Abstract
Background
Gastric cancer is one of the most common and lethal type of cancer worldwide. Infection with Helicobacter pylori (H. pylori) is recognized as the major cause of gastric cancer. However, it remains unclear the mechanism by which Helicobacter infection leads to gastric cancer. Furthermore, the underlying molecular events involved during the progression of Helicobacter infection to gastric malignancy are not well understood. In previous studies, we demonstrated that that H. felis-infected Myd88 −/− mice exhibited dramatic pathology and an accelerated progression to gastric dysplasia; however, the MyD88 downstream gene targets responsible for this pathology have not been described. This study was designed to identify MyD88-dependent genes involved in the progression towards gastric cancer during the course of Helicobacter infection.
Methods
Wild type (WT) and Myd88 deficient mice (Myd88 −/−) were infected with H. felis for 25 and 47 weeks and global transcriptome analysis performed on gastric tissue using MouseWG-6 v2 expression BeadChips microarrays. Function and pathway enrichment analyses of statistically significant, differential expressed genes (p < 0.05) were performed using the Database for Annotation, Visualization and Integrated Discovery (DAVID) online tools.
Results
Helicobacter infection affected the transcriptional profile of more genes in Myd88 −/− mice compared to WT mice. Infection of Myd88 −/− mice resulted in the differential expression of 1,989 genes at 25 weeks (1031 up and 958 downregulated). At 47 weeks post-H.felis infection, 2,162 (1140 up and 1022 downregulated) were differentially expressed. The most significant differentially upregulated gene during Helicobacter infection in Myd88 −/− mice was chitinase-like 4 (chil4), which is involved in tissue remodeling and wound healing. Other highly upregulated genes in H. felis-infected Myd88 −/− mice included, Indoleamine 2,3-Dioxygenase 1 (Ido1), Guanylate binding protein 2 (Gbp2), ubiquitin D (Ubd), β 2 -Microglobulin (B2m), CD74 antigen (Cd74), which have been reported to promote cancer progression by enhancing angiogenesis, proliferation, migration, metastasis, invasion, and tumorigenecity. For downregulated genes, the highly expressed genes included, ATPase H+/K+ transporting, alpha subunit (Atp4a), Atp4b, Mucin 5 AC (Muc5ac), Apolipoprotein A-1 (Apoa1), and gastric intrinsic factor (Gif), whose optimal function is important in maintaining gastric hemostasis and lower expression has been associated with increased risk of gastric carcinogenesis.
Conclusions
These results provide a global transcriptional gene profile during the development and progression of Helicobacter-induced gastric cancer. The data show that our mouse model system is useful for identifying genes involved in gastric cancer progression.
Similar content being viewed by others
Background
Gastric cancer is one of the most common causes of cancer-related death worldwide with an estimated 738,000 deaths each year [1]. Recently, H. pylori was recognized as the foremost cause of gastric cancer [2–7]. With an estimated half of the world’s population being infected, Helicobacter infection contributes significantly to the worldwide gastric cancer burden [7, 8]. Recognition of the factors leading up to the development and progression towards gastric cancer are critical in determination of cancer pathology. H. pylori-induced gastric carcinogenesis involves a multistep progression from normal gastric mucosa to superficial gastritis, chronic gastritis, atrophic gastritis, metaplasia, dysplasia, and finally gastric carcinoma [8, 9]. Molecular events associated with disease progression to gastric malignancy have not been elucidated. Considerable amount of confirmatory evidence shows that host immune response to H. pylori is crucial in determining gastric cancer predisposition [10–12]. We have previously shown that a key signal transduction adaptor protein, myeloid differentiation primary response gene 88 (MyD88), regulates Helicobacter-induced gastric cancer progression in a mouse model of gastric cancer [13]. We demonstrated that H. felis-infected MyD88 deficient (Myd88 −/−) mice exhibited severe gastric pathology and an accelerated progression to gastric dysplasia compared to wild type (WT) mice [13] However, the MyD88-dependent gene responsible for this pathology were not described.
MyD88 is a key adaptor molecule that is crucial in mediating innate immune signals from members of the toll-like receptor (TLR) and interleukin-1 (IL-1)/IL-18 families leading to downstream activation of nuclear factor (NF)-κB [14–16]. Consistent with involvement in these inflammatory pathways, MyD88 signaling has been associated with cancer progression, which stems from the understanding that inflammation is linked to cancer promotion [17, 18]. Studies on the role of MyD88 cancer progression have been the subject of recent intense investigations. However, the data are contradictory, which indicate that the role of MyD88 in the development and progression of inflammation-associated cancers is complex [19]. Several studies using genetic or chemical carcinogenesis models involving Myd88 deficient mice have shown MyD88 to either promote [20–27] or suppress [13, 28–34] tumor development. The complex role of MyD88 in carcinogenesis is best typified by studies in colon cancer models [22, 24, 29, 35] showing contradictory roles in the same tissue. The mechanistic basis for these opposing observation is still not fully understood and could be due to many factors including, the type of inflammation, the extent of tissue damage, and immune response elicited [35]. Further, the MyD88 dependent genes in this accelerated progression to dysplasia remain unknown. Therefore, this study was performed to identify potential genes involved in the accelerated progression of gastric cancer.
Results
Gene expression and analysis
Prior to differential gene analysis, all data from 23,015 genes with a standard deviation of less than 0.1 were used for multiple dimensional scaling (MDS) analysis (Fig. 1) to verify that Myd88 −/− and WT samples were differentiated according to gene expression in each sample with a relative p-value. Each sample is represented with distance between each one reflecting their approximate degree of correlation [36]. All genes included in the analysis had a minimum standard deviation of less than 0.1. The analysis showed that all uninfected mice were clustered together irrespective of genetic background or time point. For infected mice, WT and Myd88 −/− mice clustered distinctively separate indicating differential expression of their genes.
Multi Dimensional Scale Plot of all H. felis infected and control samples from WT and Myd88 −/− mice. Samples were separated using 8 RNA SEQ libraries based on sample relations of 23,015 genes with a standard deviation/ mean >0.1 Groups separated into infected Myd88 −/−, infected WT and uninfected samples
Statistical analysis of all 23,015 genes that went through the filtering process identified a total of 286 genes in WT and 4,151 in Myd88 −/− mice in response to H. felis infection with more genes differentially expressed at 47 than 25 weeks (Table 1). Comparing the number of upregulated genes between Myd88 −/− and WT at 47 weeks post infection, there were more upregulated genes (1140) in Myd88 −/− mice compared to WT mice (189 genes). A similar trend was observed for upregulated genes at 25 weeks and for downregulated genes at both time points, with more genes differentially expressed in Myd88 −/− than WT mice in response to H. felis infection. The number of differentiated genes at each time point in comparison to uninfected controls is illustrated in Fig. 2. Most genes overlapped between time points, however, there were a substantial number of genes that were unique to each set of time points that were differentially regulated (Fig. 2).
