Abstract
Background
Metallothionein 1H (MT1H) expression level is downregulated in several kinds of tumors, including hepatocellular cancer (HCC). However, its biological functions and underlying mechanisms in HCC is largely unknown. The current study aimed to demonstrate the expression status, biological roles and potential mechanisms of MT1H in HCC.
Methods
We investigated the expression level of MT1H in the Cancer Genome Atlas (TCGA) dataset and a panel of 12 paired tumor/non-tumor tissues. In vitro, gain-of-function experiments were performed to examine the role of MT1H on HCC cell proliferation, invasion, and migration. Using bioinformatics assay, reporter assays, quantitative real-time PCR, and western blotting, we explored the possible mechanisms underlying the role of MT1H in HCC cells. In vivo nude mice experiments were performed to assess the anti-proliferative role of MT1H in HCC.
Results
Downregulation of MT1H was observed in TCGA dataset and a panel of 12 paired tumor/non-tumor tissues. Ectopic overexpression of MT1H in HepG2 and Hep3B cells inhibited cell proliferation, invasion, and migration. Gene Set Enrichment Analysis (GSEA) showed that MT1H might involve in regulation of Wnt/β-catenin pathway. Top/Fop reporter assay confirmed that MT1H had an effect on Wnt/β-catenin signaling. Real-time PCR showed MT1H expression decreased the expression of Wnt/β-catenin target genes. Western blotting assay showed that overexpression of MT1H inhibited the nuclear translocation of β-catenin and that the Akt/GSK-3β axis mediated the modulatory role of MT1H on Wnt/β-catenin signaling in HCC. In vivo nude mice experiments demonstrated that MT1H suppressed the proliferation of HCC cells. Taken together, MT1H suppressed the proliferation, invasion and migration of HCC cells via regulating Wnt/β-catenin signaling pathway.
Conclusions
This study demonstrated that through inhibiting Wnt/β-catenin pathway, MT1H suppresses the proliferation and invasion of HCC cells. MT1H may be a potential target for HCC therapy.
Similar content being viewed by others
Background
Hepatocellular cancer (HCC), the third cause of cancer-related death worldwide, accounts for 85%-90% of all primary liver cancer [1, 2]. In China, it is the most commonly diagnosed cancer and the leading cause of cancer death in male patients younger than 60 years old [3]. Although there are various treatment options, such as surgical resection, liver transplantation, interventional treatment, and systemic therapies, the outcomes of patients with advanced stage are still extremely poor [4, 5]. Conventional cytotoxic systemic therapy is of limited benefit for unresectable or metastatic HCC [6]. Sorafenib, a molecular-targeted agent, demonstrates a modest overall survival benefit, however resistance develops eventually [6–8]. Deeper understanding of molecular carcinogenesis of HCC is urgently warranted to develop new novel target agents.
The metallothioneins (MTs) are a class of cysteine-rich, low-molecular-weight, metal-binding intracellular proteins, including four identified isoforms (MT-1, MT-2, MT-3, and MT-4) [9, 10]. In human, these proteins are encoded by a cluster of genes, which are located on chromosome 16q13 [11]. The MT-1 protein comprises many subtypes encoded by a set of MT-1 genes (MT1A, MT1B, MT1E, MT1F, MT1G, MT1H, etc.). MT-1 genes are reported to be involved in carcinogenesis in various human tumors [9]. In Fu et al’s study [12], MT1G acts as a tumor suppressor in thyroid carcinogenesis via regulating the phosphatidylinositol-3-kinase (PI3K)/Akt pathway and Rb/E2F pathway. Loss of heterozygosity (LOH) causes the downregulation of MT1F in colon cancer tissues, suggesting a tumor suppressor role for MT1F in colon cancer [13]. Of specific note, by analyzing 30 sets of online microarray data, Han et al. [14] found a consistent downregulation of MT1H in various kinds of human malignancies as compared with normal tissues, including small cell lung cancer, neuroblastoma, melanoma, B-cell lymphoma, prostate cancer, colon cancer, breast cancer, and leukemia. Furthermore, a 10- to 100-fold decrease of MT1H expression was observed in HCC in comparison with normal liver tissues, indicating a potential role of MT1H in the development and progression of HCC [14]. Nevertheless, the biological functions and underlying mechanisms of MT1H in HCC are largely unknown.
The Wnt/β-catenin signaling pathway is frequently activated during carcinogenesis, especially in HCC [15]. In the canonical Wnt pathway, Wnt binding to Fz receptor inactivates the β-catenin destruction complex of adenomatous polyposis coli (APC), axin, and glycogen synthase kinase-3 β (GSK-3β) [15]. When the Wnt pathway is activated, β-catenin is released from the complex and translocated into nucleus. The nuclear β-catenin binds to members of the lymphoid-enhancing factor/T-cell factors (LEF/TCF) family that activate target genes transcription [16]. Further delineation of the mechanisms underlying the dysregulated Wnt/β-catenin signaling in HCC is of great interest.
In the current study, we identified the biological functions of MT1H in HCC and explored the possible mechanisms. Our study suggests that MT1H plays crucial role in regulating the proliferation and invasion of HCC cells through modulating Wnt/β-catenin signaling.
Methods
Cells and culture
Human hepatoblastoma cell lines HepG2 and Hep3B were obtained from the China Infrastructure of Cell Line Resource. The cells were cultured in Dulbecco’s modified Eagles medium (DMEM) (Gibco, Grand Island, NY) containing 10% fetal bovine serum (FBS), 100 U/mL penicillin, and 100 mg/mL streptomycin (Gibco, Grand Island, NY) at 37 °C in a humidified atmosphere with 5% CO2.
