 siRNA (IDT, Coralville, IA), and/or 0–50 nM VCR (LC Lab, Woburn, MA) or equal volumes of solvent (ethanol or DMSO). We used 1 μl of interferon (Polyplus, NY) for GLI1 siRNA transfection. Cells were incubated with the inhibitor and/or VCR at 37 °C with 5% carbon dioxide for up to 72 h.
Methylthiazolyl diphenyl-tetrazolium bromide (MTT) assays
We added 10 μl of MTT reagent (5 mg/ml MTT in 1X PBS) to 0.1 ml culture media containing the cells. The mixture was incubated for 3 h at 37 °C, then we added 0.1 ml of solubilization solution (10% SDS in 0.01 M HCl) and incubated the mixture overnight at 37 °C. Absorbances at 570 and 650 nm were measured using a Ceres UV900H Di ELISA plate reader (Bio-Tek Instruments, Inc., Highland Park, VT). Background readings at 650 nm were subtracted from optical density readings at 570 nm. The experiments were completed in triplicate and an average and standard deviation calculated.
5-bromo-2′-deoxyuridine (BrdU) assays
We measured cell proliferation using a BrdU cell proliferation assay kit (MilliporeSigma, Burlington, MA). Cells were incubated with BrdU for 1 h at 37 °C following incubation for 24 h. Cells were fixed and BrdU incorporation was detected with anti-BrdU detector antibody. The signal was measured using a Ceres UV900H Di ELISA plate reader (Bio-Tek Instruments, Inc., Highland Park, VT). The experiments were completed in triplicate and an average and standard deviation calculated.
Caspase 3/7 assays
We measured apoptosis using a caspase 3/7 assay kit (Promega, Madison, WI). Cells were incubated with an equal volume of 2X caspase assay solution for 1 h at room temperature in the dark. Caspase activity was measured with a luminometer (Berthold, Oak Ridge, TN). The experiments were completed in triplicate and an average and standard deviation calculated.
Preparation of VCR-resistant cell lines
We established VCR-resistant RMS and EWS cell lines by exposing cells to serially increasing concentrations of VCR. We defined resistance as a 30-fold increase in the IC50 compared with parental cells before exposure to VCR together with persistence of resistance over several passages.
Cancer drug resistance array
qRT PCR using an 86-gene cancer drug resistance PCR array (Qiagen, Germantown, MD) was performed as described in the manual. The commercial array was modified to add GLI1 and the direct GLI1 target, ATP binding cassette subfamily B member, transporter 1 (TAP1) to the commercially available 84-gene panel. Total RNA was purified from the parental and VCR-resistant RMS cells using an RNAeasy mini kit (Qiagen, Valencia, CA). Genomic DNA was removed using on-column DNase I treatment. RNA was quantified using a Nanodrop Spectrophotometer (Thermo Scientific). Reverse transcription was carried out using 1 μg of total RNA and the RT2 First Strand kit (Qiagen). Real-Time PCR was carried out using a 7500 fast PCR machine (Applied Biosystems) as described in manufacturer’s protocol. The threshold cycle (Ct) of each well was calculated using the instrument’s software and the fold change was calculated by comparing the Ct of the parental cell line and the corresponding VCR-resistant cell line for a given gene. Any gene with a +/− 2-fold change in expression in the VCR-resistant cells compared with the corresponding parental cells was selected for further analysis. The results were shown as means and standard deviations. Statistical significance was calculated using a Student t-test and a p ≤ 0.05 was considered significant.
Electrophoretic mobility shift assays
Two microliters of Rh30 cell lysate was mixed with 5 μl of 2X binding buffer (50 mM HEPES, pH 7.5, 50 mM KCl, 5 mM MgCl2, 1 mM DTT, 20% glycerol (v/v), 50 mM poly (dI-dC), and 10 mM ZnSO4), H2O, and 0 or 1 μl (20 pmol) of unlabeled competitor oligonucleotide. The mixture was incubated at 4o C for 10 min. One microliter (155 fmol) of double stranded digoxigenin-labeled probe was added and the mixture was incubated at 4 °C for an additional 20 min. We used the following probes, each of which included a different consensus GLI1 binding site with up to 2 bp mismatches in the MVP promoter; MVP1 sense begins at nucleotide − 1710 in the MVP promoter: 5′-CATGTTGGCGAGGCTGGTCTTGAACTCCT-3′ and MVP1 antisense 5′-AGGAGTTCAAGACCAGCCTCGCCAACATG-3′, MVP2 sense begins at nucleotide − 505 in the MVP promoter: 5′- GTTTTCTATTGAACACCTATAGAGAGAGT − 3′ and MVP2 antisense 5′- ACTCTCTCTATAGGTGTTCAATAGAAAAC -3′, MVP3 sense begins at nucleotide − 438 in the MVP promoter: 5′- GTTTTCTATTGAACACCTATTCAGAGACC − 3; and MVP3 antisense 5′- GGTCTCTGAATAGGTGTTCAATAGAAAAC -3′, MVP4 sense begins at nucleotide − 233 in the MVP promoter: 5′- CCATCTCGGGCCCTCCAACTCCTCCCAGTCCCACTCCAG − 3′ and MVP4 antisense 5′- CTGGAGTGGGACTGGGAGGAGTTGGAGGGCCCGAGATGG − 3′, and MVP5 sense begins at nucleotide − 172 in the MVP promoter: 5′- AGAAACCCATGAGCACTCAGGGAGCAGTG − 3′ and MVP5 antisense 5′- CACTGCTCCCTGAGTGCTCATGGGTTTCT − 3′. The GLI consensus binding sites are underlined in the probes. The GLI1-MVP complexes were separated by electrophoresis, transferred onto nitrocellulose membranes, and bands were visualized by anti-digoxigenin antibody and chemiluminescence (Roche, Mannheim, Germany).
