Abstract
Background
Recently, microRNA-20a (miR-20a) has been reported to influence the clinical features and may have prognostic value in human cancers. The present meta-analysis assessed the prognostic role of miR-20a in various carcinomas.
Methods
Literature searches of seven electronic databases were performed for eligible articles of the prognostic role of miR-20a in human cancers. Hazard ratios (HR) for overall survival (OS), disease free survival (DFS), progression-free survival (PFS) as well as their 95% confidence intervals (95%CIs) were used to assess the influence of miR-20a expression on patient prognosis. Odds ratio (OR) and 95%CIs were applied to evaluate the correlation between miR-20a expression and clinicopathological characteristics.
Results
Based on the OS analyzed by log rank tests, there was a significant association between miR-20a levels and OS by fixed effects model. By subgroup analyses, the significance was also observed in the studies of specimen derived from blood and gastrointestinal cancer group. The independent prognostic role of miR-20a expression for the OS was observed significantly by fixed effects model. In addition, we observed significant association between miR-20a expression levels and DFS of log rank tests, DFS of cox regression. Significant relation of gender/differentiation and the expression level of miR-20a was identified.
Conclusions
Base on the findings, the elevated miR-20a expression level is related to poor prognosis of gastrointestinal cancer patients. As for other types of carcinomas, the results are still not stable and more studies are required to further identify miR-20a prognostic values. In addition, miR-20a expression level is relatively higher in women than that in men, and increased miR-20a expression level is linked to poor tumor differentiation.
Similar content being viewed by others
Background
Cancer has become the major social health problem, and it is now the leading cause of mortality worldwide due to its growing incidence each year and poor prognosis. Although new treatment approaches, such as surgery, radiotherapy and chemotherapy, have been elaborately developed, the clinical outcome of carcinomas still remains unsatisfied. One of the reasons is lack of effective biological markers help to define subgroups of patients who might benefit or not benefit from some specific treatments. Therefore, exploring potential diagnostic and prognostic biomarkers for human malignancies to guide clinical decision is crucial and urgent.
MicroRNAs (miRNA) are small noncoding molecules of with a length of approximately 18–24 nucleotides, and can negatively regulate their target genes expression [1, 2]. Many miRNAs have been identified to express abnormally in human malignancies and can play an oncogenic or anti- oncogenic role in tumor biological behaviors [3, 4]. Owing to its stability and detectability in tissues/blood, miRNA is one of the most promising biomarkers for the prognosis of human cancers [5,6,7].
MiR-20a which is one member of miR-17-92 cluster, has been identified to be closely associated with cancer proliferation [8, 9], invasion [10], metastasis [8, 9, 11] or chemotherapeutic resistance [12, 13] by recent studies. These could be largely attributed to the active biological activities of miR-20a in inference the cellular signal pathways, such as PTEN/PI3K/AKT pathway [14, 15], MAPK1/c-Myc parthway [16], ENH1/Id1 parthway [17], FAS promoter activity [18], TAKI expressions [19], FBXL5/BTG3 signaling [20], the Sonic Hedgehog pathway [21] and etc. Nevertheless, there exist inconsistencies about prognostic accuracy of miR-20a, though numerous studies have identified the associations between miR-20a and various human cancers. Wang et al. [22], Cheng et al. [23], Xu et al. [24] and Reng et al. [25] found that the high expression level of miR-20a was associated with a poor survival rates in cancer patients. But Chang et al. [26], Zhang et al. [27] and Fan et al. [28] explored an anti-tumor effect of miR-20a and patients in their studies benefited from up-regulated miR-20a. Marchini et al. [29], and Xu et al. [30] observed no statistically relationship between expression level of miR-20a and overall survivals of patients. Hence, based on the whole published relevant researches, a systematic analysis was conducted to assess the prognostic efficiency of miR-20a in human cancers as well as the association between miR-20a expression and cancer patients’ clinical characteristics.
Methods
Publication selection
An electronic search of PubMed, Web of Science (WOS), Embase in English and VIP, Wanfang, SinoMed and the China National Knowledge Infrastructure (CNKI) in Chinese was applied to select articles using the following keywords: ‘tumor’ or ‘cancer’ or ‘carcinoma’ or ‘neoplasm’ or ‘malignancies’ and ‘miRNA-20a’ or ‘miR-20a’ and ‘prognos*’, ‘surviv*’. We also retrieved articles manually from other sources to complement the results. The search was updated in July 13 2018.
Eligibility criteria
Studies from the initial researches that satisfy the criteria below were thought to be eligible. (1) studies evaluated the prognostic value of blood or tissue miR-20a level in various human cancers. (2) the relationships between miR-20a expression and patients’ survival were described; (3) Studies have sufficient data to calculate the hazard ratios (HR) and 95%confidence interval (95%CI) for survival rates or odds ratio (OR) and 95%CI for the correlation between miR-20a expression and clinicopathological characteristics. (4) there was no restrictions on the methods of detecting the miR-20a expression levels in the cancer patients by some specific methods, such as qRT-PCR, microarray or etc.
Studies were excluded if (1) patients were of benign tumors. (2) there were notthe primary carcinomas but metastatic carcinomas from other organs. (3) the articles were letters, duplicated publications, reviews or case reports. (4) literatures were published in languages other than English or Chinese.
Data extraction
To ensure the accuracy of data extraction, two authors (DH and YP) extracted data separately from the eligible studies and inconsistencies were solved by a third senior author (KM). For all enrolled studies, the following information was listed: the first author; year of publication; country; tumor type and clinical stage; number of patients included; the type of specimen; detection methods of miR-20a expression levels; follow-up time; cut-off values; survival analysis and their source of HR; HR for overall survival (OS), disease free survival (DFS), progression-Free-Survival (PFS) and relapse free survival (RFS) as well as their 95%CIs and the quality of study. What’s more, the clinicopathological characteristics of including patients were extracted from some studies which reported the data.
