Abstract
Background
While programmed death 1 (PD-1) and programmed death-ligand 1 (PD-L1) checkpoint inhibitors have activity in a proportion of patients with advanced bladder cancer, strongly predictive and prognostic biomarkers are still lacking. In this study, we evaluated PD-L1 protein expression on circulating tumor cells (CTCs) isolated from patients with muscle invasive (MIBC) and metastatic (mBCa) bladder cancer and explore the prognostic value of CTC PD-L1 expression on clinical outcomes.
Methods
Blood samples from 25 patients with MIBC or mBCa were collected at UCSF and shipped to Epic Sciences. All nucleated cells were subjected to immunofluorescent (IF) staining and CTC identification by fluorescent scanners using algorithmic analysis. Cytokeratin expressing (CK)+ and (CK)−CTCs (CD45−, intact nuclei, morphologically distinct from WBCs) were enumerated. A subset of patient samples underwent genetic characterization by fluorescence in situ hybridization (FISH) and copy number variation (CNV) analysis.
Results
CTCs were detected in 20/25 (80 %) patients, inclusive of CK+ CTCs (13/25, 52 %), CK−CTCs (14/25, 56 %), CK+ CTC Clusters (6/25, 24 %), and apoptotic CTCs (13/25, 52 %). Seven of 25 (28 %) patients had PD-L1+ CTCs; 4 of these patients had exclusively CK−/CD45−/PD-L1+ CTCs. A subset of CTCs were secondarily confirmed as bladder cancer via FISH and CNV analysis, which revealed marked genomic instability. Although this study was not powered to evaluate survival, exploratory analyses demonstrated that patients with high PD-L1+/CD45−CTC burden and low burden of apoptotic CTCs had worse overall survival.
Conclusions
CTCs are detectable in both MIBC and mBCa patients. PD-L1 expression is demonstrated in both CK+ and CK−CTCs in patients with mBCa, and genomic analysis of these cells supports their tumor origin. Here we demonstrate the ability to identify CTCs in patients with advanced bladder cancer through a minimally invasive process. This may have the potential to guide checkpoint inhibitor immune therapies that have been established to have activity, often with durable responses, in a proportion of these patients.
Similar content being viewed by others
Background
Bladder cancer is the 5th most common cancer affecting both men and women in the United States, with a rising incidence worldwide [1, 2] (http://seer.cancer.gov/statfacts/html/urinb.html). The prognosis for patients with muscle invasive (MIBC) and metastatic (mBCa) bladder cancer is poor, with median survival with cisplatin-based chemotherapy averaging 14 months in the metastatic setting [3–6]. Urothelial bladder cancers have been found to express the markers programmed death-1 (PD-1) and programmed death ligand 1 (PD-L1) [7]. Expression of PD-1 and PD-L1 on cancer cells is hypothesized to allow cancers to evade immune surveillance and eradication. The discovery of this mechanism of resistance has provided the rationale for the development of PD-1 and PD-L1 checkpoint immunotherapy.
PD-1 checkpoint immunotherapy is rapidly emerging as a promising option for pre-treated patients with advanced tumors [8–11]. Multiple monoclonal antibodies have been developed against PD-1, and its ligand, PD-L1, and are currently being evaluated in clinical trials either as monotherapy, or in combination with cytotoxic chemotherapy, anti-angiogenic agents, or other immune checkpoint inhibitors [12–14]. Complete responses have been seen in heavily pre-treated patients, with some patients garnering continued tumor regression off therapy [15]. Early phase clinical trials have yielded promising results across many tumor types, measured both by response rate and duration of tumor response [12, 13, 16–18]. As of 2016, immunotherapies targeting PD-1 or PD-L1 have been approved by the FDA for the treatment of relapsed/refractory melanoma [19], squamous cell lung cancer [20, 21], non-small cell lung cancer [22], renal cell carcinoma [23], and most recently bladder cancer [24].
While PD-1/PD-L1 blockade has activity across a number of cancers, in most studies, less than 50 % of patients respond to treatment, indicating a need for predictive biomarkers. While higher PD-1 or PD-L1 expression on tumor biopsy specimens or tumor-infiltrating lymphocytes has been correlated with an increased likelihood of response [7], the positive and negative predictive value of these assays remains modest [13, 14, 25]. In many clinical studies PD-1 and PD-L1 expression has been assessed on archived specimens and may not reflect the current state of the cancer.