Chromatin immunoprecipitation (ChIP)
ChIP was performed, using Rh41 and Ruch-2 cells and the ChIP-IT express chromatin immunoprecipitation kit (Active Motif, Carlsbad, CA). DNA and protein were cross-linked with formaldehyde for 10 min at room temperature. The DNA-protein complexes were sheared by sonication, the cell lysate was cleared by centrifugation at 14,000 x g for 10 min, and anti-c-terminal-GLI1 antibody was added (Santa Cruz Biotech, Santa Cruz, CA). Antibody-Protein-DNA complexes were precipitated with protein G magnetic beads. DNA was purified and used for PCR amplification of GLI1 binding sites. We used the following primers: MVP 4,5 sense 5′- CCTCCTGGGTTGAAGCGATT − 3′ and MVP 4,5 antisense 5′- TGCTCTTCCCTGGCAAGATG − 3′.
Co-transfection assays
We co-transfected HeLa cells with 0–1000 ng of pCMV-GLI1 effector plasmid, 200 ng of pMVP407 reporter construct (obtained from Dr. Furukawa, Kagoshima University), and 20 ng of Renilla control reporter DNA (Promega, Madison, WI), using 2–6 μl of Lipofectamine 2000 reagent (Gibco-BRL, Grand Island, NY). A total of 3 μg of DNA was transfected in each experiment and the difference was made up with pUC18 carrier DNA. Cell lysates were prepared 24 h after transfection. Twenty microliters of cell lysate was assayed by adding 100 μl of substrate solution (Promega, Madison, WI). The experiments were performed at least in triplicate and results expressed as an average with standard deviation.
Statistical analysis
Differences between groups were assessed using a Student’s t-test unless stated otherwise. p ≤ 0.05 was considered significant.
Results
RMS and EWS cell lines have active HH signal transduction pathways
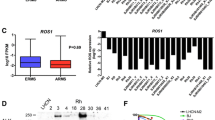
We determined the expression of HH pathway components in three ERMS cell lines (RD, Rh18, and Ruch-2), two PAX3-FOX01 fusion-positive ARMS cell lines (Rh30 and Rh41), and five EWSR1-FLI1 fusion-positive EWS cell lines (CHLA9, CHLA10, TC32, CHLA258, and TC71) by RT PCR (Table 1). All of the RMS cell lines expressed IHH, PTCH1, SMO, GLI1, GLI2, and GLI3. We demonstrated GLI1 protein in Rh18, Ruch-2, Rh30, and Rh41 cells but not RD cells by Western blot (Fig. 1a). We did not find inactivating mutations in exons 1–24 of PTCH1 in any of the RMS cell lines that we tested (RD, Rh18, Rh30 or Rh41) that could contribute to HH pathway activation and expression of GLI1 (data not shown). All of the EWS cell lines expressed PTCH1, SMO, GLI1, and GLI3 by RT PCR and GLI1 protein by Western blot (Table 1 and Fig. 1a). We demonstrated significantly higher expression of GLI1 by qRT PCR in CHLA258 and TC71 EWS cells, which were both established following chemotherapy at the time of recurrence compared with CHLA9 cells, which were established prior to any therapy (Fig. 1b). Two other non-recurrent EWS cell lines, CHLA10 and TC32, also had low GLI1 expression.
GLI1 expression and primary cilia in RMS and EWS cell lines. a Expression of GLI1 protein by Western blot in RMS and EWS cell lines. Size markers in kilodaltons (kD) are included. GLI1-amplified RMS-13 rhabdomyosarcoma cells or GLI1- amplified Rh30 cells were used as positive controls. The GAPDH control was included to show comparability of protein loading among lanes. The blots have been cropped and full-length blots are presented in Supplementary Figure 1A. b EWS cell lines that were established following recurrence (CHLA258 and TC71) had significantly higher GLI1 expression compared with cell lines established at the time of diagnosis (CHLA9, CHLA10, and TC32) by qRT PCR. c Immunofluorescence microscopy for primary cilia in RMS and EWS cell lines. Cell line names are indicated above the images. Green = pericentrin or pericentrin2 (centriole component), red = acetylated alpha tubulin (primary cilia component), and blue = DAPI nuclear staining. Primary cilia, indicated by arrows, were seen in RD, Rh30, Rh41, CHLA9, CHLA10, CHLA258, and TC32 cells. NIH3T3 cells were used as a positive control. White dotted lines indicate cell borders. Scale bars are shown. Microsoft Exel and Adobe Photoshop were used to prepare Figure 1
Since HH signaling occurs in primary cilia, we used immunofluorescence for acetylated alpha-tubulin and pericentrin or pericentrin2 to determine whether RMS and EWS cells have primary cilia (Fig. 1c). We identified primary cilia in the majority of the RMS (RD, Rh30, and Rh41) and EWS (CHLA9, CHLA10, CHLA258 and TC32) cell lines.
To assess responsiveness of RMS and EWS cells to HH ligands, we exposed RMS and EWS cells to DHH, IHH, and SHH. RD, Rh18, and Rh41 RMS cells showed up-regulation of targets of HH signaling, GLI1 and/or PTCH1, in response to HH ligands (Fig. 2a). We also saw up-regulation of the HH-target genes GLI1 and/or PTCH1 in response to HH ligands in CHLA9, CHLA10, TC32 and TC71 EWS cell lines (Fig. 2b).