Scatterplot depiction of differentially expressed genes shows significantly (P < 0.05) up- and downregulated genes in Myd88 −/− mice in response to H. felis infection (Fig. 3). A majority of genes were altered after 25 weeks of H. felis infection. Some of the new additional genes at 47 weeks post-H. felis infection included the ring finger protein 213 (Rnf213) and Furin, which have been reported to be involved in angiogenesis [37, 38] and cancer progression [39], respectively indicating their role in advanced stages of cancer progression.
Scatterplot of Differentially Expressed Genes in H. felis- infected Myd88 −/− samples at (a) 25 and (b) 47 weeks. Scatterplot represents a summary of t-tests for individual genes, depicting the Log2 fold changes and their corresponding –log10 p-values of all differentially expressed genes from microarray analysis. Genes were separated into different time points. Negative values of Log2 fold changes indicate downregulated genes. Positive Log2 fold changes indicate upregulated genes. Genes with a fold change < 2.0 and a p value <0.05 are depicted as red dots and genes not found to be significantly altered are depicted as black dots. All infected animals were normalized to uninfected control mice at the same time point
Analysis of differentially expressed genes
Tables 2 and 3 show a list of the top 50 up- and downregulated genes in Myd88 −/− mice at 25 and 47 weeks post-H. felis infection compared to uninfected controls. The most highly upregulated gene during H. felis infection in Myd88 −/− mice included Chitinase-like (chil4), which is involved in tissue remodeling and wound healing [40–42]. Many of the upregulated genes in both 25 weeks and 47 weeks post-H. felis infection involved genes in the H2 Complex (murine major MHC), particularly the class I heavy chains, H2-K and H2-D. The light chain for this MHC complex consists of the β 2 -Microglobulin (B2m) [43]. MHC class II antigen presentation including the CD74 antigen (Cd74) was another gene that was upregulated in response to H. felis infection. High expression of Cd74 has been linked to chronic inflammation and carcinogenesis in the gastrointestinal tract [44]. Another highly expressed gene was Indoleamine 2,3-Dioxygenase 1 (Ido1), which is suggested to play a role in immune tolerance and high expression in colorectal cancer and is correlated with a poor clinical outcome (reviewed in [45]. The entire list of altered genes in response to infection with H. felis including those in WT mice have been submitted and can be uploaded as an excel file in Additional files 1: Table S1 (inf vs uninfectd charts.xlsx).
STRING summary networks depicting protein- protein interactions among the top differentially expressed genes (DEGs) for both up- and downregulated genes in Myd88 −/− mice are shown in Figs. 4 and 5 for 25 and 47 weeks, respectively. Thicker lines connecting the genes indicate a stronger association between the genes. A confidence score of at least 0.70 (high) was used. One of the key central nodes in the top DEGs in Myd88 −/− mice at both 25 and 47 weeks post-infection was guanylate-binding protein 2 (Gbp2), which is considered a potential marker for esophageal squamous cell carcinoma [46].
Network characterization of selected genes at 25 weeks. STRING gene networks of interactions of DEGs with a STRING interaction confidence of 0.7 or greater (high confidence) for H. felis-infected Myd88 −/− mice at 25 weeks for both upregulated (a) and downregulated (b) genes. Known interactions are illustrated with light blue string attachments (from curated databases) and light pink/purple strings (experimentally determined). Co-expression are illustrated with black string attachments
Network characterization of selected genes at 47 weeks. STRING gene networks of interactions of DEGs with a STRING interaction confidence of 0.7 or greater (high confidence) for MyD88 −/− - H. felis infected at 47 weeks both upregulated (a) and downregulated (b) genes. Known interactions are illustrated with light blue string attachments (from curated databases) and light pink/purple strings (experimentally determined). Co-expression are illustrated with black string attachments
Functional enrichment analysis of differentially expressed genes (DEGs)
To gain insights into the biological meaning and function of the differentially expressed genes, enrichment analysis was performed using the database for annotation, visualization and integrated discovery (DAVID) online analytical tools [47–49]. Annotation according to tissue expression, molecular function, cellular component and biological processing was done using Gene Ontology (GO) [49]. Enrichment analysis was performed to identify pathways, processes and gene categories that are over-represented in the list of DEGs compared to the mouse genome. GO clustering analysis for biological processes showed that responses related to immune system processes were the most upregulated enriched process in Myd88 −/− mice at both 25 and 47 weeks in response to H. felis infection (Fig. 6a and c). For molecular functions, antigen and protein complex binding were the most enriched processes (Fig. 6b and d). Downregulated enriched processes in Myd88 −/− mice in response to H. felis infection are presented as supplementary data (Additional files 2: Figure S1). A summary of the Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway annotation shown in Fig. 7 revealed that the most enriched pathway was antigen processing and presentation. This pathway shared the majority of its genes with the other major pathways and completely engulfed the other pathways by sharing more than 90% of the genes annotated. A breakdown of up- and downregulated KEGG pathway at 25 and 47 weeks in Myd88 −/− mice in response to H. felis infection is presented as supplementary data (Additional files 3: Figure S2).