Obtainment of clinical specimens
Twelve HCC tissues (T) and their corresponding adjacent non-tumorous liver tissues (NT) were obtained from the surgery operation in the First Affiliated Hospital of Zhejiang University from Jan. 2015 to Dec. 2015. NT was defined as liver tissues more than 2 cm away from the edge of the tumor [17]. Two pathologists carried out histopathological diagnosis of the specimens independently. Soon after the tissues were collected, they were immediately snap-frozen in liquid nitrogen and stored at −80 °C for subsequent total cellular RNA extraction. The clinicopathologic characteristics of the patients are listed in Table 1. This study was performed in accordance with the ethical guidelines of the Declaration of Helsinki and was approved by the hospital’s Institutional Review Board (No. 2016397). Informed consent was obtained from each patient.
Generation of stable cell lines with MT1H overexpression
Human TrueORF Gold™ pCMV6-Entry-MT1H plasmid with a C-terminal fusion of MYC/DDK tag was purchased from OriGene Technologies (Rockville, MD). To establish stable cell lines with constitutive expression of MT1H, HepG2 and Hep3B cells were transfected with pCMV6-Entry-MT1H by Lipofectamin™ 2000 (Invitrogen, Life Technology, Carlsbad, CA) according to the manufacturer’s protocol. After selection with complete medium containing G418 (0.6 mg/mL) for 2 weeks [18], individual clones were isolated and grown separately in the presence of G418. The expression of MT1H was confirmed by Western blotting assay. A stable transfectant expressing pCMV6-Entry empty vector was established and served as the control.
Transwell invasion and migration assays
For Transwell invasion/migration assays, the indicated 2 × 104 cells suspended in 200 μL DMEM without FBS were added to Transwell insert (Millipore, Billerica, MA) with or without coated Matrigel (BD Biosciences San Diego, CA), respectively. The insert held in 24-well companion plates with DMEM containing 10% FBS. After 24 h incubation, the cells and Matrigel in the upper chambers were removed by cotton tip. Migrating and invading cells at the bottom of the filter were fixed, stained with 1.0% crystal violet solution and photographed under a light microscope. The number of migrating and invading cells was quantified by counting five fields in each chamber [19]. This experiment was repeated three separate times.
Wound healing assay
To evaluate the cell motility, the wound-healing assay was performed. Indicated cells were seeded in 6-well plates and grown into a monolayer. In order to exclude the possible role of MT1H in proliferation that may affect the result of wound healing assay, the cells were cultured in DMEM medium supplemented with lower percentage of serum (3%). A sterile p200 pipette tip was used to make a scratch and the cells were rinsed several times with medium to remove debris and unattached cells. Cells were then cultured for 36 h and the motility of cells was photographed. Migratory activity of the indicated cells was assessed by the width of the wound, and results were quantified as the percent of control [20].
MTT (3-[4, 5-dimethylthiazol-2-yl]-2, 5-diphenyl-tetrazolium bromide) assay
HepG2-Vector, HepG2-MT1H, Hep3B-Vector and Hep3B-MT1H cells were trypsinized and seeded in 96-well plates at a density of 2 × 103 cells/well. A solution of MTT (Sigma-Aldrich, St Louis, MO, 25 μL of a 5 mg/mL solution in PBS) was added and cells were incubated for another 2 h at 37 °C. Then the medium was replaced by 100 μL Dimethyl sulfoxide (DMSO, Sigma-Aldrich, St Louis, MO) and shaken at room temperature for 10 min. The plate absorbance was measured at 490 nm. This experiment was repeated three separate times.
Colony formation assay
Two hundred indicated cells per well were put in 12-well plates and cultured in complete medium for 10 days. The colonies were fixed with 4% paraformaldehyde and stained with 1% crystal violet solution [21]. This experiment was repeated three separate times.
5-Ethynyl-2′-deoxyuridine (EdU) incorporation assay
EdU incorporation assay was carried out using the Cell-Light™ EdU In Vitro kit (RiboBio, Guangzhou, China) according to the manufacturer’s instructions. Briefly, the indicated cells added with EdU (50 μM) were cultured for 2 h, and then fixed in 4% paraformaldehyde at room temperature for 30 min. TritonX (0.5%) was used to permeabilize the cells. After incubated with 1 × Apollo reaction mixture for 30 min, the cells were subsequently stained with Hoechst 33342 for 30 min at room temperature [22]. EdU positive cells were calculated with Zeiss fluorescent microscope system (Carl Zeiss, Thornwood, NY). This experiment was repeated three separate times.
Western blotting assay
Briefly, the indicated cells were washed with PBS and lysed using Radioimmunoprecipitation assay buffer (RIPA) (150 mM NaCl, 1% NP-40, 0.5% Sodium deoxycholate, 0.1% SDS, 50 mM Tris–HCl pH 7.4) and the amounts of protein were assayed using the Bradford method. Equal amount of protein was subjected to sodium dodecyl sulfate-polyacrylamide gels electrophoresis (SDS-PAGE). Proteins on the gel were transferred onto a polyvinylidene difluoride (PVDF) membrane (Millipore, MA). The membrane was blocked in 5% powdered non-fat milk in Tris-buffered saline (TBS) solution containing 0.05% Tween 20 for 1 h. Membrane was then incubated with primary antibody overnight at 4 °C, followed by incubation with secondary antibody conjugated to horseradish peroxidase at room temperature for 1 h. The signal was developed using the enhanced chemiluminescence (ECL, Advance Western blotting detection kit, Pierce, Rockford, IL) to expose an Xray film. The primary antibodies against Flag, β-catenin, lamin a, α-Tubulin (Abcam, Cambridge, UK), phospho-Akt (Ser473), Akt, phospho-GSK-3β (Ser9), and GSK-3β (Cell Signaling Technology, Beverly, MA), were used. The experiment was repeated three times to confirm the results. The intensity of the indicated bands was quantified using Quantity One software (Bio-Rad, Richmond, CA).