Modulation of HH pathway activity in RMS and EWS cells following exposure to HH ligands or a GLI1 inhibitor. aGLI1 (left panel) or PTCH1 (right panel) mRNA expression was sometimes significantly increased (indicated by *) following exposure to HH ligands in RD, Rh18, and Rh41 cells compared with their corresponding control cells (CTR) that were not treated with HH ligands. The experiments were conducted in triplicate. Averages (bars) and standard deviations (brackets) are shown. bGLI1 (left panel) or PTCH1 (right panel) mRNA expression was sometimes significantly increased (indicated by *) following exposure to HH ligands in CHLA9, CHLA10, TC32, and TC71 cells compared with their corresponding control cells (CTR) that were not treated with HH ligands. The experiments were conducted in triplicate. Averages (bars) and standard deviations (brackets) are shown. cGLI1 expression (left) measured by qRT PCR was significantly decreased, indicated by *, followed treatment with GANT61 in Rh30, Rh18, or Rh41 cells. PTCH1 expression (right) measured by qRT PCR was significantly decreased, indicated by *, following treatment with GANT61 in Rh41 cells. The experiments were conducted in triplicate. Averages (bars) and standard deviations (brackets) are shown. d MTT (left panel), BrdU (middle panel), and Caspase 3/7 (right panel) assays in RMS cell lines (listed on the x-axis). Apoptosis increased with GLI1 down-regulation with GANT61 in RMS cells. The experiments were conducted in triplicate. Averages (bars) and standard deviations (brackets) are shown. * = statistically significant difference compared with no GANT61 treatment (black bar). e MTT (left panel), BrdU (middle panel), and Caspase 3/7 (right panel) assays in EWS cell lines (listed on the x-axis). Cell viability and cell proliferation decreased and apoptosis increased with GLI1 down-regulation with GANT61 in EWS cells. The experiments were conducted in triplicate. Averages (bars) and standard deviations (brackets) are shown. * = statistically significant difference compared with no GANT61 treatment (black bar). Microsoft Exel and Adobe Photoshop were used to prepare Figure 2
To assess the effect of HH pathway up-regulation on the biology of RMS and EWS cells, we assessed cell viability (MTT assay), cell proliferation (BrdU assay), and apoptosis (Caspase 3/7 assay) in RMS and EWS cells with and without SHH ligand. Exposure to SHH did not significantly affect the results of these assays (data not shown). To assess the effect of HH pathway/GLI1 down-regulation on the biology of RMS and EWS cells, we treated the cells with the GLI inhibitor GANT61 (0–40 μM) [46]. GANT61 inhibits GLI1 function and sometimes down-regulates GLI1 and PTCH1 expression in RMS cell lines (Fig. 2c). In RMS cells, apoptosis increased in each of the cell lines following treatment with GANT61, suggesting a role for GLI1 in survival of these cells (Fig. 2d). Cell viability or cell proliferation decreased only at the highest GANT61 concentration that we tested. In EWS cells, cell viability and cell proliferation decreased, whereas apoptosis increased in each of the cell lines following treatment with GANT61, suggesting roles for GLI1 in proliferation and survival of EWS cells (Fig. 2e).
GLI1 is up-regulated in vincristine-resistant RMS and EWS cell lines
To determine if GLI1 up-regulation occurs as cells develop resistance to VCR, we established VCR-resistant RMS (Ruch-2VR, Rh30VR, and Rh41VR) and EWS (TC71VR) cell lines by exposing parental cells to serially increasing concentrations of VCR. The goal was to increase the IC50 ≥ 30-fold (Table 2). We were not able to establish VCR-resistant RD, Rh18, CHLA9, CHLA10, TC32, or CHLA258 cells that met this standard.
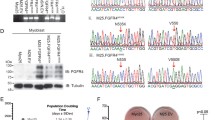
As expected, we showed that VCR-resistant cells retained viability to a greater degree than their corresponding parental cells when exposed to increasing concentrations of VCR (0–50 nM) (Fig. 3a). The VCR-resistant cell lines exhibited up-regulation of GLI1 protein compared to their respective parental cell lines (Fig. 3b).
Cell viability and GLI1 expression in VCR-resistant cell lines. a VCR-resistant cell lines retained viability to a greater extent when treated with VCR than their corresponding parental cell lines. MTT assays were conducted in triplicate. Averages (bars) and standard deviations (brackets) are shown. * = statistically significant difference in viability between the parental (black bars) and VCR-resistant (red bars) cell lines at a given VCR concentration. b Western blots showed up-regulation of GLI1 in VCR-resistant RMS (Ruch-2VR, Rh30VR, Rh41VR) (left panel) and EWS (TC71VR) (right panel) cell lines compared with their corresponding parental cells (Ruch-2, Rh30, Rh41, TC71). GAPDH was used to indicate equal loading between lanes. Size markers in kD are shown. The blots have been cropped and full-length blots are presented in Supplementary Figure 3B. Microsoft Exel and Adobe Photoshop were used to prepare Figure 3
To more globally characterize changes in gene expression in the VCR-resistant cells compared with their corresponding parental cells, we used an 86-gene cancer drug resistance array (Qiagen). GLI1 expression was significantly increased (≥2.0 fold and p ≤ 0.05) in VCR-resistant ARMS cell lines compared with their corresponding parental cells (Rh30VR cells: 2.3 fold increased and p = 0.04, Rh41VR cells: 10.1 fold increased and p = 0.0008) (Fig. 4a and b). In Ruch-2VR ERMS cells, GLI1 expression was up-regulated (p = 0.02) but did not reach the bar of a 2.0-fold increase (1.7 fold increased) (data not shown). To provide data from an additional VCR-resistant ARMS cell line we assessed expression of GLI1 in parental and VCR-resistant UKF Rhb-1 ARMS cells obtained from Dr. Cinatl (Frankfurt, Germany). Once again, we found significant up-regulation of GLI1 (2.3 fold and p = 0.0007) (data not shown). These results suggest that GLI1 potentially plays a role in the development of a multi-drug resistance phenotype in fusion-positive ARMS.