Profiles of GO enrichment analysis. Enriched Go terms are shown for H. felis-infected Myd88 −/− mice at both 25 (a, b) and 47 weeks (c, d). Biological processes are depicted in figures A and C while molecular functions are depicted in B and D. For the biological processes, the top 20 processes are shown. All scores depicted are relative scores for number of genes in each function/ process relative to the number of total genes entered into the STRING database
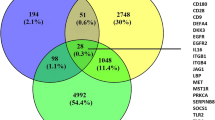
Venn Diagram showing distribution of different significantly enriched KEGG pathways. Pathways with p-value > E-04 are included. Antigen processing and presentation is the most enriched pathway overlapping the majority of genes with all other pathways. Notably, all processes are associated with inflammatory responses of the immune system
Discussion
Gastric cancer develops and progresses through a stepwise sequence of events from inflammation to atrophy, metaplasia, dysplasia, and finally to gastric cancer [50]. We previously demonstrated using a mouse model of gastric that mice deficient in MyD88 signaling exhibited dramatic pathology and an accelerated progression to gastric neoplasia in response to H. felis infection [13]. In the present study, we used microarray gene expression analysis to identify the genes involved in this progression to gastric neoplasia. Although previous studies have investigated differential gene expression in mice stomachs in response to Helicobacter infection, most have focused on H. pylori [51–53], which does not result in neoplastic changes in mice [54, 55]. The few studies that have examined gene expression profiles in mouse model of gastric cancer have used the insulin-gastrin (INS-GAS) transgenic gastric cancer mouse model [56, 57]. These mice have been shown to spontaneously develop gastric cancer even in the absence of Helicobacter infection [58]. We have previously reported that Myd88 −/− mice do not exhibit abnormal pathology in the absence of Helicobacter infection [13]. The global transcriptional profiling of mouse gastric tissue identified a large number of significant differentially expressed genes in H. felis-infected Myd88 −/− mice compared to H. felis-infected WT mice. The most over expressed gene in Myd88 −/− mice during H. felis infection at 25 weeks was Chil4. Chitinase like proteins (CLPs) have been studied in relation to other cancers yet little has been investigated in relation to gastric cancer except for our present study and a couple other studies [57, 59]. Upregulation of CLPs has been shown in a number of human cancers including brain, bone, breast, ovaries, lung, prostate, colon, thyroid, and liver [41, 60]. For gastric cancer studies, chitinase protein 3 like 1 (Chil1) was upregulated in INS-GAS mice infected with H. felis, [57]. In our present study, Chil1 was not upregulated in response to H. felis infection. However, in addition to upregulation of Chil4, another CLP, Chil3 was also significantly over expressed in H. felis-infected mice at both 25 weeks (p = 0.03) and 47 weeks (p = 0.01) (gene not listed in Tables 2 and 3, only the top 50 are listed). An abundant over expression of Chil1, Chil4 as well as Chil3 has been reported in early preneoplastic stage in the epidermis [61]. Overall, CLPs have been implicated to play a role in chronic inflammation, tissue remodeling, and wound healing [40]. Up-regulation of genes involved in tissue remodeling is noteworthy because chronic inflammation and subsequent damage to the gastric epithelium has been suggested to play an important role in cancer development and progression [62]. During chronic inflammation, the resulting prolonged tissue damage creates a loss of control over normal tissue repair mechanisms resulting in persistent hyper-tissue repair, which is accompanied with sustained proliferation [63] and ultimately advancing to precancerous lesions. Lost tissue is then replaced with stem and progenitor cells that are under a continuous stimulus of proliferation, leading to the accumulation of replacement cells with dysregulated and altered signaling pathways [63]. Further, studies investigating associations between chronic inflammation, tissue repair and carcinogenesis highlight the potential of these cellular changes in inducing both pro-oncogenic and tumor suppressor pathways [62, 64–67]. Our study, in addition to the work done by Li et al. [59] and Takaishi and Wang [57] show a need for further investigation into the role of CLPs in gastric carcinogenesis.
Other upregulated genes included Cd74, B2m, and interferon (IFN) induced genes such as GTPases (interferon gamma induced GTPase, lgtp, immune mediated GTPase family M member 2, lrgm2), Guanylate binding protein 2 (Gbp2), and transcription factor interferon regulatory factor 1 (Irf1). Cd74 or invariant chain (Ii) protein is a chaperone molecule responsible for regulating antigen presentation of MHC II molecules. It has been linked to chronic inflammation and carcinogenesis in the gastrointestinal tract [44]. Further, Cd74 was also shown to play a role as a receptor for migration inhibitory factor (MIF), a molecule reported to have pro-carcinogenic effects on gastric epithelial cells [68]. IFNs are known to activate signal transducer and activator of transcription 3 (STAT3) [69, 70] signaling leading to epithelial proliferation and inhibition of apoptosis [71, 72]. Currently not much is known about IFNs in gastric cancer. Ubiquitin D (Ubd), which is associated with progression of colon cancer [73], was also significantly expressed genes in Myd88 −/− in response to H. felis infection.
For downregulated genes, the significantly expressed ones included, ATPase H+/K+ transporting, alpha subunit (Atp4a), Atp4b, Mucin 5 AC (Muc5ac), apolipoprotein A-1 (Apoa1), and gastric intrinsic factor (Gif). The genes, Atp4a and Atp4b encode gastric H+/K + − ATPase alpha and beta subunits, respectively. Gastric H+/K + − ATPase alpha and beta subunits are expressed in parietal cells [74] and their loss has been associated with gastric dysplasia [58]. We observed downregulation of Atp4a and Atp4b in response to infection with H. felis, which may represent a loss of parietal cells that has been shown to precede gastric dysplasia. These results are in line with those observed in another fast progressing gastric cancer model involving the use of INS-GAS mice [58]. Muc5ac, which encodes gastric M1 mucin [75] has been reported to play a role in gastric carcinogenesis [76] was also downregulated in response to infection with H. felis in Myd88 −/− mice. Progression of gastric lesions has been reported to be associated with the gradual decrease in expression of Muc5ac [57, 77–79] followed by the transformation of the gastric epithelium [80] resulting in gastric dysplasia. Another highly downregulated gene we found in Myd88 −/− mice infected with H. felis was Apoa1, which was also reported to be downregulated in a fast progressing gastric cancer mouse model [57]. Proteomics approach in human gastric cancer also showed downregulation of Apoa1 [81], but its role in gastric carcinogenesis is unknown. In the lung, downregulation of Apoa1 was associated with an increased risk of lung cancer [82]. Data from Apoa1-deficient mice suggest antitumorigenic properties of Apoa1 via modulation of the immune system [83]. Gastric intrinsic factor (Gif), another downregulated gene is secreted by parietal cells and is required for Vitamin B12 absorption [84]. Concomitantly, the downregulation of Gif results in vitamin B12 deficiency (pernicious anemia). Gastric intrinsic factor was downregulated in H. felis-infected Myd88 −/− mice at both 25 and 47 weeks post-infection. A previous study using INS-GAS mice infected with H. felis also reported downregulation of Gif [57]. In human gastric cancer, Gif was one of the genes found by SAGE analysis to be downregulated [85]. Further, studies have shown an increased risk of gastric cancer in pernicious anemic patients [86].
We found a number of new genes in our fast progressing gastric mouse model, i.e., Myd88 −/− mice infected with H. felis. The genes included up- and downregulated genes, which had not been previously linked to Helicobacter-related gastric carcinogenesis including BPI fold containing family B member 1 (Bpifb1) and proteasome subunit beta 8 (Psmb8). These genes have been linked to cancer-related processes including, apoptosis and in some cases other cancers as well as prognosis indicators [87, 88]. Psmb8 was shown to be significantly up regulated in cancers such as bladder, breast, kidney, lung, uterine, and head and neck [89]. A recent study by Kwon, et al. [90], which was published during the writing of our manuscript reported that Psmb8 may be a potential marker for prognosis in gastric cancer. Bpifb1 may be involved in the innate immune response particularly in response to bacterial exposure. The protein encoded for by Bpifb1 binds bacterial lipopolysaccharide (LPS) as well as modulates the cellular response to LPS [91]. Bpifb1 has been found to be overexpressed in mucous cells of salivary gland tumors of papillary cystadenocarcinoma [87]. Future studies using a gastric culture organoid system will validate these genes and some of the important novel genes we identified for their role in rapid progression of Helicobacter-induced gastric cancer.