Obtaining solid tumor tissue biopsy specimens involves an invasive, technically challenging procedure posing risks to the patient. Instead, circulating tumor cell (CTC) isolation and analysis from peripheral blood samples may provide a fairly non-invasive approach to identify biomarkers and serially monitor response to treatment. Here, we present an assay for PD-L1 protein expression on peripherally collected CTCs [26, 27] and evaluate the incidence of circulating PD-L1+ CTCs in blood samples from patients with bladder cancer.
Methods
Cell culture and preparation of cell line control slides
Authenticated cell line cells H820 (lung cancer), Colo205 (colon cancer), A549 (lung cancer), SU-DHL-1 (lymphoma), H441 (lung cancer) and H23 (lung cancer), were purchased from ATCC and cultured in RPMI 1640 media supplemented with 10 % fetal bovine serum. Where applicable, cells were treated for 24 h with 100 ng/mL IFN-γ (R&D Systems, Minneapolis, MN). Cell line cells were then detached and spiked into healthy donor (HD) blood, which was then processed per Epic Sciences standard operating procedure [28, 29]. Briefly, red blood cells were lysed using ammonium chloride solution and the remaining nucleated cells were plated onto glass slides at a density of 3 million cells per slide. Slides were then stored in a − 80 °C biorepository until used for immunofluorescence (IF) staining and analysis.
Patient blood sample processing
Blood samples were collected from 25 bladder cancer patients who consented to an IRB-approved protocol at UCSF. Ten mL of whole blood was collected from each patient in Cell Free DNA BCT tubes (Streck, Omaha, NE) and shipped to Epic Sciences at ambient temperature for processing. Red blood cells were lysed using ammonium chloride solution, and nucleated cells were purified for direct deposition onto glass slides (at a density of 3 million cells per slide) and subsequent storage in the − 80 °C biorepository.
PD-L1 IF staining and analysis
Slides created from cell line control (CLC)-spiked HD samples or bladder cancer patient samples were subjected to automated IF staining for cytokeratin (CK), CD45 (hematopoietic marker) and PD-L1 (clone E1L3N, Cell Signaling Technology). Stained slides were analyzed with fluorescent scanners and morphology algorithms for the identification of traditional CK+ CTCs, CTC clusters, apoptotic CTCs and CK−CTCs morphologically distinct from hematological cells [26]. Trained classifiers conducted final classification of CTC subpopulations based on morphological parameters and biomarker expression. CLC slides were stained in parallel with patient samples. Threshold for PD-L1 cell positivity in patient samples was set to 95 % specificity of negative control CLC signal (i.e., 95 % negative control cell line cells lie below threshold).
Patient sample testing
CTC detection and classification on the Epic Sciences platform has been described previously [28, 29]. In brief, slides created from bladder cancer patient samples underwent automated IF staining for CK, CD45, and PD-L1, and counterstained with DAPI to visualize nuclei. Up to two slides were stained and evaluated per patient sample with fluorescent scanners and morphology algorithms for the identification of CTCs, CTC clusters, and apoptotic CTCs. A more thorough description of CTC types identified on the Epic Sciences platform has been published previously [26]. Briefly, all CTCs are strictly CD45−and have intact nuclei, with the exception of apoptotic CTCs, which are defined by their fragmented nuclei. CTCs are classified by the presence or absence of CK staining, and whether they are single CTCs or a CTC cluster. CK−CTCs must also exhibit distinctive nuclear morphology compared to neighboring white blood cells (WBCs) consistent with malignant origin. Apoptotic CTCs were defined as CK+/CD45−CTCs with DAPI pattern of chromosomal condensation and/or nuclear fragmentation and blebbing. A more detailed description of characterization of apoptotic CTCs has been published previously [26, 30]. California-licensed Clinical Laboratory Cytologists conduct final quality control of CTC subpopulation classification.
Fluorescence in situ hybridization
Slide coordinates of every CTC are recorded during the Epic Sciences CTC enumeration process, from which CTCs were relocated and analyzed by fluorescence in situ hybridization (FISH) for specific genetic alterations. One patient with CK−/PD-L1+ CTCs was selected and tested for genetic alterations by FISH. Coverslips were removed, IF staining attenuated, and cells were fixed and dehydrated. After dehydration, a probe solution (Cymogen Dx, Irvine, CA) selected for bladder cancer to assess polyploidy and gross genomic alterations in identified CTCs (targeting the CEP3, CEP7, CEP10, 5p15 DNA sequences of interest) was applied to each slide. After hybridization, slides were then washed to remove the unbound probe, counterstained with DAPI, and mounted with an anti-fade mounting medium. Adjacent patient WBCs were used as internal negative controls for endogenous genetic status for each cell analyzed.