Immunofluorescence
Indicated cells were grown on coverslip (Fisher Scientific, Pittsburgh, PA) on 24-well plate were fixed with 4% paraformaldehyde and permeabilized with 0.1% Triton-X in PBS. β-catenin were detected by incubation with specific anti-β-catenin antibody (Abcam, Cambridge, UK) overnight at 4 °C. A secondary antibody (donkey anti-rabbit rhodamine red) (Jackson Immunoresearch Laboratories, West Grove, PA) was used before staining the nuclei with 1 μg/mL DAPI (Sigma-Aldrich, St. Louis). Pictures were acquired by using a fluorescence microscope (Olympus, Tokyo, Japan).
Quantitative real-time PCR (qPCR) analysis
Total RNA was extracted from the cells or tissues using Trizol reagent (Invitrogen, Life Technology, Carlsbad, CA), and reverse transcription was conducted as follows with random primers and Moloney murine leukemia virus reverse transcriptase (Promega, Madison, Wisconsin) in accordance with the manufacturer’s protocol. The original amount of the specific transcripts were measured by real-time PCR according to a standard protocol with a SYBR Green PCR kit (Roche Diagnostics, Indianapolis, IN) using the ABI Prism 7000 sequence detection system (Applied Biosystems, Foster City, CA). For each gene and sample, three independent assays were conducted and the comparative Ct method was performed to calculate the relative abundance of mRNA compared with that of the endogenous reference control β-actin. The primers used were listed in Table 2.
Nuclear protein extraction
Nuclear protein extraction was carried out using Active Motif Nuclear Extraction Kit (Active Motif, Carlsbad, CA) according to the manufacture’s protocol. Lamin A was used as nuclear fraction controls [23].
Luciferase reporter assay
The Top/Fop flash plasmid system (Upstate, Lake Placid, NY) was used to evaluate the transcriptional activity of TCF. Topflash and Fopflash plasmids were co-transfected in the indicated cells using Lipofectamin™ 2000 (Invitrogen, Life Technology, Carlsbad, CA). The luciferase values were measured after 48 h incubation, using Dual-Luciferase reporter assay system (Promega, Madison, WI) according to the manufacturer’s instructions [24]. The experiment was repeated three times to confirm the results.
In vivo tumorigenicity assay
HepG2-Vector and HepG2-MT1H (1.5 × 106 cells) were injected subcutaneously into the right flanks of female BALB/c athymic nude mice (4–6 weeks of age), five mice per group [25]. The tumor volume was measured routinely using a caliper every 5 days, and was calculated with the formula 0.5 × (length × width2). At 30 days after injection, all mice were sacrificed and tumors were weighted. This experiment was performed in accordance with the institutional ethical guidelines for animal experiment.
Gene set enrichment analysis (GSEA)
GSEA (http://software.broadinstitute.org/gsea/index.jsp) was performed as previously described [26]. We used LIHC RNA-seq data generated by the Cancer Genome Atlas (TCGA) and sorted the samples into the top and bottom quartiles of MT1H expression (high and low MT1H expression, respectively). Analyzed gene sets (PID_WNT_CANONICAL_PATHWAY [27] and WNT_SIGNALING (http://software.broadinstitute.org/gsea/msigdb/cards/WNT_SIGNALING.html) were downloaded from GSEA and the genes are listed in Additional file 1: Table S1. To assess the enrichment, the absolute value of the GSEA metric (Pearson correlation) was considered for deregulated genes.
Statistical analysis
All statistical analyses were performed using the SPSS 13.0 package (SPSS International, Chicago, IL, USA). Continuous variables were presented as mean ± SD and analyzed using the Student’s t-test. The relationship between MT1H expression and the expression of Wnt/β-catenin target genes was analyzed by Chi-square tests. P < 0.05 was considered statistically significant.
Results
MT1H expression in human HCCs
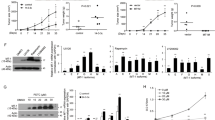
To explore the role of MT1H in HCC, we analyzed the MT1H expression in large datasets derived from TCGA databases [26]. TCGA is a collaboration of the National Cancer Institute (NCI) and National Human Genome Research Institute (NHGRI) [26]. TCGA contains multi-dimensional and comprehensive maps of the key genomic changes in various types of cancer, including HCC. The results from TCGA RNASeq2 data showed that in 50 pairs of HCC tissues (T) and their corresponding adjacent non-tumorous liver tissues (NT), MT1H is significantly downregulated in most (48/50) HCCs (Fig. 1a). The clinical data of these 50 patients are shown in Additional file 2: Table S2. Furthermore, we expanded our investigation to clinical samples in order to assess the expression level of MT1H in 12 human HCCs by qRT-PCR. As shown in Fig. 1b, MT1H expression level was found to be markedly decreased in all HCC tumors as compared with the adjacent non-tumorous liver tissues. These results suggest that MT1H expression is downregulated in human HCCs and may play a role in hepatocellular carcinogenesis.