Conclusions
In this study, we have identified genes that are involved in the rapid progression of Helicobacter-induced gastric cancer that are also potentially regulated by MyD88. The identification of these important genes could potentially serve as targets for disease prevention. In addition, we show that our model is a useful mouse model system to identify genes involved in gastric cancer progression.
Methods
Animals
Six- to ten- week-old wild type (WT) and MyD88 deficient (Myd88 −/−) mice in the C57BL/6 background were used in this study. WT mice were purchased from The Jackson Laboratory (Bar Harbor, ME). Myd88 −/− mice were from our breeding colony originally provided by Dr. Akira (Osaka University, Japan). All mice were housed together before infection with H. felis and for the duration of the study. The Institutional Animal Care and Use Committee at the University of California, San Diego, approved all animal procedures and performed using accepted veterinary standards.
Bacterial growth conditions
Helicobacter felis, strain CS1 (ATCC 49179) was purchased from American Type Culture Collection (Manassas, VA). H. felis was routinely maintained on solid medium, Columbia agar (Becton Dickinson, MD) supplemented with 5% laked blood under microaerophilic conditions (5% O2, 10% CO2, 85% N2) at 37 °C and passaged every 2–3 days as described previously [13, 92]. Prior to mouse infections, H. felis was cultured in liquid medium, brain heart infusion broth (BHI, Becton Dickinson) supplemented with 10% fetal calf serum and incubated at 37 °C under microaerophilic conditions for 48 h. Spiral bacteria were enumerated using a Petroff-Hausser chamber before infections.
Mouse infections
A well-characterized cancer mouse model, which involves infecting C57BL/6 mice with H. felis (strain CS1), a close relative of the human gastric pathogen H. pylori was used in this study. Mice were inoculated with 109organisms in 300 μL of BHI by oral gavage three times at 2-day intervals as previously described [13, 92]. Control mice received BHI only. At 25 and 47 weeks post-infection, mice were euthanized and the stomachs removed under aseptic conditions and processed for assessment of gene expression.
RNA extraction and oligonucleotide microarray hybridization
Total RNA was extracted from gastric tissue obtained from H. felis-infected and uninfected WT and Myd88 −/− mice. Stomach tissue sections of 12 mice i.e., two uninfected controls and four infected WT (6) and MyD88 −/− (6) per time point (25 and 47 weeks) were analyzed. RNA was extracted from each section using the RNeasy miniprep kit (Qiagen) according to the manufacturer’s instructions followed by digestion with DNase 1 to remove genomic DNA. RNA concentration was determined using a NanoDrop spectrophotometer (NanoDrop Technologies, Inc., Waltham, MA). Double stranded cDNA and biotin-labeled cRNA were synthesized following the recommended Illumina protocol. Integrity of purified cRNAs was assessed on an Agilent 2100 Bioanalyzer prior to hybridization. 1.5 μg of labeled cRNA was hybridized to MouseWG-6 v2 Expression BeadChips genome wide arrays, which analyzes 25,600 transcripts (Illumina, San Diego, CA) using recommended Illumina reagents and protocols.
Identification of differentially expressed genes
Probe profiles (each row corresponding to a given probe and different columns for each sample) were exported from Genome Studio v1.8 (Illumina). The resultant tab-delimited file (Additional file 4, Probe_Profile_f.txt) was used as input for the Bioconductor lumi v2.18 R package (http://bioconductor.org/packages/release/bioc/html/lumi.html) [93]. Sample information is provided in Additional file 5 (sampleInfotxt.txt). Following quality assessment [density plot of intensity, cumulative density (CD), MA (transformed data onto log ratios and mean average) and pairwise MDS plots], the data were transformed using the vst (variance-stabilizing transformation) algorithm [94] then normalized using the Robust Spline Normalization (rsn) algorithm [94]. A second round of quality control of the normalized data was done to ensure data quality. After normalization genes with minimal variance across samples and genes that were not expressed in any sample (determined by detection calls) were removed prior to differential gene expression using linear models as implemented in limma [95]. We compared gene expression profiles of uninfected and infected mice at 25 and 47 weeks post infection. All mice used were on the C57BL/6 background. In addition, after normalization, significantly expressed genes were identified through volcano plot generation on the R - platform. Genes exhibiting statistically significant differential mean expression values (p < 0.05) were subjected to hierarchical clustering.
Enrichment analysis and pathway generation
To analyze KEGG pathway enrichment, official gene symbols for differentially expressed genes were submitted to DAVID (Database for Annotation, Visualization and Integrated Discovery) (http://david.abcc.ncifcrf.gov). This tool allows for identification of over expression within a set of genes as compared to the entire genome of a specific animal [47]. In addition, it allows for the demonstration of Gene Ontology (GO) terms as well as pathway and functional processes (molecular functions, biological expression, and cellular expression) enrichment. Functionally enriched gene networks, KEGG Pathways and ontology terms were identified. The mouse, Mus musculus complete genome was used as background genes. All functionally annotated genes presented in gene networks or pathways had p values < 0.05.
Gene network analysis
Gene networks were built using data on protein-protein interactions from EMBL STRING (Search Tool for the Retrieval of Interacting Genes/Proteins) database (http://string-db.org). This search tool is used to identify interactions correlated to expression data and/or literature citations among other criteria [96]. All gene connections created using STRING had a combined confidence score higher than 0.7 as previously described [97], Garcia-Alonso, 2014 #613}.
Statistical analysis
Statistics were done on the R platform using the Limma package from Bioconductor. To control for multiple testing, the False Discovery Rate (FDR) method was used with a cutoff for statistical significance of P values of < 0.05 and a log fold expression of 2. Differentially expressed genes were determined at 25 and 47 weeks after removing background differences from both Myd88 −/− and WT mice by comparing infected to uninfected mice in the same background.