Figures of HR and its corresponding 95% CI of univariate and/or multivariate analyses could be directly obtained from some of studies, whereas others only showed Kaplan-Meier curves without specific data. For these researches, we extracted necessary data from Kaplan-Meier curves by Engauge Digitizer version 9.8 and then input the extracted survival rates at specific time points into the spreadsheet constructed by Tierney et al. [31] to acquire the HR and its corresponding 95%CIs.
Quality assessment
All the included studies were retrospective and non-randomized studies. We applied the Newcastle-Ottawa Scale (NOS) for evaluating the quality [32]. The NOS scores ranged from 0 to 9, and score more than 6 was regarded as high quality. Three authors (DH, YP and KM) assess the quality independently and any disagreement was resolved by discussion.
Statistical analysis
The PRISMA checklists and their guidelines were strictly followed during the whole procedure of the study [33, 34]. The meta-analysis was conducted with software version 14.0 (Stata Corporation, College Station, TX, USA). The pooled HRs and their 95%CIs were used to assess the impact of miR-20a expression levels on clinical prognosis for OS, DFS and PFS. The adjusted HRs (95 %CIs) for OS and DFS were also calculated using data extracted from the cox regression. HRs larger than 1 denoted poorer prognosis in patients with increased miR-20a expression. The fixed effects model and the random pooling model were both used in the analyses. P less than 0.05 or the 95%CI did not overlap with 1 indicated statistically significant. The heterogeneity among studies was calculated by the Chi square-based Q test and I2 statistics. P value less than 0.10 for the Q test or I2 larger than 50% was considered as significant heterogeneity. Subgroup analyses stratified by population (Chinese and Italian), sample size (< 150 and ≥ 150), NOS scores (< 8 and ≥ 8), specimen (blood and tissues) and tumor category (gastrointestinal cancer and non-gastrointestinal cancer) were carried out. The sensitivity analysis also managed to assess the stability of the results by omitting each study in turn. Publication bias was estimated by visually evaluating the asymmetry of the funnel plot. What’s more, Egger’s linear regression test and Begg’s funnel plot test were applied to offer quantitative evidence of publication bias. The odds ratios (ORs) and corresponding 95%CIs were also computed to detect the relation of miR-20a expression to clinicopathological characteristics. All P values were two tailed.
Results
Characteristics of the enrolled studies
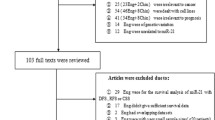
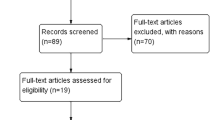
The article retrieval strategy was shown in Fig. 1. The initial search identified a total of 1662 papers, 266 of which were removed due to duplications. Titles/abstracts of there maining 1396 publications were reviewed, 1374 being excluded and leaving 22 as candidate literatures. After a full text evaluation, a total of 14 studies were finally included [22,23,24,25,26,27,28,29,30, 35,36,37,38,39].
The articles were published between 2011 and 2018. The total number of subjects included in the current meta-analysis was 1822, and the sample size ranged from 52 to 544 with a mean value of 130.1. Twelve enrolled studies were carried out in China, and the other two studies were conducted in Europe. There were eight types of cancer in the included papers, with three studies for squamous cell carcinoma, three studies for gastric cancer, two studies for NSCLC, two studies for ovarian cancer, one study for colorectal cancer, one study for glioblastoma, one study for multiple myeloma and one study for hepatocellular carcinoma. There were 10, 4, 2, 1 studies containing HR and its corresponding 95% CI for OS, DFS, PFS and RFS, respectively. Thirteen studies measured the miR-20a expression level by qRT-PCR (Real-time Polymerase Chain Reaction), while two studies also applied the methods of miRNA array besides qRT-PCR and one study used the nCounter Human v2 miRNA Expression Assay described in its article [38]. The main information of the included studies was performed in Table 1.
The association between miR-20a expression levels and overall survival (OS)
Ten enrolled studies including 1497 patients investigated the relation of miR-20a expression levels to the prognostic parameters (OS) using log rank tests, resulting in the univariate data. Generally, there was a significant association between miR-20a levels and OS (HR = 1.26, CI: 1.06–1.50, Fig. 2a), however, a significant heterogeneity was observed among the researches (I2 = 89.30%, P < 0.10, Table 2). Whereby, the random pooling model was applied in succession and the significance was vanished (HR = 0.99, CI: 0.56–1.75, Table 2), indicating that the heterogeneity influenced the results significantly. Then subgroup analyses were conducted by factors including population (Chinese and Italian), sample size (≥150 and < 150), NOS scores (≥8 and < 8), specimen (blood and tissues) and tumor category (gastrointestinal cancer and non-gastrointestinal cancer), so as to diminish the heterogeneity. As a consequence, the heterogeneity was controlled successfully in the group with specimen derived from blood (I2 = 0.00%, P = 0.878, Table 2) and the corresponding significance was obvious (HR = 1.93, CI: 1.54–2.41, Fig. 2e). Also, the gastrointestinal cancer group revealed eliminated heterogeneity as well (I2 = 0.00%, P = 0.402, Fig. 2f), and the relation of miR-20a levels to OS was also significant (HR = 1.85, CI: 1.43–2.40, Fig. 2f). Moreover, significant associations were observed between miR-20a expression levels and OS in the studies with Chinese samples (HR = 1.31, CI: 1.10–1.56, Fig. 2b), sample size greater than or equal to 150 (HR = 1.77, CI: 1.34–2.34, Fig. 2c), NOS scores greater than or equal to 8 (HR = 1.51, CI: 1.24–1.83, Fig. 2d) or less than 8 (HR = 0.59, CI: 0.40–0.88, Fig. 2d) and specimen derived from tissues (HR = 0.69, CI: 0.53–0.90, Fig. 2e) by fixed pooling model, while there were no significances identified in these groups, when the random pooling model was applied (Table 2). And the heterogeneities within the subgroups were still significant, except for the groups of gastrointestinal cancer as well as samples derived from tissues.