Low expression level of MT1H in HCC patients. a The expression of MT1H in 50 pairs of primary HCC tumors versus paired non-tumorous liver tissues mined from RNAseqV2 data set on TCGA. b The log2Tumor/Non-tumorous change of MT1H expression in 12 paired fresh HCC tissues and the adjacent non-cancerous liver tissues
MT1H suppresses the growth of HepG2 and Hep3B cells
To examine the potential role of MT1H in hepatocellular carcinogenesis, we sought to assess the effect of MT1H on the growth of HCC cells. HepG2 and Hep3B cells were stably transfected with full length Flag-tagged MT1H and immunoblots were performed to confirm the overexpression of MT1H (Fig. 2a). As shown in Fig. 2b, ectopic overexpression of MT1H in HepG2 and Hep3B cells significantly inhibited the MTT response as compared with that of empty vector control cells. Next, we examined the effects of enforced expression of MT1H on the colony forming capability of HCC cells. As shown in Fig. 2c, enforced expression of MT1H in HepG2 and Hep3B cells decreased more than 50% in colony numbers as compared with the control cells. Moreover, in Edu incorporation assay, the DNA synthesis capability was significantly decreased in MT1H-overexpressed cells (Fig. 2d). The results of these experiments demonstrate that enforced expression of MT1H in HCC cells is correlated with impaired cell growth and DNA synthesis.
MT1H plays a pivotal role in HCC cell proliferation. a Expression of MT1H-Flag in indicated cells. Protein extracts from indicated cells were immunoblotted with antibody for Flag. α-Tubulin as a loading control. b HepG2-Vector, HepG2-MT1H, Hep3B-Vector, and HepG3B-MT1H cells was measured by MTT response. Error bars indicate standard deviation (Student’s t-test; *, P < 0.05). c Colony number of MT1H overexpressing HepG2 and Hep3B cells was significantly decreased as compared with respective vector-control. Error bars indicate standard deviation (Student’s t-test; *, P < 0.05). d Edu incorporation analysis indicates that MT1H suppresses the proliferation of HCC cells. Error bars indicate standard deviation (Student’s t-test; *, P < 0.05)
MT1H suppresses tumor cell invasiveness and motility
The effect of MT1H overexpression on invasiveness and migration ability of HepG2 and Hep3B cells was studied. The matrigel-coated transwell assay was performed and the number of MT1H overexpressed cells in the bottom well was significantly reduced compared with the control cells, which indicated that MT1H may suppress the cell invasion (Fig. 3a and b, left). Likewise, the matrigel-uncoated transwell assay showed that ectopic overexpression of MT1H significantly impaired the migration capability of hepatoma cells as compared with the vector control cells (Fig. 3a and b, right). Furthermore, scratch and wound healing assay indicated that enforced expression of MT1H significantly inhibited the migration of HepG2 and Hep3B (Fig. 3c and d). Taken together, these results suggest that MT1H can affect the invasiveness and migration of human HCC cells.
MT1H suppresses the invasion and migration of HCC cells. a and b (left) Matrigel invasion assay showed that invading number of MT1H overexpressing HCC cells was lower than vector-controls. Error bars indicate standard deviation (Student’s t-test; *, P < 0.05). a and b (right) Transwell migration assay showed that MT1H expression inhibited the migration capability of HCC cells. Error bars indicate standard deviation (Student’s t-test; *, P < 0.05). c and d Wound healing assay indicated that MT1H expression inhibited HepG2 and Hep3B cell migration
MT1H suppresses the Wnt/β-catenin signaling via inhibiting nuclear translocation of β-catenin
Next, we sought to explore the driving mechanisms behind the effects of MT1H on HCC cell proliferation, invasion as well as migration. We used LIHC RNAseq2 data generated by the TCGA and sorted the samples into the top and bottom quartiles of MT1H expression (high and low MT1H expression, respectively). By using TCGA dataset and the GSEA assay, we found that Wnt/β-catenin pathway related genes were significantly enriched in the HCC patients with low MT1H expression (Fig. 4a). Moreover, we analyzed whether the association between the MT1H expression and Wnt/β-catenin target genes (http://web.stanford.edu/group/nusselab/cgi-bin/wnt/) expression can be found in TCGA data. We categorized the Wnt/β-catenin target genes’ expression into two groups: low expression group (less than the median; n = 50) and high expression group (greater than the median; n = 50). As shown in Additional file 3: Figure S1a and Additional file 4: Table S3, significantly higher levels of POU5F1, DKK1, NOS2, MSL1, MYCBP, NRCAM, LEF1, and PPARD transcripts were found in HCC tissues versus adjacent non-tumorous liver tissues. Taken together, these data suggest that MT1H plays a potential role in regulating β-catenin signaling in HCC.