Abbreviations
- (NF)-κB:
-
Nuclear factor kappa B
- Apoa1:
-
Apolipoprotein A-1
- Atp4a:
-
ATPase H+/K+ alpha subunit
- B2m:
-
β 2 -microglobulin
- Cd74:
-
CD74 antigen
- Chil4:
-
Chitinase-like 4
- DAVID:
-
Database for Annotation, Visualization and Integrated Discovery
- Gbp2:
-
Guanylate binding protein 2
- Gif:
-
Gastric intrinsic factor
- GO:
-
Gene ontology
- Ido1:
-
Indoleamine2, 3- dioxygenase 1
- IFN:
-
Interferon
- IL-1/IL-8:
-
Interleukin-1/-8
- INS-GAS:
-
Insulin-gastrin transgenic gastric mice
- KEGG:
-
Kyoto Encyclopedia of Genes and Genomes
- MDS:
-
Multi dimensional scaling
- MIF:
-
Migration inhibitory factor
- Muc5ac:
-
Mucin 5 AC
- Rnf213:
-
Ring finger protein 213
- STAT3:
-
Signal transducer and activator of transcription 3
- TLR:
-
Toll like receptor
- Ubd:
-
Ubiquitin d
References
Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011;61(2):69–90.
Asaka M. Helicobacter pylori infection--from gastritis to stomach cancer. Nihon Naika Gakkai Zasshi. 1996;85(9):1540–5.
Forman D. Helicobacter pylori infection: a novel risk factor in the etiology of gastric cancer. J Natl Cancer Inst. 1991;83(23):1702–3.
Schistosomes, liver flukes and Helicobacter pylori. IARC Working Group on the Evaluation of Carcinogenic Risks to Humans. Lyon, 7–14 June 1994. IARC Monogr Eval Carcinog Risks Hum. 1994;61:1–241.
Nomura A, Stemmermann GN, Chyou PH, Kato I, Perez-Perez GI, Blaser MJ. Helicobacter pylori infection and gastric carcinoma among Japanese Americans in Hawaii. N Engl J Med. 1991;325(16):1132–6.
Parsonnet J, Friedman GD, Vandersteen DP, Chang Y, Vogelman JH, Orentreich N, Sibley RK. Helicobacter pylori infection and the risk of gastric carcinoma. N Engl J Med. 1991;325(16):1127–31.
Peek Jr RM, Blaser MJ. Helicobacter pylori and gastrointestinal tract adenocarcinomas. Nat Rev Cancer. 2002;2(1):28–37.
Fox JG, Rogers AB, Whary MT, Ge Z, Ohtani M, Jones EK, Wang TC. Accelerated progression of gastritis to dysplasia in the pyloric antrum of TFF2 −/− C57BL6 x Sv129 Helicobacter pylori-infected mice. Am J Pathol. 2007;171(5):1520–8.
Correa P. Chronic gastritis: a clinico-pathological classification. Am J Gastroenterol. 1988;83(5):504–9.
Ernst PB, Gold BD. The disease spectrum of Helicobacter pylori: the immunopathogenesis of gastroduodenal ulcer and gastric cancer. Annu Rev Microbiol. 2000;54:615–40.
Gonzalez CA, Sala N, Capella G. Genetic susceptibility and gastric cancer risk. Int J Cancer. 2002;100(3):249–60.
Nardone G. Review article: molecular basis of gastric carcinogenesis. Aliment Pharmacol Ther. 2003;17 Suppl 2:75–81.
Banerjee A, Thamphiwatana S, Carmona EM, Rickman B, Doran KS, Obonyo M. Deficiency of the myeloid differentiation primary response molecule MyD88 leads to an early and rapid development of Helicobacter-induced gastric malignancy. Infect Immun. 2014;82(1):356–63.
O’Neill LA, Bowie AG. The family of five: TIR-domain-containing adaptors in Toll-like receptor signalling. Nat Rev Immunol. 2007;7(5):353–64.
Janssens S, Beyaert R. A universal role for MyD88 in TLR/IL-1R-mediated signaling. Trends Biochem Sci. 2002;27(9):474–82.
Akira S, Takeda K. Toll-like receptor signalling. Nat Rev Immunol. 2004;4(7):499–511.
Balkwill F, Mantovani A. Inflammation and cancer: back to Virchow? Lancet. 2001;357(9255):539–45.
Mantovani A, Allavena P, Sica A, Balkwill F. Cancer-related inflammation. Nature. 2008;454(7203):436–44.
Pradere JP, Dapito DH, Schwabe RF. The Yin and Yang of Toll-like receptors in cancer. Oncogene. 2014;33(27):3485–95.
Lee SH, Hu LL, Gonzalez-Navajas J, Seo GS, Shen C, Brick J, Herdman S, Varki N, Corr M, Lee J, et al. ERK activation drives intestinal tumorigenesis in Apc(min/+) mice. Nat Med. 2010;16(6):665–70.
Naugler WE, Sakurai T, Kim S, Maeda S, Kim K, Elsharkawy AM, Karin M. Gender disparity in liver cancer due to sex differences in MyD88-dependent IL-6 production. Science. 2007;317(5834):121–4.
Rakoff-Nahoum S, Medzhitov R. Regulation of spontaneous intestinal tumorigenesis through the adaptor protein MyD88. Science. 2007;317(5834):124–7.
Swann JB, Vesely MD, Silva A, Sharkey J, Akira S, Schreiber RD, Smyth MJ. Demonstration of inflammation-induced cancer and cancer immunoediting during primary tumorigenesis. Proc Natl Acad Sci U S A. 2008;105(2):652–6.
Uronis JM, Muhlbauer M, Herfarth HH, Rubinas TC, Jones GS, Jobin C. Modulation of the intestinal microbiota alters colitis-associated colorectal cancer susceptibility. PLoS One. 2009;4(6):e6026.
Maeda Y, Echizen K, Oshima H, Yu L, Sakulsak N, Hirose O, Yamada Y, Taniguchi T, Jenkins BJ, Saya H, et al. Myeloid Differentiation Factor 88 Signaling in Bone Marrow-Derived Cells Promotes Gastric Tumorigenesis by Generation of Inflammatory Microenvironment. Cancer Prev Res (Phila). 2016;9(3):253–63.
Tye H, Kennedy CL, Najdovska M, McLeod L, McCormack W, Hughes N, Dev A, Sievert W, Ooi CH, Ishikawa TO, et al. STAT3-driven upregulation of TLR2 promotes gastric tumorigenesis independent of tumor inflammation. Cancer Cell. 2012;22(4):466–78.
Kennedy CL, Najdovska M, Tye H, McLeod L, Yu L, Jarnicki A, Bhathal PS, Putoczki T, Ernst M, Jenkins BJ. Differential role of MyD88 and Mal/TIRAP in TLR2-mediated gastric tumourigenesis. Oncogene. 2014;33(19):2540–6.
Garrett WS, Punit S, Gallini CA, Michaud M, Zhang D, Sigrist KS, Lord GM, Glickman JN, Glimcher LH. Colitis-associated colorectal cancer driven by T-bet deficiency in dendritic cells. Cancer Cell. 2009;16(3):208–19.