The association between miR-20a expression levels and a overall survival, subgroup analyses of b population (Chinese and Italian) c sample sizes (≥150 and < 150), d NOS scores (≥8 and < 8), e specimen (blood and tissues), f tumor category (gastrointestinal cancer and non-gastrointestinal cancer) by fixed effects model
Based on the results above, meta regression was further performed, but there was no significant contribution identified to greatly influence the variation of HRs (p = 0.894 for population, p = 0.405 for sample size, p = 0.829 for NOS scores, p = 0.621 for specimen, p = 0.350 for tumor category, respectively, Table 2). Moreover, the sensitivity analysis was performed and no studies seemed to have great impacts on the significance of the results (Fig. 3a). In addition, funnel plots, Begg’s rank correlation and Egger’s weighted regression method were implemented to evaluate the publication bias. Though the efficacy of these methods might be limited due to the insufficient studies amount, we still chose those methods for lack of alternatives. The funnel plot of all seven studies reported symmetric and the Begg’s, Egger’s tests revealed no significant publication bias (P = 0.721, P = 0.213, respectively). The sensitivity analysis within the gastrointestinal cancer group also revealed that no studies could significantly impacted the results, indicating the stableness and reliability of the results (Fig. 3b).
The independent role of miR-20a expression level as prognostic indicator
Seven researches containing 1252 patients implemented the cox multivariate regression to evaluate the prognostic value of miR-20a expression levels in cancer patients, adjusting other factors. The significant relation of miR-20a expression to the OS (HR = 1.52, CI: 1.24–1.85, Fig. 4a) was observed by fixed pooling model. However, the significance was vanished by random pooling model (HR = 1.07, CI: 0.47–2.44, Table 3) and the heterogeneity was relatively obvious (I2 = 93.40%, P < 0.10, Table 3). Similarly, to reduce heterogeneity, subgroup analyses were applied and the homogeneity was reached within the studies of samples derived from blood (I2 = 9.60%, P = 0.331, Table 3). And the significant association was identified between miR-20a expression levels and OS within the group of samples derived from blood (HR = 1.87, CI: 1.47–2.37, Fig. 4e). In addition, the significant relations of miR-20a expression levels to OS were also recognized within the studies of Chinese samples (HR = 1.59, CI: 1.29–1.94, Fig. 4b), sample size greater than or equal to 150 (HR = 2.16, CI: 1.62–2.88, Fig. 4c), NOS scores greater than or equal to 8 (HR = 2.19, CI: 1.74–2.77, Fig. 4d) or less than 8 (HR = 0.52, CI: 0.35–0.77, Fig. 4d) and the patients of gastrointestinal cancer (HR = 2.41, CI: 1.77–3.28, Fig. 4f) by fixed effects model, which turned out to be of no significance within those subgroups by random pooling model (Table 3). Sensitivity analyses revealed no studies had significant impacts on the results (Fig. 3c). Furthermore, no obvious publication bias was identified among the four studies (P = 0.230 for Begg’s test and P = 0.287 for Egger’s test, respectively). Moreover, meta regression was further performed to identify the underlying factors contributing to the variation of HRs. As a result, it suggested that nearly all of the subgroup factors except NOS scores had contributed to the between-study variance (p = 0.020 for population, p = 0.011 for sample size, p = 0.107 for NOS scores, p = 0.040 for specimen, p = 0.012 for tumor category, respectively). Normalizing all the significant factors observed, the estimate of between-study variance, Tau-squared (tau2), plummeted from 1.1191 to 0, indicating that these factors completely explained the between-study variance. Also, the residual heterogeneity was diminished (I2 = 0.00%).
The independent role of miR-20a as a prognostic detector for the overall survivals in patients of carcinoma, a overall survivals, subgroup analyses of b population (Chinese and Italian) c sample sizes (≥150 and < 150), d NOS scores (≥8 and < 8), e specimen (blood and tissues), f tumor category (gastrointestinal cancer and non-gastrointestinal cancer) by fixed effects model
The relation of miR-20a expression levels to DFS and PFS
Four studies reported disease-free survival (DFS), of which two studies applied only log rank tests, while others also utilized cox multivariate regression. Thus, after pooling the HR, we observed significant association between miR-20a expression levels and DFS of log rank tests (HR = 1.99, CI: 1.52–2.61, Fig. 5a), DFS of cox regression (HR = 2,41, CI: 1.88–3.09, Fig. 5c) by fixed effects model. However, the heterogeneities were rather obvious (I2 = 67.80%, P = 0.078, Fig. 5a; I2 = 83.6%, P = 0.000, respectively Fig. 5c). Nevertheless, the significance for data extracted from log rank tests and cox regression still existed by random pooling model (HR = 2.33, CI: 1.22–4.46, Fig. 5b; HR = 2.88, CI: 1.38–5.99, Fig. 5d), indicating the stability of the results. Furthermore, owing to limited number of statistics from log rank tests, the sensitivity analysis was only applied to the analyses with data extracted from cox regression, revealing that no studies had great impacts on the results (Fig. 3d). However, the investigation of publication bias identified an outlier (Fig. 5e), Cheng et al. [23] After removing this study, the heterogeneity was completely eliminated (I2 = 0.00%, P = 0.688, Figure5F) and the significance of association between miR-20a expression levels and DFS was not altered (HR = 1.90, CI: 1.45–2.49, Fig. 5f).There were only two studies containing 174 patients revealed the available PFS statistics. However, the prognostic value of miR-20a expression levels to PFS were completely different (Gao et al. [39] HR = 2.25, CI 1.18–4.30; Marchini et al. [29] HR = 0.36, CI 0.16–0.80). Due to insufficient data, pooling effects would be lack of efficiency and the heterogeneity was significant (I2 = 91.8%, P < 0.10). Thus, more relevant studies reporting the prognostic effects of miR-20a to PFS are required to perform the analysis.