MT1H regulates Wnt/β-catenin signaling in HCC cells. a GSEA analysis showed that MT1H might has a regulative role in Wnt/β-catenin signaling in HCC cells. b qRT-PCR analysis was used to detect the mRNA levels of MYC, MMP7, and LEF1 in indicated cells. Error bars indicate standard deviation (Student’s t-test; *, P < 0.05). c Wnt/β-catenin signaling activity was assessed by TOP/FOP flash dual-luciferase reporter assay in indicated cells. Error bars indicate standard deviation (Student’s t-test; *, P < 0.05). d Immunofluorescence of β-catenin distribution in indicated cells. e Western blot analysis of nuclear extraction from indicated cells. f Western blot analysis was used to detected the level of p-Akt (Ser 473), total-Akt, p-GSK-3β (Ser 9), and total-GSK-3β in indicated cells. α-Tubulin as a loading control
Next, qPCR was performed and the results showed that ectopic overexpression of MT1H in HepG2 and Hep3B cells significantly suppressed the levels of MYC, MMP7, and LEF1 (Fig. 4b). To further confirm whether MT1H regulates the Wnt/β-catenin signaling, luciferase assay of β-catenin reporter (TOP/FOP) was conducted. As shown in Fig. 4c, MT1H inhibited the Wnt/β-catenin transcriptional activity in HepG2 and Hep3B cells. As expected and shown in Fig. 4d, e, and Additional file 3: Figure S1b, β-catenin nuclear translocation was significantly attenuated in MT1H over-expressed HCC cells in comparison with the control cells. Next, we explored the mechanisms of MT1H regulating the β-catenin signaling. We found that the phosphorylation of Akt was significantly inhibited by MT1H overexpression (Fig. 4f and Additional file 3: Figure S1c). It has been well known that Akt can lead to phosphorylated inactivation of GSK-3β, and thereby resulting in β-catenin stabilization and nuclear transportation. As expected, the dephosphorylation of Akt reduced phosphorylation of GSK-3β (Fig. 4f and Additional file 3: Figure S1c), suggesting the Akt/GSK-3β axis mediates the modulatory role of MT1H on Wnt/β-catenin signaling in HCC. Collectively, these data indicate that Wnt/β-catenin signaling is a target downstream of MT1H in HCC.
MT1H overexpression attenuates tumorigenicity in nude mice
To further assess the effect of MT1H overexpression on tumorigenicity in vivo, we subcutaneously injected HepG2-Vector and HepG2-MT1H cells into the nude mice. Intriguingly, as shown in Fig. 5a and b, the tumors formed by HepG2-MT1H cells were significantly smaller than that of control cells. The average tumor weight of the MT1H- transfected cells inoculated mice was significantly decreased as compared with that of control cells, at day 30 (Fig. 5c).
MT1H suppresses tumorigenicity in vivo. a Tumor growth curves measured after subcutaneous injection of HepG2-Vector and HepG2-MT1H. The tumor volume was calculated every 5 days. Error bars indicate standard deviation (Student’s t-test; *, P < 0.05, n = 5). b Photographs of dissected tumors from nude mice. c Weight of tumors from mice with HepG2-Vector or HepG2-MT1H implantation. Error bars indicate standard deviation (Student’s t-test; *, P < 0.05, n = 5)
Discussion
In the present study, we found significantly downregulated MT1H in HCC tissues, which is consistent with previous study [14]. Moreover, MT1H plays an anti-proliferative and anti-invasive role in HCC cells. MT1H in HCC can inhibit Wnt/β-catenin signaling, therefore suppresses HCC progression.
MTs are demonstrated to be involved in various physiological and pathological processes, such as protection against oxidative damage, cell proliferation, and apoptosis [11, 28]. Of note, the important roles of MTs in cancer development and progression have been emphasized [29–32]. MT1H, one member of MT-1 genes, has been proved of decreased expression in HCC tissue [14]. Consistently, the results in current study from TCGA database and paired fresh tumor/non-tumor tissues showed significantly decreased MT1H expression in HCC tissues. The significant downregulation of MT1H in HCC implicates its potential roles in the development and progression of HCC. As expected, our results indicated that ectopic overexpression of MT1H may inhibit the growth of HCC cells. Moreover, MT1H involves in the invasiveness of HCC cells.
The mechanisms underlying the downregulation of MTs have been investigated in malignant diseases [12–14, 29, 33–40]. The downregulation of MT1F in colon cancer is mainly caused by loss of heterozygosity (LOH) [13]. In addition, a direct relation between p53 and MT-1A and MT-2A was found in epithelial cancer cells [41]. Whether these mechanisms or other mechanism are involved in the downregulation of MT1H in HCC need to be further explored. Delineating the precise molecular mechanisms for the altered expression of MT1H in HCC will put new insights into the understanding of HCC tumorigenesis.
Given that Wnt/β-catenin pathway plays an essential role in tumorigenesis, together with our bioinformatics analysis implicated that Wnt/β-catenin signaling might involve in the function of MT1H in HCC. It is important to explore the tumor suppressor role of MT1H mediated by Wnt/β-catenin signaling in HCC. We found that MT1H suppressed β-catenin nuclear translocation and transcriptional activity in HCC. Wnt/β-catenin signaling has demonstrated to have important effects on proliferation and invasion of tumor cells. The activated Wnt/β-catenin pathway has been observed in at least one-third HCCs and most of them have mutations in the β-catenin gene [42]. Therefore, it is rational to deduce that tumor suppressive role of MT1H on HCC is attributed, at least partly, to the inhibition of Wnt/β-catenin signaling.
Akt can lead to phosphorylated inactivation of GSK-3β, which in turn results in β-catenin stabilization and nuclear transportation [43]. Of note, Akt signaling has been implicated in mediating the biological functions of MTs [12, 44, 45]. These prompt us to investigate whether MT1H regulates Wnt/β-catenin via Akt/GSK-3β axis. Intriguingly, our results implicate that Akt/GSK-3β axis mediates the modulatory role of MT1H on Wnt/β-catenin signaling. These provide new insights into regulatory networks for dysregulated Akt activation in HCC. Nevertheless, whether MT1H regulates Akt by direct or indirect mechanisms needs to be further explored. Moreover, Han et al. have reported that MT1H interacted with euchromatin histone methyltransferase 1 (EHMT1) and enhanced its methyltransferase activity on histone 3, thereby involved the tumorigenesis of prostate cancer [14]. It remains to be determined whether this mechanism or other mechanisms may also be involved in the role of MT1H in HCC.