Gene expression changes in VCR-resistant RMS and EWS cells compared with parental cells by PCR array. a Rh30VR cells showed significant changes (at least 2-fold up-regulated or down-regulated, and p ≤ 0.05) in gene expression in 30 genes, including GLI1, that were included in the 86 gene drug resistance panel compared with parental cells. The experiment was completed in triplicate. Averages (bars) and standard deviations (brackets) are shown. * indicates p ≤ 0.05 and fold change is indicated on the y-axis. b Rh41VR cells showed significant changes (at least 2-fold up-regulated or down-regulated, and p ≤ 0.05) in gene expression in 35 genes, including GLI1, that were included in the 86 gene drug resistance panel compared with the parental cells. The experiment was completed in triplicate. Averages (bars) and standard deviations (brackets) are shown. * indicates p ≤ 0.05 and fold change is indicated on the y-axis. c TC71VR cells showed significant changes (at least 2-fold up-regulated or down-regulated, and p ≤ 0.05) in gene expression in 12 genes, including GLI1, that were included in the 86 gene drug resistance panel compared with the parental cells. The experiment was completed in triplicate. Averages (bars) and standard deviations (brackets) are shown. * indicates p ≤ 0.05 and fold change is indicated on the y-axis. Microsoft Exel and Adobe Photoshop were used to prepare Figure 4
Expression of only 2 additional genes was also significantly up-regulated (≥2 fold and p ≤ 0.05) in both the Rh30VR and Rh41VR ARMS cell lines compared with their corresponding parental cells: ATP-binding cassette sub-family B member 1 (MDR1) (Rh30 cells: 13,307 fold increased and p = 0.002, Rh41 cells: 1755 fold increased and p = 0.02) and major vault protein (MVP) (Rh30 cells: 2.4 fold increased and p = 0.002, Rh41 cells: 8.8 fold increased and p = 0.008) (Fig. 4a and b). MDR1 is a known target of GLI1 [26]. Gel mobility shifts and chromatin immunoprecipitation showed interaction of GLI1 with a consensus GLI binding site in the MVP promoter (Fig. 5a and b). Cotransfection assays showed that GLI1 up-regulates reporter gene expression through the MVP promoter (Fig. 5c), establishing MVP as another target of GLI1 during development of drug resistance. Four genes were significantly down-regulated (≥2 fold and p ≤ 0.05) in both the Rh30VR and Rh41VR ARMS cell lines compared with their corresponding parental cells: CYP1A1 (2.6 fold decreased and p = 0.01), CYP2D6 (2.8 fold decreased and p = 0.008), cyclin E1 (CCNE1) (4.4 fold decreased and p = 0.008), and ERBB3 (6.8 fold decreased and p = 0.006).
MVP is a direct transcriptional target of GLI1. a Electrophoretic mobility shift assays were completed using Rh30 cell lysate and 5 different probes that span the MVP promoter region and include each of the GLI consensus binding sites. Shifted bands (indicated by arrows) were visualized by anti-digoxigenin antibody and chemiluminescence for MVP probe 5 (lane marked -). Cold competitor was used to show specificity of binding in the lanes marked +. The gel has been cropped and the full-length gel is presented in Supplementary Figure 5A. b Chromatin immunoprecipitation was performed using Ruch-2 (left panel) and Rh41 (right panel) cells. PCR-amplified DNA bands are indicated in each of the cell lines by the arrows. IgG was used as negative control and anti-RNA polymerase2 (alpha-RNAP2) was used as positive control for the ChIP. Alpha-GLI1 = anti-c-terminal-GLI1 antibody. MVP 4,5 = PCR primers spanned the region from MVP probe 4 through MVP probe 5, which were also used in 5A. The gels have been cropped and full-length gels are presented in Supplementary Figure 5B. c Cotransfection assays were completed by transfecting HeLa cells with 0–1000 ng pCMV-GLI1 (black bars) or pCMV-control (yellow bars) effector plasmids, 200 ng of pMVP407 reporter construct and 20 ng of Renilla control reporter DNA. The experiments were performed in triplicate and results are expressed as an average with standard deviation. * indicates statistically significantly increased reporter activity using a given amount of effector pCMV-GLI1 DNA compared with control effector DNA (p ≤ 0.05). Microsoft Exel and Adobe Photoshop were used to prepare Figure 5
GLI1 was significantly up-regulated (≥2 fold and p ≤ 0.05) in TC71VR cells compared with parental cells (2.2 fold increased and p = 0.01) together with 6 other genes: MDR1 (4029 fold increased and p = 0.0002), transporter 1, ATP-binding cassette subfamily B member (TAP1) (2.8 fold and p = 0.003), fibroblast growth factor 2 (FGF2) (4.7 fold and p = 0.03), insulin growth factor 1 receptor (IGF1R) (2.0 fold and p = 0.01), estrogen receptor 2 (ESR2) (6.6 fold and p = 0.01), and tumor necrosis factor receptor superfamily member 11A (TNFRSF11A) (3.2 fold and p = 0.02) (Fig. 4c). Five genes were significantly down-regulated (≥2 fold and p ≤ 0.05) in the TC71VR cells compared with parental cells: aryl hydrocarbon receptor nuclear translocator (ARNT) (2.1 fold decreased and p = 0.01), CYP2B6 (4.2 fold decreased and p = 0.02), ERBB2 (4.2 fold decreased and p = 0.02), ERBB3 (16.7 fold decreased and p = 0.01), and retinoic acid receptor beta (RARb) (2.8 fold decreased and p = 0.01). Taken together, the VCR-resistant RMS and EWS cell lines that we established showed up-regulation of GLI1.
Modulation of GLI1 activity affects sensitivity of VCR-resistant RMS and EWS cell lines to VCR
Next, we tested whether SMO inhibition with vismodgeb or GLI1 inhibition either through pharmacologic inhibition or GLI1 siRNA, enhanced sensitivity of VCR-resistant RMS and EWS cell lines to treatment with VCR. Treatment of VCR-resistant ARMS or ERMS cells (Ruch-2VR, Rh30VR, Rh41VR, and UKF Rhb-1 cells) or VCR-resistant EWS cells (TC71VR) with VCR together with GANT61 significantly decreased cell viability (MTT assay) at doses that did not reduce cell viability individually (Fig. 6a and b). Treatment of Rh41VR cells with VCR together with vismodegib reduced cell viability to a much smaller extent than seen with GANT61 (Fig. 6a). Treatment of Ruch-2VR ARMS cells and TC71VR EWS cells with VCR together with GLI1 siRNA also significantly decreased cell viability at doses that did not reduce cell viability individually. However, treatment of Rh30VR and Rh41VR ARMS cells with GLI1 siRNA alone impacted cell viability even at the lowest concentration we tested (Fig. 6c and d).