The association between miR-20a expression levels and DFS extracted from log rank tests by a fixed effects model and b random pooling model, cox regression by c fixed effects model and d random pooling model, f cox regression by fixed effects model after removing the outlier; e publication bias evaluation for DFS
Correlations between miR-20a levels and clinicopathological features among various carcinomas
There are seven articles containing 1103 patients of various carcinomas investigated the association between miR-20a expression levels and different clinic characteristics. As shown in Table 4, there were significant relations observed among gender (OR = 0.69, CI: 0.51–0.92), differentiation (OR = 1.50, CI: 1.06–2.13) by fixed effects model to the expression level of miR-20a. However, the significance was altered by random pooling model. Moreover, there were no significance identified in the association between age (OR = 1.44, CI: 0.81–2.57), tumor sizes (OR = 0.77, CI: 0.44–1.36), lymph node metastasis (OR = 1.03, CI = 0.48–2.21) or TNM stage (OR = 1.15, CI: 0.51–2.58) and the expression level of miR-20a. The heterogeneity was absent in the analysis of age (I2 = 0.00%, P = 0.373) and moderate in the analysis of gender (I2 = 30.70%, P = 0.194) but obvious in the analyses of tumor sizes (I2 = 49.20%, P = 0.116), lymph node metastasis (I2 = 54.30%, P = 0.112), TNM stage (I2 = 74.40%, P = 0.004) and differentiation (I2 = 59.10%, P = 0.032). Sensitivity analysis and investigation of publication bias were applied to each clinic characteristic analysis. The sensitivity analysis of the gender identified a study, Huang et al. [35], which had significant impact on the pooling results (Fig. 6a). After remove the outlier, the heterogeneity was greatly decreased from 30.70 to 0.00%, and also there was a significant association between miR-20a expression and gender (HRs = 0.61, CI: 0.45–0.83, Fig. 6c). In addition, the sensitivity analysis recognized another outlier in the characteristics analysis of differentiation, which was Fan et al. [28] (Fig. 6b). The removal of Fan et al. did diminish the heterogeneity (I2 = 36.20%, P = 0.180, Fig. 6d), and a significant relation was identified of miR-20a expression levels to the degrees of differentiation (HRs = 1.73, CI: 1.19–2.51, Fig. 6d). Moreover, the publication bias was identified in the analysis of TNM stages (P = 0.086 for Begg test, P = 0.059 for Egger test, respectively). By the Begg’s plot (Fig. 6e), we identified the outlier, Wang et al. [22] The heterogeneity was reduced in an extent (I2 = 51.7%, P = 0.102), but the pooling result was not significantly altered (HRs = 0.80, CI: 0.43–1.50, Fig. 6f).
Sensitivity analyses for the pooling relation of miR-20a expression levels to clinicopathological characteristics, such as a gender, b tumor differentiation degree; the association between miR-20a expression levels and c gender, d tumor differentiation degree or f TNM stages without the outlier; e publication bias evaluation for the studies reporting TNM stages
Discussion
MiR-20a, which has been detected to be aberrantly expressed in many malignancies, may play as a novel biomolecule in tumor progression [40,41,42]. Several genes or signal pathways have been discovered to be targeted by miR-20a in tumor biological behavior, such as KIF26B in osteosarcoma [43], RB1CC1/FIP200 in breast cancer [44], LIMK1 in anaplastic thyroid cancer [45], ABL2 in prostate cancer [42] and etc. There are three systematic reviews (Zheng et al., Li et al., Troiano et al.) summarizing the prognostic value of miR-20a expression in specific type of tumor recently: Zheng et al. [46] and Li et al. [47] generalized that the up-regulated expression of miR-20a was correlated with a poor prognosis in gastrointestinal cancer and cervical cancer, respectively. Nevertheless, Troiano et al. [48] came to an opposite conclusion in oral squamous cell carcinoma against the former two articles. A number of studies have been carried out to illustrate the prognostic role of miR-20a in cancer, but the underlying value of miR-20a for survival rates in various cancer patients remains unclear.
The current meta-analysis intended to explore the association between the expression level of miR-20a and human cancer prognosis. To our knowledge, this is the most comprehensive meta-analysis providing insights into the clinical value of miR-20a in various types of human cancers currently. Fourteen papers including 1822 patients were recruited in this meta-analysis. Ten studies containing 1497 patients reported the statistics of OS as a result of log rank tests. By the pooling strategy, we identified that the elevated miR-20a expression was linked to poor prognosis of cancer patients. Then subgroup analyses were implemented to eliminate the potential sources of heterogeneity. Consequently, the homogeneity was reached within the groups of gastrointestinal cancer and samples derived from blood, and the OS of gastrointestinal cancer group was found to be greatly associated with the miR-20a expression levels. In addition, seven articles including 1252 patients contained the data of HRs derived from cox multivariate regression of survival analysis. The cox regression [49] has been proved to be effective in the survival analysis, because it evaluates the contribution of each factor independently by adjusting others. Thus, the results always indicate the independent effects of each factor on the clinic outcome. However, subgroup analyses found that the significance might be vanished in gastrointestinal cancer group when the pooling strategy was changed from fixed effects model to random pooling model. As shown in Table 3, the subgroup of gastrointestinal cancer recruited only two studies and the results might not be reliable. In addition, by retrieving the studies, we found that the relation of miR-20a levels to OS was both significant and consistent (Wang et al. [22] HR = 1.58, CI 1.10–2.25; Cheng et al. [23] HR = 8.22, CI 4.47–15.12), which means that it is still safe to draw the conclusion that overexpression of miR-20a is linked to poor prognosis of gastrointestinal cancer patients. Moreover, meta regression illustrated that the contribution of various factors included population, sample size, specimen and tumor category. By adjusting all the identified factors, the between-study variance was completely explained and furthermore, the remaining heterogeneity was diminished.