Salcedo R, Worschech A, Cardone M, Jones Y, Gyulai Z, Dai RM, Wang E, Ma W, Haines D, O’HUigin C, et al. MyD88-mediated signaling prevents development of adenocarcinomas of the colon: role of interleukin 18. J Exp Med. 2010;207(8):1625–36.
Gibson DL, Ma C, Bergstrom KS, Huang JT, Man C, Vallance BA. MyD88 signalling plays a critical role in host defence by controlling pathogen burden and promoting epithelial cell homeostasis during Citrobacter rodentium-induced colitis. Cell Microbiol. 2008;10(3):618–31.
Ochi A, Nguyen AH, Bedrosian AS, Mushlin HM, Zarbakhsh S, Barilla R, Zambirinis CP, Fallon NC, Rehman A, Pylayeva-Gupta Y, et al. MyD88 inhibition amplifies dendritic cell capacity to promote pancreatic carcinogenesis via Th2 cells. J Exp Med. 2012;209(9):1671–87.
Araki A, Kanai T, Ishikura T, Makita S, Uraushihara K, Iiyama R, Totsuka T, Takeda K, Akira S, Watanabe M. MyD88-deficient mice develop severe intestinal inflammation in dextran sodium sulfate colitis. J Gastroenterol. 2005;40(1):16–23.
Brown SL, Riehl TE, Walker MR, Geske MJ, Doherty JM, Stenson WF, Stappenbeck TS. Myd88-dependent positioning of Ptgs2-expressing stromal cells maintains colonic epithelial proliferation during injury. J Clin Invest. 2007;117(1):258–69.
Pull SL, Doherty JM, Mills JC, Gordon JI, Stappenbeck TS. Activated macrophages are an adaptive element of the colonic epithelial progenitor niche necessary for regenerative responses to injury. Proc Natl Acad Sci U S A. 2005;102(1):99–104.
Salcedo R, Cataisson C, Hasan U, Yuspa SH, Trinchieri G. MyD88 and its divergent toll in carcinogenesis. Trends Immunol. 2013;34(8):379–89.
Khan J, Simon R, Bittner M, Chen Y, Leighton SB, Pohida T, Smith PD, Jiang Y, Gooden GC, Trent JM, et al. Gene expression profiling of alveolar rhabdomyosarcoma with cDNA microarrays. Cancer Res. 1998;58(22):5009–13.
Scholz B, Korn C, Wojtarowicz J, Mogler C, Augustin I, Boutros M, Niehrs C, Augustin HG. Endothelial RSPO3 Controls Vascular Stability and Pruning through Non-canonical WNT/Ca(2+)/NFAT Signaling. Dev Cell. 2016;36(1):79–93.
Kobayashi H, Matsuda Y, Hitomi T, Okuda H, Shioi H, Matsuda T, Imai H, Sone M, Taura D, Harada KH, et al. Biochemical and Functional Characterization of RNF213 (Mysterin) R4810K, a Susceptibility Mutation of Moyamoya Disease, in Angiogenesis In Vitro and In Vivo. J Am Heart Assoc. 2015;4(7):e002146.
Page RE, Klein-Szanto AJ, Litwin S, Nicolas E, Al-Jumaily R, Alexander P, Godwin AK, Ross EA, Schilder RJ, Bassi DE. Increased expression of the pro-protein convertase furin predicts decreased survival in ovarian cancer. Cell Oncol. 2007;29(4):289–99.
Lee CG, Da Silva CA, Dela Cruz CS, Ahangari F, Ma B, Kang MJ, He CH, Takyar S, Elias JA. Role of chitin and chitinase/chitinase-like proteins in inflammation, tissue remodeling, and injury. Annu Rev Physiol. 2011;73:479–501.
Coffman FD. Chitinase 3-Like-1 (CHI3L1): a putative disease marker at the interface of proteomics and glycomics. Crit Rev Clin Lab Sci. 2008;45(6):531–62.
Bretscher A, Edwards K, Fehon RG. ERM proteins and merlin: integrators at the cell cortex. Nat Rev Mol Cell Biol. 2002;3(8):586–99.
Gussow D, Rein R, Ginjaar I, Hochstenbach F, Seemann G, Kottman A, Ploegh HL. The human beta 2-microglobulin gene. Primary structure and definition of the transcriptional unit. J Immunol. 1987;139(9):3132–8.
Borghese F, Clanchy FI. CD74: an emerging opportunity as a therapeutic target in cancer and autoimmune disease. Expert Opin Ther Targets. 2011;15(3):237–51.
Chang M, Muller A, Pendergast G. Review on IDO1 (indoleamine 2,3-dioxygenase 1), with data on DNA, on the protein encoded, and where the gene is implicated. Atlas Genet Cytogenet Onccology Haematol. 2010;14(2):141–8.
Guimaraes DP, Oliveira IM, de Moraes E, Paiva GR, Souza DM, Barnas C, Olmedo DB, Pinto CE, Faria PA, De Moura Gallo CV, et al. Interferon-inducible guanylate binding protein (GBP)-2: a novel p53-regulated tumor marker in esophageal squamous cell carcinomas. Int J Cancer. 2009;124(2):272–9.
Dennis Jr G, Sherman BT, Hosack DA, Yang J, Gao W, Lane HC, Lempicki RA. DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biol. 2003;4(5):3.
da Huang W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4(1):44–57.
Huang DW, Sherman BT, Tan Q, Kir J, Liu D, Bryant D, Guo Y, Stephens R, Baseler MW, Lane HC, et al. DAVID Bioinformatics Resources: expanded annotation database and novel algorithms to better extract biology from large gene lists. Nucleic Acids Res. 2007;35(Web Server issue):W169–75.
Correa P. A human model of gastric carcinogenesis. Cancer Res. 1988;48(13):3554–60.
Mueller A, Merrell DS, Grimm J, Falkow S. Profiling of microdissected gastric epithelial cells reveals a cell type-specific response to Helicobacter pylori infection. Gastroenterology. 2004;127(5):1446–62.
Rad R, Brenner L, Krug A, Voland P, Mages J, Lang R, Schwendy S, Reindl W, Dossumbekova A, Ballhorn W, et al. Toll-like receptor-dependent activation of antigen-presenting cells affects adaptive immunity to Helicobacter pylori. Gastroenterology. 2007;133(1):150–63. e153.
Vivas JR, Regnault B, Michel V, Bussiere FI, Ave P, Huerre M, Labigne A, MM DE, Touati E. Interferon gamma-signature transcript profiling and IL-23 upregulation in response to Helicobacter pylori infection. Int J Immunopathol Pharmacol. 2008;21(3):515–26.