Effect of modulation of GLI1 activity on VCR sensitivity of RMS and EWS cells. a GANT61 enhanced sensitivity of VCR-resistant RMS cells to VCR. Vismodegib enhanced sensitivity of Rh41VR cells to VCR to a smaller extent GANT61. The experiments were performed in triplicate and results are expressed as an average with standard deviation. * indicates statistically significant differences compared with treatment with VCR alone (black bars) (p ≤ 0.05). b GANT61 enhanced sensitivity of VCR-resistant EWS cells to VCR. The experiments were performed in triplicate and results are expressed as an average with standard deviation. * indicates statistically significant differences compared with treatment with VCR alone (black bars) (p ≤ 0.05). cGLI1 siRNA enhanced sensitivity of Ruch-2VR (top panel) RMS cells to VCR. Rh30VR (middle panel) and Rh41VR (bottom panel) cells were sensitive to GLI1 siRNA without VCR. The experiments were performed in triplicate and results are expressed as an average with standard deviation. * indicates statistically significant differences compared with VCR alone (black bars) (p ≤ 0.05). dGLI1 siRNA enhanced sensitivity of TC71VR EWS cells to VCR. The experiments were performed in triplicate and results are expressed as an average with standard deviation. * indicates statistically significant differences compared with VCR alone (black bars) (p ≤ 0.05). Microsoft Exel and Adobe Photoshop were used to prepare Figure 6
In summary, these experiments demonstrate that GLI1 up-regulation contributes to VCR-resistance of RMS and EWS cell lines and suggest that targeting GLI1 may benefit patients with RMS or EWS by reducing multidrug resistance.
Discussion
We have shown that ERMS, ARMS, and EWS cell lines express HH pathway components and that most of the cell lines have primary cilia, the organelle in which HH signal transduction occurs. We chose an in vitro approach to assess HH responsiveness and roles for GLI1 in drug resistance using well characterized RMS and EWS cell lines. Some of the cell lines (RD, Rh18, Rh41, CHLA9, CHLA10, TC32, and TC71) showed evidence of HH responsiveness with up-regulation of GLI1 and/or PTCH1 through the canonical pathway even when non-canonical mechanisms of activation of GLI1 may be present, such as EWS cell lines with a EWSR1-FLI1 fusion. This suggests that both canonical and non-canonical mechanisms may lead to GLI1 expression in RMS and EWS cells and that paracrine HH-signaling may also contribute to the behavior of these tumors in vivo. We saw the largest effects with DHH or SHH for RMS cell lines and with SHH for EWS cell lines. We did not see up-regulation of GLI1 or PTCH1 following exposure to HH ligands in Rh30 and CHLA258 cells, both of which have high basal levels of GLI1 expression. Rh30 RMS cells have GLI1-amplification and CHLA258 EWS cells, which were established at the time of recurrence, have a higher level of GLI1 expression than the other EWS cell lines. Presumably, non-canonical mechanisms drive high constitutive levels of GLI1 expression in both of these cell lines. Although EWS cell lines that were established at the time of recurrence and presumably have some degree of drug resistance showed higher levels of GLI1 expression than a EWS cell line established at the time of diagnosis, we could not make a similar comparison for the RMS cell lines. The information we had regarding the RMS cell lines included whether they were established before or after receiving any therapy and not whether they were established before or after recurrence. Previously treated cells could remain sensitive to chemotherapy and may not recur.
We analyzed the role of GLI1 in RMS and EWS cell lines in vitro by up-regulating HH signaling or down-regulating GLI1 activity with the small molecule inhibitor GANT61. It has been previously reported that GANT61 effects in RMS cell lines are specifically mediated through GLI1 inhibition [33]. Although we did not see effects of SHH exposure on cell viability or proliferation, we did see increased apoptosis in RMS and EWS cells and reduced viability and proliferation in EWS cells with GANT61 treatment, suggesting fundamental roles for GLI1 in the biology of these sarcoma cell lines.
Our focus was to determine whether up-regulation of GLI1 occurs as cells develop resistance to VCR. We chose VCR because it is a P-glycoprotein multi-drug resistance substrate and chemotherapeutic agent that is used for treatment of RMS and EWS [1,2,3,4]. We established VCR-resistant RMS and EWS cell lines, which we defined as an increase in the IC50 of ≥30 fold. We were able to achieve this in four (Ruch-2VR, Rh30VR, Rh41VR, and TC71VR cells) of ten cell lines that we attempted. We focused our analysis of the drug resistance array results in RMS cell lines on genes whose expression was altered in the same direction in resistant cells compared with the corresponding parental cells in more than one cell line. We were only able to establish a single VCR-resistant EWS cell line, which limits conclusions that can be made in EWS.
We saw up-regulation of GLI1 in all of the VCR-resistant cells by RT PCR array, although the magnitude of change did not reach the bar of 2.0-fold and p ≤ 0.05 for Ruch-2VR ERMS cells (1.7 fold and p = 0.02). The only other gene in the 86-gene drug resistance array that was up-regulated in Rh30VR, Rh41VR, and TC71VR cells was the known direct GLI1 target and multidrug resistance gene, MDR1. Only one gene was down-regulated in Rh30VR, Rh41VR, and TC71VR cells, the receptor tyrosine kinase, ERBB3. ERBB3, a member of the epidermal growth factor receptor family, is often aberrantly expressed and/or activated in cancers and has been associated with drug resistance [47]. It is unclear why this receptor tyrosine kinase is down-regulated in the VCR-resistant RMS and EWS cells compared with their parental cells. Curiously, ERBB3 localizes to 12q13, the same genomic region as GLI1, raising the possibility of complex co-regulation. Based on up-regulation of MVP in both VCR-resistant ARMS cell lines (Rh30VR and Rh41VR), we demonstrated that MVP is a direct transcriptional target of GLI1 using gel mobility shift assays, chromatin immunoprecipitation, and cotransfection assays. Down-regulation of CYP1A1 and CYP2D6 was also seen in both of the ARMS cell lines (Rh30VR and Rh41VR) possibly based on a decreased need for detoxifying enzymes in the presence of active drug efflux pumps.