Upregulation of miR-20a has been found to inhibit the proliferation, invasion and migration of cancer cells [16, 50, 51]. Whereby, overexpression of miR-20a has been reported to promote migration and invasion of various cancers [52,53,54]. The regulation of miR-20a to cancer cells verified from different cancers, and even for the same kind of carcinoma such as breast cancer, the results are controversial [16, 52]. But the result is consistent among gastrointestinal cancers, such as colorectal cancer [55], gastric cancer [56] and etc. Though the mechanism of how miR-20a induces unfavorable outcome of gastrointestinal cancers is still not clarified, there are several potential explanations. It has been reported that miR-20a/LRIG1 axis might regulate gastric cancer drug resistance through EGFR-mediated PI3K/AKT and MAPK/ERK signaling [57]. Also, miR-20a has been found to be able to repressed the expression of cylindromatosis, leading to activation of the NF-κB pathway and the downstream targets, livin and survivin, which potentially induced GC chemoresistance [58]. In addition, knockdown of miR-20a enhanced sensitivity of colorectal cancer cells to cisplatin through the ROS/ASK1/JNK pathway [59]. Besides, overexpression of miR-20a could induce gastric cancer progression by miR-20a (miR-17)-FBXO31-CyclinD1 pathway [60]. Based on our findings, we could conclude that the elevated miR-20a expression level is associated with poor prognosis of gastrointestinal cancer patients. But for the other types of carcinomas, the results were still not stable and more studies including normalized research conditions (such as specimens, miRNA cut-off values, miRNA detection methods, etc.) were required to further identify miR-20a prognostic value.
As for the clinical features, seven articles enrolled in our analysis including 1103 patients have evaluated the relation of miR-20a to the clinic characteristics. Significant association between miR-20a expression levels and gender or differentiation by fixed effects model was identified. However, due to rather high heterogeneity, the results were not stable and the significances were vanished by random pooling model. Appling sensitivity analyses, we identified two studies (Huang et al. [35] and Fan et al. [28]) that had great impact on the results for the gender and differentiation groups, respectively. After removing them, the heterogeneity completely disappeared for gender group and largely reduced for differentiation group. Moreover,the association between miR-20a expression levels and gender or differentiation was significantly recognized and the results were stable and reliable. As there was still significant heterogeneity existing within other groups that we could not identify the specific sources, further relevant researches were demanded to enrich the results and improve the reliability. Based on the findings, it suggested that women were more likely to develop elevated miR-20a expression, and increased miR-20a expression levels were linked to poor tumor differentiation.
A few limits shall be claimed in this analysis. First of all, the papers language was restricted to English and Chinese and may cause the bias due to lack of other populations. Second, the HRs and its corresponding 95%CI of Yang et al. [37] extracted by the Kaplan-Meier Curves with Engauge Digitizer 9.8 and calculated in the spreadsheet calculator designed by Tierney JF et al. [31] was not consistent to the significance claimed in the original article. Three independent authors (XD, KM and LT) had extracted the data from Yang et al. [37] for several times using the methods described above whose accuracy had been proved by many researches [61,62,63]. The extracted statistics were always harmonious. But they were different from the significance of the original survival curves. The bias demanded prevention by better precise data extracting methods or improving qualities of the recruited studies. Third, the cutoff values of the expression levels of miR-20a were not precisely acknowledged among some studies included. Fourth, the number of recruited studies was relatively insufficient. More associated researches should be performed and enrolled for this analysis, so as to improve the stability and reliability of the findings.
To enlarge the enrolled studies for the meta-analysis, research checklist for cohort study on http://www.equator-network.org/ is recommended to perform further researches on prognostic values and clinical correlation of certain biomarkers for a specific cancer, then these researches can conform with the inclusion criteria. Also, cut-off value that defines high/low expression of biomarkers (RNAs or genes) should be clearly demonstrated and unified. Besides, the HRs and confidence intervals of log rank test or cox regression should be presented in the form of specific figures, otherwise, the required figures can only be extracted by the Kaplan-Meier Curves with methods described previously, which may induce potential bias.
Conclusions
Base on the findings, we conclude that the elevated miR-20a expression level is related to poor prognosis of gastrointestinal cancer patients. As for other types of carcinomas, the results are still not stable and more studies including normalized research conditions are required to further identify miR-20a prognostic values. In addition, miR-20a expression level is relatively higher in women than that in men, and increased miR-20a expression level is linked to poor differentiation.
Abbreviations
- 95%CIs:
-
95% confidence intervals
- CNKI:
-
The China National Knowledge Infrastructure
- DFS:
-
Disease-free survival
- HR:
-
Hazard ratio
- miRNAs:
-
microRNAs
- NOS:
-
The Newcastle-Ottawa Scale
- OS:
-
Overall survival
- PFS:
-
Progression-free survival
- qRT-PCR:
-
Real-time Polymerase Chain Reaction
- RFS:
-
Recurrence-free survival;tau2, tau-squared
- WOS:
-
Web of Science
References
Lau NC, Lim LP, Weinstein EG, Bartel DP. An abundant class of tiny RNAs with probable regulatory roles in Caenorhabditis elegans. Science. 2001;294(5543):858–62.