Conclusions
In summary, our data showed that through inhibiting Wnt/β-catenin pathway, MT1H could suppress the proliferation and invasion of HCC cells. MT1H might be a potential new target for HCC therapy. Ongoing studies are to explore the molecular mechanism of downregulated MT1H and the interaction between MT1H and Akt/GSK-3β/β-catenin signaling.
Abbreviations
- APC:
-
Adenomatous polyposis coli
- BCLC:
-
Barcelona Clinic Liver Cancer
- DKK1:
-
Dickkopf WNT Signaling Pathway Inhibitor 1
- DMEM:
-
Dulbecco minimum essential medium
- DMSO:
-
Dimethyl sulfoxide
- ECL:
-
Enhanced chemiluminescence
- EDU:
-
5-Ethynyl-2′-deoxyuridine
- FBS:
-
Fetal bovine serum
- GSEA:
-
Gene set enrichment analysis
- GSK3beta:
-
Glycogen synthase kinase-3 beta
- HCC:
-
Hepatocellular cancer
- LEF:
-
Lymphoid-enhancing factor
- LOH:
-
Loss of heterozygosity
- MMP:
-
Matrix metallopeptidase
- mRNA:
-
messenger RNA
- MSL1:
-
Male specific lethal 1
- MT:
-
Metallothionein
- MT1H:
-
Metallothionein 1H
- MTT:
-
3-[4, 5-dimethylthiazol-2-yl]-2, 5-diphenyl-tetrazolium bromide
- MYC:
-
V-myc avian myelocytomatosis viral oncogene
- MYCBP:
-
MYC binding protein
- NOS2:
-
Nitric oxide synthase 2
- NRCAM:
-
Neuronal cell adhesion molecule
- PCR:
-
Polymerase chain reaction
- PI3K:
-
Phosphatidylinositol-3-kinase
- POU5F1:
-
POU class 5 homeobox 1
- PPARD:
-
Peroxisome proliferator activated receptor delta
- PVDF:
-
Polyvinylidene difluoride
- RIPA:
-
Radioimmunoprecipitation assay buffer
- SDS-PAGE:
-
Sodium dodecyl sulfate-polyacrylamide gels
- TBS:
-
Tris-buffered saline
- TCF:
-
T-cell factors
- TCGA:
-
The Cancer Genome Atlas
References
Siegel RL, Miller KD, Jemal A. Cancer statistics, 2015. CA Cancer J Clin. 2015;65:5–29.
Schutte K, Schulz C, Malfertheiner P. Hepatocellular Carcinoma: Current Concepts in Diagnosis, Staging and Treatment. Gastrointest Tumors. 2014;1:84–92.
Chen W, Zheng R, Baade PD, Zhang S, Zeng H, Bray F, Jemal A, Yu XQ, He J. Cancer statistics in China, 2015. CA Cancer J Clin. 2016;66:115–32.
Forner A, Llovet JM, Bruix J. Hepatocellular carcinoma. Lancet. 2012;379:1245–55.
Akinleye A, Furqan M, Mukhi N, Ravella P, Liu D. MEK and the inhibitors: from bench to bedside. J Hematol Oncol. 2013;6:27.
Deng GL, Zeng S, Shen H. Chemotherapy and target therapy for hepatocellular carcinoma: New advances and challenges. World J Hepatol. 2015;7:787–98.
Bertino G, Demma S, Ardiri A, Proiti M, Gruttadauria S, Toro A, Malaguarnera G, Bertino N, Malaguarnera M, Malaguarnera M, Di Carlo I. Hepatocellular carcinoma: novel molecular targets in carcinogenesis for future therapies. Biomed Res Int. 2014;2014:203693.
You A, Cao M, Guo Z, Zuo B, Gao J, Zhou H, Li H, Cui Y, Fang F, Zhang W, Song T, Li Q, Zhu X, Yin H, Sun H, Zhang T. Metformin sensitizes sorafenib to inhibit postoperative recurrence and metastasis of hepatocellular carcinoma in orthotopic mouse models. J Hematol Oncol. 2016;9:20.
Thirumoorthy N, Shyam Sunder A, Manisenthil Kumar K, Senthil Kumar M, Ganesh G, Chatterjee M. A review of metallothionein isoforms and their role in pathophysiology. World J Surg Oncol. 2011;9:54.
Takahashi S. Molecular functions of metallothionein and its role in hematological malignancies. J Hematol Oncol. 2012;5:41.
Cherian MG, Jayasurya A, Bay BH. Metallothioneins in human tumors and potential roles in carcinogenesis. Mutat Res. 2003;533:201–9.
Fu J, Lv H, Guan H, Ma X, Ji M, He N, Shi B, Hou P. Metallothionein 1G functions as a tumor suppressor in thyroid cancer through modulating the PI3K/Akt signaling pathway. BMC Cancer. 2013;13:462.
Yan DW, Fan JW, Yu ZH, Li MX, Wen YG, Li DW, Zhou CZ, Wang XL, Wang Q, Tang HM, Peng ZH. Downregulation of metallothionein 1F, a putative oncosuppressor, by loss of heterozygosity in colon cancer tissue. Biochim Biophys Acta. 2012;1822:918–26.