We believe that canonical and non-canonical mechanisms contribute to GLI1 up-regulation and therefore focused on the effect of GLI1 inhibition rather than SMO inhibition on VCR sensitivity. In support of this decision, the GLI1 inhibitor GANT61 reversed VCR resistance more effectively in Rh41VR cells than the SMO inhibitor vismodegib. We were able to show that GLI1 down-regulation, either by GANT61 or GLI1 siRNA, in the VCR-resistant cell lines enhanced their sensitivity to VCR in vitro, supporting a role for GLI1 in RMS and EWS VCR resistance. Although statistically significant, the magnitude of the effect was somewhat limited when using our experimental conditions for GLI1 siRNA in Ruch-2VR and TC71VR cells. Rh30VR and Rh41VR cells were sensitive to GLI1 siRNA treatment even at the lowest concentrations tested, suggesting that viability of these cell lines is dependent on GLI1. This work supports the importance of developing ways to inhibit the GLI1 transcription factor, which may include targeting GLI1 expression or GLI1-coactivator interactions [48, 49].
Conclusions
In this study we showed that RMS and EWS cell lines express HH pathway components and are often responsive to HH ligands, with up-regulation of GLI1 and/or PTCH1 expression. HH signaling appears to play roles in cell survival in RMS and cell survival as well as proliferation in EWS cells. Up-regulation of GLI1 expression occurred in most cells as they developed resistance to VCR. In addition to MDR1, the ATP-binding cassette family member MVP is also a direct target of GLI1 and can also contribute to VCR resistance. Treatment of vincristine-resistant cells with the GLI1 small molecule inhibitor GANT61 or GLI1 siRNA together with VCR significantly decreased cell viability at doses that did not reduce viability individually. These experiments demonstrate that GLI1 up-regulation contributes to VCR resistance in RMS and EWS cell lines and suggest that targeting GLI1 may benefit patients with RMS or EWS by reducing multidrug resistance.
Availability of data and materials
The datasets used and/or analyzed during this study are available from the corresponding author on reasonable request.
Abbreviations
- DAPI:
-
4′,6-diamidino-2-phenyindole
- BrdU:
-
5-bromo-2′-deoxyuridine
- ARMS:
-
Alveolar rhabdomyosarcoma
- ARNT:
-
Aryl hydrocarbon receptor nuclear translocator
- MDR1:
-
ATP-binding cassette sub-family B member 1
- TAP1:
-
ATP binding cassette subfamily B member, transporter 1
- ChIP:
-
Chromatin immunoprecipitation
- CTR:
-
Control cells
- CCNE1:
-
Cyclin E1
- DHH:
-
Desert hedgehog
- ERMS:
-
Embryonal rhabdomyosarcoma
- ESR2:
-
Estrogen receptor 2
- EWS:
-
Ewing sarcoma
- FGF2:
-
Fibroblast growth factor 2
- HH:
-
Hedgehog
- IHH:
-
Indian hedgehog
- IGF1R:
-
Insulin growth factor 1 receptor
- kD:
-
Kilodaltons
- MVP:
-
Major vault protein
- MTT:
-
Methylthiazolyl diphenyl-tetrazolium bromide
- PTCH:
-
Patched
- qRT PCR:
-
Quantitative reverse transcriptase polymerase chain reaction
- RARb:
-
Retinoic acid receptor beta
- RT PCR:
-
Reverse transcriptase polymerase chain reaction
- RMS:
-
Rhabdomyosarcoma
- SMO:
-
Smoothened
- SHH:
-
Sonic hedgehog
- SUFU:
-
Suppressor of Fused
- Ct:
-
Threshold cycle
- TNFRSF11A:
-
Tumor necrosis factor receptor superfamily member 11A
- VCR:
-
Vincristine
References
Pappo AS, Anderson JR, Crist WM, Wharam MD, Breitfeld PP, Hawkins D, Raney RB, Womer RB, Parham DM, Qualman SJ, et al. Survival after relapse in children and adolescents with rhabdomyosarcoma: a report from the Intergroup Rhabdomyosarcoma Study Group. J Clin Oncol. 1999;17:3487–93.
Mascarenhas L, Lyden ER, Breitfeld PP, Walterhouse DO, Donaldson SS, Paidas CN, Parham DM, Anderson JR, Meyer WH, Hawkins DS. Randomized phase II window trial of two schedules of irinotecan with vincristine in patients with first relapse or progression of rhabdomyosarcoma: a report from the Children’s Oncology Group. J Clin Oncol. 2010;28:4658–63.
Leavey PJ, Mascarenhas L, Marina N, Chen Z, Krailo M, Miser J, Brown K, Tarbell N, Bernstein ML, Granowetter L, et al. Prognostic factor for patients with Ewing sarcoma (EWS) at first recurrence following multi-modality therapy: a report from the Children’s Oncology Group. Pediatr Blood Cancer. 2008;51:334–8.
Stahl M, Ranft A, Paulussen M, Bölling T, Vieth V, Bielack S, Görtitz I, Braun-Munzinger G, Hardes J, Jürgens H, et al. Risk of recurrence and survival after relapse in patients with Ewing sarcoma. Pediatr Blood Cancer. 2011;57:549–53.
Ingham PW. From Drosophila segmentation to human cancer therapy. Development. 2018;145(21):dev168898.
Briscoe J, Thérond PP. The mechanisms of hedgehog signaling and its roles in development and disease. Nat Rev Mol Cell Biol. 2013;14:416–29.
Hui CC, Angers S. Gli proteins in development and disease. Annu Rev Cell Dev Biol. 2011;27:513–37.
Wong SY, Reiter JF. The primary cilium at the crossroads of mammalian hedgehog signaling. Curr Top Dev Biol. 2008;85:2225–60.
Hooper JE, Scott MP. Communicating with hedgehogs. Nat Rev Mol Cell Biol. 2005;6:306–17.