Lee RC, Ambros V. An extensive class of small RNAs in Caenorhabditis elegans. Science. 2001;294(5543):862–4.
Catto JW, Alcaraz A, Bjartell AS, De Vere White R, Evans CP, Fussel S, Hamdy FC, Kallioniemi O, Mengual L, Schlomm T, et al. MicroRNA in prostate, bladder, and kidney cancer: a systematic review. Eur Urol. 2011;59(5):671–81.
Andorfer CA, Necela BM, Thompson EA, Perez EA. MicroRNA signatures: clinical biomarkers for the diagnosis and treatment of breast cancer. Trends Mol Med. 2011;17(6):313–9.
Filipowicz W, Bhattacharyya SN, Sonenberg N. Mechanisms of post-transcriptional regulation by microRNAs: are the answers in sight? Nat Rev Genet. 2008;9(2):102–14.
Chen X, Ba Y, Ma L, Cai X, Yin Y, Wang K, Guo J, Zhang Y, Chen J, Guo X, et al. Characterization of microRNAs in serum: a novel class of biomarkers for diagnosis of cancer and other diseases. Cell Res. 2008;18(10):997–1006.
Calin GA, Croce CM. MicroRNA signatures in human cancers. Nat Rev Cancer. 2006;6(11):857–66.
Zhao S, Yao D, Chen J, Ding N, Ren F. MiR-20a promotes cervical cancer proliferation and metastasis in vitro and in vivo. PLoS One. 2015;10(3):e0120905.
Zhou J, Liu R, Luo C, Zhou X, Xia K, Chen X, Zhou M, Zou Q, Cao P, Cao K. MiR-20a inhibits cutaneous squamous cell carcinoma metastasis and proliferation by directly targeting LIMK1. Cancer Biol Ther. 2014;15(10):1340–9.
Kang HW, Wang F, Wei Q, Zhao YF, Liu M, Li X, Tang H. miR-20a promotes migration and invasion by regulating TNKS2 in human cervical cancer cells. FEBS Lett. 2012;586(6):897–904.
Li JY, Zhang Y, Zhang WH, Jia S, Kang Y, Zhu XY. Differential distribution of miR-20a and miR-20b may underly metastatic heterogeneity of breast cancers. Asian Pac J Cancer Prev. 2012;13(5):1901–6.
Chai H, Liu M, Tian R, Li X, Tang H. miR-20a targets BNIP2 and contributes chemotherapeutic resistance in colorectal adenocarcinoma SW480 and SW620 cell lines. Acta Biochim Biophys Sin. 2011;43(3):217–25.
Wei J, Qi X, Zhan Q, Zhou D, Yan Q, Wang Y, Mo L, Wan Y, Xie D, Xie J, et al. miR-20a mediates temozolomide-resistance in glioblastoma cells via negatively regulating LRIG1 expression. Biomed Pharmacother. 2015;71:112–8.
Zhang Y, Zheng L, Ding Y, Li Q, Wang R, Liu T, Sun Q, Yang H, Peng S, Wang W, et al. MiR-20a induces cell Radioresistance by activating the PTEN/PI3K/Akt signaling pathway in hepatocellular carcinoma. Int J Radiat Oncol Biol Phys. 2015;92(5):1132–40.
Jiang Y, Chang H, Chen G. Effects of microRNA-20a on the proliferation, migration and apoptosis of multiple myeloma via the PTEN/PI3K/AKT signaling pathway. Oncol Lett. 2018;15(6):10001–7.
Si W, Shen J, Du C, Chen D, Gu X, Li C, Yao M, Pan J, Cheng J, Jiang D, et al. A miR-20a/MAPK1/c-Myc regulatory feedback loop regulates breast carcinogenesis and chemoresistance. Cell Death Differ. 2018;25(2):406–20.
Qiu H, Liu N. MicroRNA-17-92 regulates myoblast proliferation and differentiation by targeting the ENH1/Id1 signaling axis. Cell Death Differ. 2016;23(10):1658–69. PMID: 27315298.
Yang Y, Huang G, Zhou Z, Fewell JG, Kleinerman ES. miR-20a regulates FAS expression in osteosarcoma cells by modulating FAS promoter activity and can be therapeutically targeted to inhibit lung metastases. Mol Cancer Ther. 2018;17(1):130–9.
Yuan G, Zhao Y, Wu D, Gao C, Jiao Z. miRNA-20a upregulates TAK1 and increases proliferation in osteosarcoma cells. Future Oncol(London, England). 2018;14(5):461–9.
Xiong Y, Sun F, Dong P, Watari H, Yue J, Yu MF, Lan CY, Wang Y, Ma ZB. iASPP induces EMT and cisplatin resistance in human cervical cancer through miR-20a-FBXL5/BTG3 signaling. J Exp Clin Cancer Res. 2017;36(1):48.
Uziel T, Karginov FV, Xie S, Parker JS, Wang YD, Gajjar A, He L, Ellison D, Gilbertson RJ, Hannon G, et al. The miR-17~92 cluster collaborates with the sonic hedgehog pathway in medulloblastoma. Proc Natl Acad Sci U S A. 2009;106(8):2812–7.
Wang M, Gu H, Wang S, Qian H, Zhu W, Zhang L, Zhao C, Tao Y, Xu W. Circulating miR-17-5p and miR-20a: molecular markers for gastric cancer. Mol Med Rep. 2012;5(6):1514–20.
Cheng D, Zhao S, Tang H, Zhang D, Sun H, Yu F, Jiang W, Yue B, Wang J, Zhang M, et al. MicroRNA-20a-5p promotes colorectal cancer invasion and metastasis by downregulating Smad4. Oncotarget. 2016;7(29):45199–213.