Smythies LE, Waites KB, Lindsey JR, Harris PR, Ghiara P, Smith PD. Helicobacter pylori-induced mucosal inflammation is Th1 mediated and exacerbated in IL-4, but not IFN-gamma, gene-deficient mice. J Immunol. 2000;165(2):1022–9.
van Doorn NE, Namavar F, Sparrius M, Stoof J, van Rees EP, van Doorn LJ, Vandenbroucke-Grauls CM. Helicobacter pylori-associated gastritis in mice is host and strain specific. Infect Immun. 1999;67(6):3040–6.
Kobayashi M, Lee H, Schaffer L, Gilmartin TJ, Head SR, Takaishi S, Wang TC, Nakayama J, Fukuda M. A distinctive set of genes is upregulated during the inflammation-carcinoma sequence in mouse stomach infected by Helicobacter felis. J Histochem Cytochem. 2007;55(3):263–74.
Takaishi S, Wang TC. Gene expression profiling in a mouse model of Helicobacter-induced gastric cancer. Cancer Sci. 2007;98(3):284–93.
Wang TC, Dangler CA, Chen D, Goldenring JR, Koh T, Raychowdhury R, Coffey RJ, Ito S, Varro A, Dockray GJ, et al. Synergistic interaction between hypergastrinemia and Helicobacter infection in a mouse model of gastric cancer. Gastroenterology. 2000;118(1):36–47.
Li ML, Zhang JC, Li SG, Wu WG, Rao LH, Dong P, Gu J, Lu JH, Zhang L, Ding QC, et al. Characteristic gene expression profiles in the progression from normal gastric epithelial cells to moderate gastric epithelial dysplasia and to gastric cancer. Chin Med J (Engl). 2012;125(10):1777–83.
Johansen JS. Studies on serum YKL-40 as a biomarker in diseases with inflammation, tissue remodelling, fibroses and cancer. Dan Med Bull. 2006;53(2):172–209.
Qureshi AM, Hannigan A, Campbell D, Nixon C, Wilson JB. Chitinase-like proteins are autoantigens in a model of inflammation-promoted incipient neoplasia. Genes Cancer. 2011;2(1):74–87.
Nickoloff BJ, Ben-Neriah Y, Pikarsky E. Inflammation and cancer: is the link as simple as we think? J Invest Dermatol. 2005;124(6):x–xiv.
Perryman SV, Sylvester KG. Repair and regeneration: opportunities for carcinogenesis from tissue stem cells. J Cell Mol Med. 2006;10(2):292–308.
Greten FR, Eckmann L, Greten TF, Park JM, Li ZW, Egan LJ, Kagnoff MF, Karin M. IKKbeta links inflammation and tumorigenesis in a mouse model of colitis-associated cancer. Cell. 2004;118(3):285–96.
Pikarsky E, Porat RM, Stein I, Abramovitch R, Amit S, Kasem S, Gutkovich-Pyest E, Urieli-Shoval S, Galun E, Ben-Neriah Y. NF-kappaB functions as a tumour promoter in inflammation-associated cancer. Nature. 2004;431(7007):461–6.
Beachy PA, Karhadkar SS, Berman DM. Tissue repair and stem cell renewal in carcinogenesis. Nature. 2004;432(7015):324–31.
Beachy PA, Karhadkar SS, Berman DM. Mending and malignancy. Nature. 2004;431(7007):402.
Leng L, Metz CN, Fang Y, Xu J, Donnelly S, Baugh J, Delohery T, Chen Y, Mitchell RA, Bucala R. MIF signal transduction initiated by binding to CD74. J Exp Med. 2003;197(11):1467–76.
Grivennikov SI, Karin M. Dangerous liaisons: STAT3 and NF-kappaB collaboration and crosstalk in cancer. Cytokine Growth Factor Rev. 2010;21(1):11–9.
Mitani Y, Takaoka A, Kim SH, Kato Y, Yokochi T, Tanaka N, Taniguchi T. Cross talk of the interferon-alpha/beta signalling complex with gp130 for effective interleukin-6 signalling. Genes Cells. 2001;6(7):631–40.
Jarnicki A, Putoczki T, Ernst M. Stat3: linking inflammation to epithelial cancer - more than a “gut” feeling? Cell Div. 2010;5:14.
Wang H, Lafdil F, Kong X, Gao B. Signal transducer and activator of transcription 3 in liver diseases: a novel therapeutic target. Int J Biol Sci. 2011;7(5):536–50.
Benson AK, Kelly SA, Legge R, Ma F, Low SJ, Kim J, Zhang M, Oh PL, Nehrenberg D, Hua K, et al. Individuality in gut microbiota composition is a complex polygenic trait shaped by multiple environmental and host genetic factors. Proc Natl Acad Sci U S A. 2010;107(44):18933–8.
Shin JM, Munson K, Vagin O, Sachs G. The gastric HK-ATPase: structure, function, and inhibition. Pflugers Arch. 2009;457(3):609–22.
Bara J, Chastre E, Mahiou J, Singh RL, Forgue-Lafitte ME, Hollande E, Godeau F. Gastric M1 mucin, an early oncofetal marker of colon carcinogenesis, is encoded by the MUC5AC gene. Int J Cancer. 1998;75(5):767–73.
Machado JC, Nogueira AM, Carneiro F, Reis CA, Sobrinho-Simoes M. Gastric carcinoma exhibits distinct types of cell differentiation: an immunohistochemical study of trefoil peptides (TFF1 and TFF2) and mucins (MUC1, MUC2, MUC5AC, and MUC6). J Pathol. 2000;190(4):437–43.
Song JY, Kim BW, Lee AW, Lee KY, Chung IS, Lee BI, Choi H, Ji JS, Chae HS, Choi KY. Expression of MUC5AC and Trefoil Peptide 1 (TFF1) in the Subtypes of Intestinal Metaplasia. Clin Endosc. 2012;45(2):151–4.
Sun Y, Li JY, He JS, Zhou LX, Chen K. Tissue microarray analysis of multiple gene expression in intestinal metaplasia, dysplasia and carcinoma of the stomach. Histopathology. 2005;46(5):505–14.
Zhang HK, Zhang QM, Zhao TH, Li YY, Yi YF. Expression of mucins and E-cadherin in gastric carcinoma and their clinical significance. World J Gastroenterol. 2004;10(20):3044–7.
Carrato C, Balague C, de Bolos C, Gonzalez E, Gambus G, Planas J, Perini JM, Andreu D, Real FX. Differential apomucin expression in normal and neoplastic human gastrointestinal tissues. Gastroenterology. 1994;107(1):160–72.