Ruiz i Altaba A, Sánchez P, Dahmane N. Gli and hedgehog in cancer: tumours, embryos and stem cells. Nat Rev Cancer. 2002;2:361–72.
Gu D, Xie J. Non-canonical Hh signaling in cancer-current understanding and future directions. Cancers (Basel). 2015;7:1684–98.
Yoon JW, Gilbertson R, Iannaccone S, Iannaccone P, Walterhouse D. Defining a role for sonic hedgehog pathway activation in desmoplastic medulloblastoma by identifying GLI1 target genes. Int J Cancer. 2009;124:109–19.
Das S, Samant RS, Shevde LA. Nonclassical activation of hedgehog signaling enhances multidrug resistance and makes cancer cells refractory to smoothened-targeting hedgehog inhibition. J Biol Chem. 2013;288:11824–33.
Varnat F, Duquet A, Malerba M, Zbinden M, Mas C, Gervaz P, Ruiz i Altaba A. Human colon cancer epithelial cells harbour active HEDGEHOG-GLI signalling that is essential for tumour growth, recurrence, metastasis and stem cell survival and expansion. EMBO Mol Med. 2009;1:338–51.
Sims-Mourtada J, Izzo JG, Apisarnthanarax S, Wu TT, Malhotra U, Luthra R, Liao Z, Komaki R, van der Kogel A, Ajani J, et al. Hedgehog: an attribute to tumor regrowth after chemoradiotherapy and a target to improve radiation response. Clin Cancer Res. 2006;12:6565–72.
Cui D, Xu Q, Wang K, Che X. Gli1 is a potential target for alleviating multidrug resistance of gliomas. J Neurol Sci. 2010;288:156–66.
Queiroz KC, Ruela-de-Sousa RR, Fuhler GM, Aberson HL, Ferreira CV, Peppelenbosch MP, Spek CA. Hedgehog signaling maintains chemoresistance in myeloid leukemic cells. Oncogene. 2010;29:6314–22.
Sims-Mourtada J, Izzo JG, Ajani J, Chao KS. Sonic hedgehog promotes multiple drug resistance by regulation of drug transport. Oncogene. 2007;26:5674–9.
Whitson RJ, Lee A, Urman NM, Mirza A, Yao CY, Brown AS, Li JR, Shankar G, Fry MA, Atwood SX, et al. Noncanonical hedgehog pathway activation through SRF-MKL1 promotes drug resistance in basal cell carcinomas. Nat Med. 2018;24:271–81.
Faião-Flores F, Alves-Fernandes DK, Pennacchi PC, Sandri S, Vicente AL, Scapulatempo-Neto C, Vazquez VL, Reis RM, Chauhan J, Goding CR, et al. Targeting the hedgehog transcription factors GLI1 and GLI2 restores sensitivity to vemurafenib-resistant human melanoma cells. Oncogene. 2017;36:1849–61.
Della Corte CM, Bellevicine C, Vicidomini G, Vitagliano D, Malapelle U, Accardo M, Fabozzi A, Fiorelli A, Fasano M, Papaccio F, et al. SMO gene amplification and activation of the hedgehog pathway as novel mechanisms of resistance to anti-epidermal growth factor receptor drugs in human lung cancer. Clin Cancer Res. 2015;21:4686–97.
Gan GN, Eagles J, Keysar SB, Wang G, Glogowska MJ, Altunbas C, Anderson RT, Le PN, Morton JJ, Frederick B, et al. Hedgehog signaling drives radioresistance and stroma-driven tumor repopulation in head and neck squamous cancers. Cancer Res. 2014;74:7024–36.
Liu Z, Xu J, He J, Zheng Y, Li H, Lu Y, Qian J, Lin P, Weber DM, Yang J, Yi Q. A critical role of autocrine sonic hedgehog signaling in human CD138+ myeloma cell survival and drug resistance. Blood. 2014;124:2061–71.
Zahreddine HA, Culjkovic-Kralijacic B, Assouline S, Gendron P, Romeo AA, Morris SJ, Cormack G, Jaquith JB, Cerchietti L, Cocolakis E, et al. The sonic hedgehog factor GLI1 imparts drug resistance through inducible glucuronidation. Nature. 2014;511:90–3.
Keysar SB, Le PN, Anderson RT, Morton JJ, Bowles DW, Paylor JJ, Vogler BW, Thorburn J, Fernandez P, Glogowska MJ, et al. Hedgehog signaling alters reliance on EGF receptor signaling and mediates anti-EGFR therapeutic resistance in head and neck cancer. Cancer Res. 2013;73:3381–92.
Chen Y, Bieber MM, Teng NN. Hedgehog signaling regulates drug sensitivity by targeting ABC transporters ABCB1 and ABCG2 in epithelial ovarian cancer. Mol Carcinog. 2014;53:625–34.
Singh RR, Kunkalla K, Qu C, Schlette E, Neelapu SS, Samaniego F, Vega F. ABCG2 is a direct transcriptional target of hedgehog signaling and involved in stroma-induced drug tolerance in diffuse large B-cell lymphoma. Oncogene. 2011;30:4874–86.
Ragazzini P, Gamberi G, Pazzaglia L, Serra M, Magagnoli G, Ponticelli F, Ferrari C, Ghinelli C, Alberghini M, Bertoni F, et al. Amplification of CDK4, MDM2, SAS and GLI genes in leiomyosarcoma, alveolar and embryonal rhabdomyosarcoma. Histol Histopathol. 2004;19:401–11.
Pressey JG, Anderson JR, Crossman DK, Lynch JC, Barr FG. Hedgehog pathway activity in pediatric embryonal rhabdomyosarcoma and undifferentiated sarcoma: a report from the Children’s Oncology Group. Pediatr Blood Cancer. 2011;57:930–8.
Zibat A, Missiaglia E, Rosenberger A, Pritchard-Jones K, Shipley J, Hahn H, Fulda S. Activation of the hedgehog pathway confers a poor prognosis in embryonal and fusion gene-negative alveolar rhabdomyosarcoma. Oncogene. 2010;29:6323–30.