Xu X, Zhu S, Tao Z, Ye S. High circulating miR-18a, miR-20a, and miR-92a expression correlates with poor prognosis in patients with non-small cell lung cancer. Cancer Med. 2018;7(1):21–31. PMID: 29266846.
Ren H. The associations between expression levels of miR-20a, let-7a in ovarian serous cystadenocarcinoma tissues and the prognosis of patients. Shandong Med J. 2016;56(07):47–9.
Chang CC, Yang YJ, Li YJ, Chen ST, Lin BR, Wu TS, Lin SK, Kuo MY, Tan CT. MicroRNA-17/20a functions to inhibit cell migration and can be used a prognostic marker in oral squamous cell carcinoma. Oral Oncol. 2013;49(9):923–31.
Zhang L, Xiang P, Han X, Wu L, Li X, Xiong Z. Decreased expression of microRNA-20a promotes tumor progression and predicts poor prognosis of cutaneous squamous cell carcinoma. Int J Clin Exp Pathol. 2015;8(9):11446–51.
Fan MQ, Huang CB, Gu Y, Xiao Y, Sheng JX, Zhong L. Decrease expression of microRNA-20a promotes cancer cell proliferation and predicts poor survival of hepatocellular carcinoma. J Exp Clin Cancer Res. 2013;32(1):21.
Marchini S, Cavalieri D, Fruscio R, Calura E, Garavaglia D, Fuso Nerini I, Mangioni C, Cattoretti G, Clivio L, Beltrame L, et al. Association between miR-200c and the survival of patients with stage I epithelial ovarian cancer: a retrospective study of two independent tumour tissue collections. Lancet Oncol. 2011;12(3):273–85.
Xu XL, Jiang YH, Feng JG, Su D, Chen PC, Mao WM. MicroRNA-17, microRNA-18a, and microRNA-19a are prognostic indicators in esophageal squamous cell carcinoma. Ann Thorac Surg. 2014;97(3):1037–45.
Tierney JF, Stewart LA, Ghersi D, Burdett S, Sydes MR. Practical methods for incorporating summary time-to-event data into meta-analysis. Trials. 2007;8:16.
Stang A. Critical evaluation of the Newcastle-Ottawa scale for the assessment of the quality of nonrandomized studies in meta-analyses. Eur J Epidemiol. 2010;25(9):603–5.
Vrabel M. Preferred reporting items for systematic reviews and Meta-analyses. Oncol Nurs Forum. 2015;42(5):552–4.
Liberati A, Altman DG, Tetzlaff J, Mulrow C, Gotzsche PC, Ioannidis JP, Clarke M, Devereaux PJ, Kleijnen J, Moher D. The PRISMA statement for reporting systematic reviews and meta-analyses of studies that evaluate health care interventions: explanation and elaboration. PLoS Med. 2009;6(7):e1000100.
Huang D, Wang H, Liu R, Li H, Ge S, Bai M, Deng T, Yao G, Ba Y. miRNA27a is a biomarker for predicting chemosensitivity and prognosis in metastatic or recurrent gastric cancer. J Cell Biochem. 2014;115(3):549–56.
Sanfiorenzo C, Ilie MI, Belaid A, Barlesi F, Mouroux J, Marquette CH, Brest P, Hofman P. Two panels of plasma microRNAs as non-invasive biomarkers for prediction of recurrence in resectable NSCLC. PLoS One. 2013;8(1):e54596.
Yang R, Fu Y, Zeng Y, Xiang M, Yin Y, Li L, Xu H, Zhong J, Zeng X. Serum miR-20a is a promising biomarker for gastric cancer. Biomed Rep. 2017;6(4):429–34.
Zhao H, Shen J, Hodges TR, Song R, Fuller GN, Heimberger AB. Serum microRNA profiling in patients with glioblastoma: a survival analysis. Mol Cancer. 2017;16(1):59.
Xiao Gao LC. The role of miRNA-17-92 cluster in multiple myeloma. In: The thirteenth National Conference on experimental hematology: 2011. China: Wuhan; 2011. p. 2.
Du Y, Zhu M, Zhou X, Huang Z, Zhu J, Xu J, Cheng G, Shu Y, Liu P, Zhu W, et al. miR-20a enhances cisplatin resistance of human gastric cancer cell line by targeting NFKBIB. Tumour Biol J Int Soc Oncodev Biol Med. 2016;37(1):1261–9.
Zhao S, Yao DS, Chen JY, Ding N. Aberrant expression of miR-20a and miR-203 in cervical cancer. Asian Pac J Cancer Prev. 2013;14(4):2289–93.
Qiang XF, Zhang ZW, Liu Q, Sun N, Pan LL, Shen J, Li T, Yun C, Li H, Shi LH. miR-20a promotes prostate cancer invasion and migration through targeting ABL2. J Cell Biochem. 2014;115(7):1269–76.
Pu Y, Yi Q, Zhao F, Wang H, Cai W, Cai S. MiR-20a-5p represses multi-drug resistance in osteosarcoma by targeting the KIF26B gene. Cancer Cell Int. 2016;16:64.
Li S, Qiang Q, Shan H, Shi M, Gan G, Ma F, Chen B. MiR-20a and miR-20b negatively regulate autophagy by targeting RB1CC1/FIP200 in breast cancer cells. Life Sci. 2016;147:143–52.
Xiong Y, Zhang L, Kebebew E. MiR-20a is upregulated in anaplastic thyroid cancer and targets LIMK1. PLoS One. 2014;9(5):e96103.
Zheng Q, Chen C, Guan H, Kang W, Yu C. Prognostic role of microRNAs in human gastrointestinal cancer: a systematic review and meta-analysis. Oncotarget. 2017;8(28):46611–23.