Ryu JW, Kim HJ, Lee YS, Myong NH, Hwang CH, Lee GS, Yom HC. The proteomics approach to find biomarkers in gastric cancer. J Korean Med Sci. 2003;18(4):505–9.
Borgquist S, Butt T, Almgren P, Shiffman D, Stocks T, Orho-Melander M, Manjer J, Melander O. Apolipoproteins, lipids and risk of cancer. Int J Cancer. 2016;138(11):2648–56.
Zamanian-Daryoush M, Lindner D, Tallant TC, Wang Z, Buffa J, Klipfell E, Parker Y, Hatala D, Parsons-Wingerter P, Rayman P, et al. The cardioprotective protein apolipoprotein A1 promotes potent anti-tumorigenic effects. J Biol Chem. 2013;288(29):21237–52.
Hoedemaeker PJ, Abels J, Wachters JJ, Arends A, Nieweg HO. Further investigations about the site of production of Castle’s gastric intrinsic factor. Lab Invest. 1966;15(7):1163–73.
Lee JY, Eom EM, Kim DS, Ha-Lee YM, Lee DH. Analysis of gene expression profiles of gastric normal and cancer tissues by SAGE. Genomics. 2003;82(1):78–85.
Kokkola A, Sjoblom SM, Haapiainen R, Sipponen P, Puolakkainen P, Jarvinen H. The risk of gastric carcinoma and carcinoid tumours in patients with pernicious anaemia. A prospective follow-up study. Scand J Gastroenterol. 1998;33(1):88–92.
Vargas PA, Speight PM, Bingle CD, Barrett AW, Bingle L. Expression of PLUNC family members in benign and malignant salivary gland tumours. Oral Dis. 2008;14(7):613–9.
Yang HZ, Cui B, Liu HZ, Mi S, Yan J, Yan HM, Hua F, Lin H, Cai WF, Xie WJ, et al. Blocking TLR2 activity attenuates pulmonary metastases of tumor. PLoS One. 2009;4(8):e6520.
Rouette A, Trofimov A, Haberl D, Boucher G, Lavallee VP, D’Angelo G, Hebert J, Sauvageau G, Lemieux S, Perreault C. Expression of immunoproteasome genes is regulated by cell-intrinsic and -extrinsic factors in human cancers. Sci Rep. 2016;6:34019.
Kwon CH, Park HJ, Choi YR, Kim A, Kim HW, Choi JH, Hwang CS, Lee SJ, Choi CI, Jeon TY, et al. PSMB8 and PBK as potential gastric cancer subtype-specific biomarkers associated with prognosis. Oncotarget. 2016;7(16):21454–68.
Shin OS, Uddin T, Citorik R, Wang JP, Della Pelle P, Kradin RL, Bingle CD, Bingle L, Camilli A, Bhuiyan TR, et al. LPLUNC1 modulates innate immune responses to Vibrio cholerae. J Infect Dis. 2011;204(9):1349–57.
Obonyo M, Rickman B, Guiney DG. Effects of myeloid differentiation primary response gene 88 (MyD88) activation on Helicobacter infection in vivo and induction of a Th17 response. Helicobacter. 2011;16(5):398–404.
Du P, Kibbe WA, Lin SM. Lumi: a pipeline for processing Illumina microarray. Bioinformatics. 2008;24(13):1547–8.
Lin SM, Du P, Huber W, Kibbe WA. Model-based variance-stabilizing transformation for Illumina microarray data. Nucleic Acids Res. 2008;36(2):e11.
Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, Smyth GK. Limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43(7):e47.
Jensen LJ, Kuhn M, Stark M, Chaffron S, Creevey C, Muller J, Doerks T, Julien P, Roth A, Simonovic M, et al. STRING 8--a global view on proteins and their functional interactions in 630 organisms. Nucleic Acids Res. 2009;37(Database issue):D412–6.
Franceschini A, Szklarczyk D, Frankild S, Kuhn M, Simonovic M, Roth A, Lin J, Minguez P, Bork P, von Mering C, et al. STRING v9.1: protein-protein interaction networks, with increased coverage and integration. Nucleic Acids Res. 2013;41(Database issue):D808–15.
Acknowledgements
The authors thank the Biomedical Genomics Laboratory (BIOGEM), UCSD for microarray analysis.
Funding
This work was supported in part by grants from the National Institutes of Health (NIH) U54CA132384, U54CA132379, R21CA188752, and R21AI115273-02.
Availability of data and materials
All data analyzed during this study are included in this published article and as supplementary material.
Authors’ contributions
MO and KD conceived and designed the study; AS carried out the experiments; IL performed database searches; IL and MM analyzed the microarray data; MO helped draft the manuscript; IL wrote the manuscript and was edited by MO. All authors read and approved the final manuscript.
Competing interests
The authors declare that they have no competing interests.
Consent for publication
Not applicable.
Ethics approval and consent to participate
Animals were studied under a research protocol approved by the Institutional Care and Use Committee (IACUC) of the University of California, San Diego.
Author information
Authors and Affiliations
Corresponding author
Additional files
Additional files 1:Table S1.
Excel file: Data of upregulated and downregulated genes in Myd88 −/− and WT mice at 25 and 47 weeks. (XLSX 4782 kb)
Additional files 2:Figure S1.
Downregulated biological processes and molecular functions in Myd88 −/− mice. Enriched Go terms are shown at both 25 (A, B) and 47 weeks (C, D). Biological processes are depicted in figures A and C while molecular functions are depicted in B and D. These functions were identified using STRING functional annotation tool. Relative scores were calculated using the number of genes found within each process relative to the total number of genes entered into the annotation tool. The top 20 Biological Processes are shown. (PPTX 482 kb)
Additional files 3:Figure S2.
KEGG Pathways for both up and downregulated Genes at 25 and 47 weeks in Myd88 −/− mice. KEGG pathway analysis of up- and downregulated genes in Myd88 −/− mice at 25 (Additional files 3: Figure S2a and b) and 47 weeks (Additional files 3: Figure S2c and d). Significantly enriched pathways (p < 0.05) are presented in each pie graph. (PPTX 1888 kb)
Additional files 4:
Text File: Probe Profile of Microarray Data used in analysis of gene expression. (TXT 30459 kb)
Additional files 5:
Text file: Sample Info file listing name and ID of each mouse used in the analysis. (TXT 533 bytes)
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Lozano-Pope, I., Sharma, A., Matthias, M. et al. Effect of myeloid differentiation primary response gene 88 on expression profiles of genes during the development and progression of Helicobacter-induced gastric cancer. BMC Cancer 17, 133 (2017). https://doi.org/10.1186/s12885-017-3114-y
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12885-017-3114-y