Srivastava RK, Kaylani SZ, Edrees N, Li C, Talwelkar SS, Xu J, Palle K, Pressey JG, Athar M. GLI inhibitor GANT-61 diminishes embryonal and alveolar rhabdomyosarcoma growth by inhibiting Shh/AKT-mTOR axis. Oncotarget. 2014;5:12151–65.
Kawabata N, Ijiri K, Ishidou Y, Yamamoto T, Nagao H, Nagano S, Maeda S, Komiya S, Setoguchi T. Pharmacological inhibition of the hedgehog pathway prevents human rhabdomyosarcoma cell growth. Int J Oncol. 2011;39:899–906.
Tostar U, Toftgård R, Zaphiropoulos PG, Shimokawa T. Reduction of human embryonal rhabdomyosarcoma tumor growth by inhibition of the hedgehog signaling pathway. Genes Cancer. 2010;1:941–51.
Almazán-Moga A, Zarzosa P, Molist C, Velasco P, Pyczek J, Simon-Keller K, Girlat I, Vidal I, Navarro N, Segura MF, et al. Ligand-dependent hedgehog pathway activation in rhabdomyosarcoma: the oncogenic role of the ligands. Br J Cancer. 2017;117:1314–25.
Zwerner JP, Joo J, Warner KL, Christensen L, Hu-Lieskovan S, Triche TJ, May WA. The EWS/FLI1 oncogenic transcription factor deregulates GLI1. Oncogene. 2008;27:3282–91.
Beauchamp E, Bulut G, Abaan O, Chen K, Merchant A, Matsui W, Endo Y, Rubin JS, Toretsky J, Uren A. Gli1 is a direct transcriptional target of EWS-FLI1 oncoprotein. J Biol Chem. 2009;274:9074–82.
Tostar U, Malm CJ, Meis-Kindblom JM, Kindblom LG, Toftgård R, Undén AB. Deregulation of the hedgehog signaling pathway: a possible role for the PTCH and SUFU genes in human rhabdomyoma and rhabdomyosarcoma development. J Pathol. 2006;208:17–25.
Hahn H, Wojnowski L, Zimmer AM, Hall J, Miller G, Zimmer A. Rhabdomyosarcomas and radiation hypersensitivity in a mouse model of Gorlin syndrome. Nat Med. 1998;4:619–22.
Lee Y, Kawagoe R, Sasai K, Li Y, Russel HR, Curran T, McKinnon PJ. Loss of suppressor-of-fused function promotes tumorigenesis. Oncogene. 2007;26:6442–7.
Svärd J, Rozell B, Toftgård R, Teglund S. Tumor suppressor gene co-operativity in compound Patched1 and suppressor of fused heterozygous mutant mice. Mol Carcinog. 2009;48:408–19.
Barr FG, Duan F, Smith LM, Gustafson D, Pitts M, Hammond S, Gastier-Foster JM. Genomic and clinical analyses of 2p24 and 12q13-q14 amplification in alveolar rhabdomyosarcoma: a report from the Children’s Oncology Group. Genes Chromosomes Cancer. 2009;48:661–72.
Weber-Hall S, Anderson J, McManus A, Abe S, Nojima T, Pinkerton R, Pritchard-Jones K, Shipley J. Gains, losses, and amplification of genomic material in rhabdomyosarcoma analyzed by comparative genomic hybridization. Cancer Res. 1996;56:3220–4.
Kelleher FC, Cain JE, Healy JM, Watkins DN, Thomas DM. Prevailing importance of the hedgehog signaling pathway and the potential for treatment advancement in sarcoma. Pharmacol Ther. 2012;136:153–68.
Joo J, Christensen L, Warner K, States L, Kang HG, Vo K, Lawlor ER, May WA. GLI1 is a central mediator of EWS/FLI1 signaling in Ewing tumors. PLoS One. 2009;4(10):e7608.
Oue T, Yoneda A, Uehara S, Yamanaka H, Fukuzawa M. Increased expression of the hedgehog signaling pathway in pediatric solid malignancies. J Pediatr Surg. 2010;45:387–92.
Lauth M, Bergström A, Shimokawa T, Toftgård R. Inhibition of GLI-mediated transcription and tumor cell growth by small-molecule antagonists. Proc Natl Acad Sci U S A. 2007;104:8455–60.
Lee Y, Ma J, Lyu H, Huang J, Kim A, Liu B. Role of erbB3 receptors in cancer therapeutic resistance. Acta Biochim Biophys Sin Shanghai. 2014;46:190–8.
Yoon JW, Lamm M, Iannaccone S, Higashiyama N, Leong KF, Iannaccone P, Walterhouse D. p53 modulates the activity of the GLI1 oncogene through interactions with the shared coactivator TAF9. DNA Repair (Amst). 2015;34:9–17.
Taylor R, Long J, Yoon JW, Childs R, Sylvestersen KB, Nielsen ML, Leong KF, Iannaccone S, Walterhouse DO, Robbins DJ, et al. Regulation of GLI1 by cis DNA elements and epigenetic marks. DNA Repair (Amst). 2019;79:10–21.
Acknowledgements
Not applicable.
Funding
All of this work was supported together by a Gold in September (G9) Center of Excellence Grant, the Pediatric Cancer Research Foundation, and the Megan Bugg Foundation.
Author information
Authors and Affiliations
Contributions
JWY designed the experiments, conducted the experiments, analyzed the data, and was a major contributor in writing the manuscript. ML analyzed the data. CC conducted the experiments for Fig. 2. PI analyzed the data and was a major contributor in writing the manuscript. DW designed the experiments, analyzed the data, and was a major contributor in writing the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
None of the human cancer cell lines that were used in this study required ethics approval for use.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Yoon, J., Lamm, M., Chandler, C. et al. Up-regulation of GLI1 in vincristine-resistant rhabdomyosarcoma and Ewing sarcoma. BMC Cancer 20, 511 (2020). https://doi.org/10.1186/s12885-020-06985-0
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12885-020-06985-0