Han YC, Zheng ZL, Zuo ZH, Yu YP, Chen R, Tseng GC, Nelson JB, Luo JH. Metallothionein 1 h tumour suppressor activity in prostate cancer is mediated by euchromatin methyltransferase 1. J Pathol. 2013;230:184–93.
Klaus A, Birchmeier W. Wnt signalling and its impact on development and cancer. Nat Rev Cancer. 2008;8:387–98.
Lin HH, Feng WC, Lu LC, Shao YY, Hsu CH, Cheng AL. Inhibition of the Wnt/beta-catenin signaling pathway improves the anti-tumor effects of sorafenib against hepatocellular carcinoma. Cancer Lett. 2016;381:58–66.
Zheng L, Yang W, Wu F, Wang C, Yu L, Tang L, Qiu B, Li Y, Guo L, Wu M, Feng G, Zou D, Wang H. Prognostic significance of AMPK activation and therapeutic effects of metformin in hepatocellular carcinoma. Clin Cancer Res. 2013;19:5372–80.
Campbell RA, Bhat-Nakshatri P, Patel NM, Constantinidou D, Ali S, Nakshatri H. Phosphatidylinositol 3-kinase/AKT-mediated activation of estrogen receptor alpha: a new model for anti-estrogen resistance. J Biol Chem. 2001;276:9817–24.
Valster A, Tran NL, Nakada M, Berens ME, Chan AY, Symons M. Cell migration and invasion assays. Methods. 2005;37:208–15.
Kobayashi Y, Hiraga T, Ueda A, Wang L, Matsumoto-Nakano M, Hata K, Yatani H, Yoneda T. Zoledronic acid delays wound healing of the tooth extraction socket, inhibits oral epithelial cell migration, and promotes proliferation and adhesion to hydroxyapatite of oral bacteria, without causing osteonecrosis of the jaw, in mice. J Bone Miner Metab. 2010;28:165–75.
Zhang Q, Yan HB, Wang J, Cui SJ, Wang XQ, Jiang YH, Feng L, Yang PY, Liu F. Chromatin remodeling gene AT-rich interactive domain-containing protein 1A suppresses gastric cancer cell proliferation by targeting PIK3CA and PDK1. Oncotarget. 2016;7:46127–41.
Miao HL, Pan ZJ, Lei CJ, Wen JY, Li MY, Liu ZK, Qiu ZD, Lin MZ, Chen NP, Chen M. Knockdown of GPC3 inhibits the proliferation of Huh7 hepatocellular carcinoma cells through down-regulation of YAP. J Cell Biochem. 2013;114:625–31.
Ning BF, Ding J, Yin C, Zhong W, Wu K, Zeng X, Yang W, Chen YX, Zhang JP, Zhang X, Wang HY, Xie WF. Hepatocyte nuclear factor 4 alpha suppresses the development of hepatocellular carcinoma. Cancer Res. 2010;70:7640–51.
Xu S, Wen Z, Jiang Q, Zhu L, Feng S, Zhao Y, Wu J, Dong Q, Mao J, Zhu Y. CD58, a novel surface marker, promotes self-renewal of tumor-initiating cells in colorectal cancer. Oncogene. 2015;34:1520–31.
Zhang S, Shan C, Kong G, Du Y, Ye L, Zhang X. MicroRNA-520e suppresses growth of hepatoma cells by targeting the NF-kappaB-inducing kinase (NIK). Oncogene. 2012;31:3607–20.
Lapointe J, Li C, Giacomini CP, Salari K, Huang S, Wang P, Ferrari M, Hernandez-Boussard T, Brooks JD, Pollack JR. Genomic profiling reveals alternative genetic pathways of prostate tumorigenesis. Cancer Res. 2007;67:8504–10.
Schaefer CF, Anthony K, Krupa S, Buchoff J, Day M, Hannay T, Buetow KH. PID: the Pathway Interaction Database. Nucleic Acids Res. 2009;37:D674–9.
Theocharis SE, Margeli AP, Koutselinis A. Metallothionein: a multifunctional protein from toxicity to cancer. Int J Biol Markers. 2003;18:162–9.
Kanda M, Nomoto S, Okamura Y, Nishikawa Y, Sugimoto H, Kanazumi N, Takeda S, Nakao A. Detection of metallothionein 1G as a methylated tumor suppressor gene in human hepatocellular carcinoma using a novel method of double combination array analysis. Int J Oncol. 2009;35:477–83.
Sun X, Niu X, Chen R, He W, Chen D, Kang R, Tang D. Metallothionein-1G Facilitates Sorafenib Resistance through Inhibition of Ferroptosis. Hepatology. 2016;64:488–500.
Mehrian-Shai R, Yalon M, Simon AJ, Eyal E, Pismenyuk T, Moshe I, Constantini S, Toren A. High metallothionein predicts poor survival in glioblastoma multiforme. BMC Med Genomics. 2015;8:68.
Tian ZQ, Xu YZ, Zhang YF, Ma GF, He M, Wang GY. Effects of metallothionein-3 and metallothionein-1E gene transfection on proliferation, cell cycle, and apoptosis of esophageal cancer cells. Genet Mol Res. 2013;12:4595–603.
Pan Y, Huang J, Xing R, Yin X, Cui J, Li W, Yu J, Lu Y. Metallothionein 2A inhibits NF-kappaB pathway activation and predicts clinical outcome segregated with TNM stage in gastric cancer patients following radical resection. J Transl Med. 2013;11:173.