Li MY, Hu XX. Meta-analysis of microRNA expression profiling studies in human cervical cancer. Med Oncol (Northwood, London, England). 2015;32(6):510.
Troiano G, Mastrangelo F, Caponio VCA, Laino L, Cirillo N, Lo Muzio L. Predictive prognostic value of tissue-based MicroRNA expression in Oral squamous cell carcinoma: a systematic review and Meta-analysis. J Dent Res. 2018;97(7):759–66.
van Houwelingen HC, Putter H. Comparison of stopped cox regression with direct methods such as pseudo-values and binomial regression. Lifetime Data Anal. 2015;21(2):180–96.
Hu J, Ge W, Xu J. HPV 16 E7 inhibits OSCC cell proliferation, invasion, and metastasis by upregulating the expression of miR-20a. Tumour Biol J Int Soc Oncodev Biol Med. 2016;37(7):9433–40.
Wang Y, Zhao YR, Zhang AY, Ma J, Wang ZZ, Zhang X. Targeting of miR-20a against CFLAR to potentiate TRAIL-induced apoptotic sensitivity in HepG2 cells. Eur Rev Med Pharmacol Sci. 2017;21(9):2087–97.
Bai X, Han G, Liu Y, Jiang H, He Q. MiRNA-20a-5p promotes the growth of triple-negative breast cancer cells through targeting RUNX3. Biomed Pharmacother. 2018;103:1482–9.
Liu Y, Han S, Li Y, Liu Y, Zhang D, Li Y, Zhang J. MicroRNA-20a contributes to cisplatin-resistance and migration of OVCAR3 ovarian cancer cell line. Oncol Lett. 2017;14(2):1780–6.
Chen Y, Wang X, Cheng J, Wang Z, Jiang T, Hou N, Liu N, Song T, Huang C. MicroRNA-20a-5p targets RUNX3 to regulate proliferation and migration of human hepatocellular cancer cells. Oncol Rep. 2016;36(6):3379–86.
Longqiu Y, Pengcheng L, Xuejie F, Peng Z. A miRNAs panel promotes the proliferation and invasion of colorectal cancer cells by targeting GABBR1. Cancer Med. 2016;5(8):2022–31.
Zhang Y, Han T, Wei G, Wang Y. Inhibition of microRNA-17/20a suppresses cell proliferation in gastric cancer by modulating UBE2C expression. Oncol Rep. 2015;33(5):2529–36.
Zhou L, Li X, Zhou F, Jin Z, Chen D, Wang P, Zhang S, Zhuge Y, Shang Y, Zou X. Downregulation of leucine-rich repeats and immunoglobulin-like domains 1 by microRNA-20a modulates gastric cancer multidrug resistance. Cancer Sci. 2018;109(4):1044–54.
Zhu M, Zhou X, Du Y, Huang Z, Zhu J, Xu J, Cheng G, Shu Y, Liu P, Zhu W, et al. miR-20a induces cisplatin resistance of a human gastric cancer cell line via targeting CYLD. Mol Med Rep. 2016;14(2):1742–50.
Zhang L, He L, Zhang H, Chen Y. Knockdown of MiR-20a enhances sensitivity of colorectal Cancer cells to cisplatin by increasing ASK1 expression. Cell Physiol Biochem Int J Exp Cell Physiol Biochem Pharmacol. 2018;47(4):1432–41.
Zhang X, Kong Y, Xu X, Xing H, Zhang Y, Han F, Li W, Yang Q, Zeng J, Jia J, et al. F-box protein FBXO31 is down-regulated in gastric cancer and negatively regulated by miR-17 and miR-20a. Oncotarget. 2014;5(15):6178–90.
Malouf R, Ashraf A, Hadjinicolaou AV, Doree C, Hopewell S, Estcourt LJ. Comparison of a therapeutic-only versus prophylactic platelet transfusion policy for people with congenital or acquired bone marrow failure disorders. Cochrane Database Syst Rev. 2018;5:Cd012342.
Moreno Roig E, Yaromina A, Houben R, Groot AJ, Dubois L, Vooijs M. Prognostic role of hypoxia-inducible factor-2alpha tumor cell expression in Cancer patients: a meta-analysis. Front Oncol. 2018;8:224.
Ai L, Mu S, Hu Y. Prognostic role of RDW in hematological malignancies: a systematic review and meta-analysis. Cancer Cell Int. 2018;18:61.
Acknowledgments
We would like to thank all the people who helped us in the current study.
Funding
This study was supported by the National Key Research and Development Program of China (2016YFC1100100) and the Major Research Plan of National Natural Science Foundation of China (No.91649204). Funding bodies have no role in the study design, study setting, analysis, or writing of the manuscript.
Availability of data and materials
The authors declare that all data supporting the findings of this study are available within the article and the enrolled articles for meta-analysis.
Author information
Authors and Affiliations
Contributions
DH and YP contributed equally to this work and shall share first authorship. Study design and basic data extraction: YP, DH, ZS. Data extraction from the Kaplan–Meier curves: XD, KM, LT. Evaluation of studies qualities: DH, YP, KM. Data analysis: YP, XD, KM. Manuscript composition: DH, YP, LT, DJ. Manuscript revision: ZS, KM, DJ. Tables Drafting: DH, XD, DJ. Figures Drafting: YP, KM, DJ. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Additional information
Donghua Huang and Yizhong Peng shall share first authorship.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Huang, D., Peng, Y., Ma, K. et al. MiR-20a, a novel promising biomarker to predict prognosis in human cancer: a meta-analysis. BMC Cancer 18, 1189 (2018). https://doi.org/10.1186/s12885-018-4907-3
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12885-018-4907-3