Otsuka T, Hamada A, Iguchi K, Usui S, Hirano K. Suppression of metallothionein 3 gene expression by androgen in LNCaP prostate cancer cells. Biomed Rep. 2013;1:614–8.
Liang GY, Lu SX, Xu G, Liu XD, Li J, Zhang DS. Expression of metallothionein and Nrf2 pathway genes in lung cancer and cancer-surrounding tissues. World J Surg Oncol. 2013;11:199.
Kim HG, Kim JY, Han EH, Hwang YP, Choi JH, Park BH, Jeong HG. Metallothionein-2A overexpression increases the expression of matrix metalloproteinase-9 and invasion of breast cancer cells. FEBS Lett. 2011;585:421–8.
Lai Y, Lim D, Tan PH, Leung TK, Yip GW, Bay BH. Silencing the Metallothionein-2A gene induces entosis in adherent MCF-7 breast cancer cells. Anat Rec (Hoboken). 2010;293:1685–91.
Pontes HA, de Aquino Xavier FC, da Silva TS, Fonseca FP, Paiva HB, Pontes FS, dos Santos Pinto Jr D. Metallothionein and p-Akt proteins in oral dysplasia and in oral squamous cell carcinoma: an immunohistochemical study. J Oral Pathol Med. 2009;38:644–50.
Lim D, Jocelyn KM, Yip GW, Bay BH. Silencing the Metallothionein-2A gene inhibits cell cycle progression from G1- to S-phase involving ATM and cdc25A signaling in breast cancer cells. Cancer Lett. 2009;276:109–17.
Ferrario C, Lavagni P, Gariboldi M, Miranda C, Losa M, Cleris L, Formelli F, Pilotti S, Pierotti MA, Greco A. Metallothionein 1G acts as an oncosupressor in papillary thyroid carcinoma. Lab Invest. 2008;88:474–81.
Fan LZ, Cherian MG. Potential role of p53 on metallothionein induction in human epithelial breast cancer cells. Br J Cancer. 2002;87:1019–26.
Gedaly R, Galuppo R, Daily MF, Shah M, Maynard E, Chen C, Zhang X, Esser KA, Cohen DA, Evers BM, Jiang J, Spear BT. Targeting the Wnt/beta-catenin signaling pathway in liver cancer stem cells and hepatocellular carcinoma cell lines with FH535. PLoS One. 2014;9:e99272.
Park NR, Cha JH, Jang JW, Bae SH, Jang B, Kim JH, Hur W, Choi JY, Yoon SK. Synergistic effects of CD44 and TGF-beta1 through AKT/GSK-3beta/beta-catenin signaling during epithelial-mesenchymal transition in liver cancer cells. Biochem Biophys Res Commun. 2016;477:568–74.
Zhou S, Yin X, Zheng Y, Miao X, Feng W, Cai J, Cai L. Metallothionein prevents intermittent hypoxia-induced cardiac endoplasmic reticulum stress and cell death likely via activation of Akt signaling pathway in mice. Toxicol Lett. 2014;227:113–23.
Luo K, Long H, Xu B, Luo Y. Metallothionein ameliorates burn sepsis partly via activation of Akt signaling pathway in mice: a randomized animal study. World J Emerg Surg. 2015;10:53.
Acknowledgements
Not applicable.
Funding
Zhejiang Provincial Natural Science Fund (LY13H160007 and LY15H160018), and Zhejiang Medicines & Health Science and Technology Project (201348801) supported this research.
Availability of data and materials
All data generated or analyses during this study are included in this article and its Additional files.
Authors’ contributions
ZY and JL performed and analyzed the experiments and draft the manuscript. ZX and ZJ designed and analyzed the experiments and revised the drafting of the manuscript. XC and HY analyzed the data and helped to prepare the manuscript. XN designed the experiment. All authors have read and approved the final manuscript.
Competing interests
The authors declare that they have no competing interests.
Consent for publication
Not applicable.
Ethics approval and consent to participate
The use of clinical specimens and clinical data were reviewed and approved by the institutional ethics committee of the First Affiliated Hospital of Zhejiang University School of Medicine (Reference No. 2016397). All patients provided informed consent to participate in this study. All mouse experiments were performed in accordance with the Principles of Laboratory Animal Care of the National Institutes of Health and were approved by the institutional ethics committee of the First Affiliated Hospital of Zhejiang University School of Medicine (Reference No. 2016340).
Author information
Authors and Affiliations
Corresponding authors
Additional files
Additional file 1: Table S1.
Gene sets downloaded from the GSEA. (XLSX 9 kb)
Additional file 2: Table S2.
The clinical data of patients analyzed in Fig. 1a. (XLSX 25 kb)
Additional file 3: Figure S1.
a. Significantly higher levels of POU5F1, DKK1, NOS2, MSL1, MYCBP, NRCAM, LEF1, and PPARD transcripts in HCC tissues versus adjacent non-tumorous liver tissues. b and c. Quantification of indicated protein levels normalized to control using Bio-Rad Quantity One software. Error bars indicate standard deviation (Student’s t-test; *, P < 0.05). (TIF 379 kb)
Additional file 4: Table S3.
The clinical data of patients with low or high MT1H expression. (XLSX 16 kb)
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Zheng, Y., Jiang, L., Hu, Y. et al. Metallothionein 1H (MT1H) functions as a tumor suppressor in hepatocellular carcinoma through regulating Wnt/β-catenin signaling pathway. BMC Cancer 17, 161 (2017). https://doi.org/10.1186/s12885-017-3139-2
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12885-017-3139-2