Cell Isolation, amplification, and next-generation sequencing
Non-apoptotic individual CTCs were relocated on the slide based on a mathematical algorithm that converts the original CTC positions (x and y coordinates) computed during the scanning procedure into a new set of x, y references compatible with the Nikon TE2000 inverted immunofluorescent microscope used for cell capture. Single cells were captured using an Eppendorf TransferMan NK4 micromanipulator. Cells were deposited into PCR tubes using 1 μL of TE buffer and single cell whole genome amplification (WGA) was performed using SeqPlex Enhanced (Sigma) according to the manufacturer’s instructions with minor modifications. For patient copy number variation (CNV) analysis, 9 CK−/PD-L1+ CTCs from one patient were sequenced. Post-WGA, DNA concentrations were determined by UV/Vis. Next-Generation Sequencing (NGS) libraries were constructed using NEBNext Ultra DNA Library Prep Kit for Illumina (NEB) from 100 ng of WGA DNA as per manufacturer recommendation with minor modifications. After NGS library preparation, library concentrations and size distributions were determined with NEBNext Library Quant Kit for Illumina (NEB) and Fragment Analyzer (Advanced Analytical). Equinanomolar concentrations from each library were pooled and sequenced on an Illumina NextSeq 500 using a High Output Paired-End 2 × 150 format (PE 2 × 150).
Genome wide copy number variation analysis was performed using Epic single cell CNV analysis pipeline. Briefly, raw sequencing data (FASTQ) were aligned to hg38 human reference genome from UCSC (http://hgdownload.soe.ucsc.edu/goldenPath/hg38/bigZips/) using Burrows-Wheeler Aligner (BWA, http://bio-bwa.sourceforge.net). Binary alignment map files (BAM) were filtered for quality (MAPQ 30) to keep only the reads that have one or just a few “good” hits to the reference sequence. Hg38 human genome was divided into ~3000 bins of 1 million base pair and reads were counted within each bin for each sample. For each sample, read counts per bin were normalized against white blood cell controls, and the ratios were log2 transformed before plotted against genome positions.
Clinical data collection and survival analysis
All patients consented to participate in this IRB-approved research study prior to providing peripheral blood samples for analysis. All patient identifiers were removed prior to analysis by Epic Sciences. Clinical data extracted from their charts were maintained and tracked on a secure database. Overall survival was defined as the length of time from the date of the blood draw till death or last follow-up, and was calculated for patients who maintained follow-up at UCSF. Survival analysis was performed using the log-rank test and time-to-event curves were plotted using the Kaplan-Meier method.
Results
Assessment of PD-L1 antibody specificity
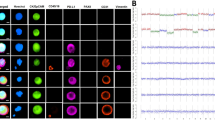
To evaluate the specificity of the PD-L1 CTC assay, anti-PD-L1 antibody and species-matched isotype controls were tested on high PD-L1 expressing and negative control cell lines (H820 and Colo205, respectively; Fig. 1a). Membrane-localized staining was observed in H820 cells stained with anti-PD-L1, whereas no staining was observed in negative control cell lines or with isotype control antibody. To further evaluate the specificity of the assay for PD-L1 protein, Colo205, A549, and SU-DHL-1 cell lines were treated with interferon gamma (IFN-γ), a known cytokine that induces PD-L1 expression [31] (Fig. 1b). Negative (Colo205) or low (A549, SU-DHL-1) PD-L1-expressing cell lines were selected specifically to observe if IFN-γ was sufficient to up-regulate PD-L1 protein expression. As detected by the PD-L1 CTC assay, IFN-γ treatment increased PD-L1 expression in both Colo205 and A549 cells compared to untreated cells, however, PD-L1 expression in SU-DHL-1 cells remained unchanged. This observed insensitivity to IFN-γ by SU-DHL-1 could be due to suppression of cytokine signaling 3 (SOCS3) protein, known to be highly expressed in SU-DHL-1, which inhibits the cytokine signaling required for IFN-γ mediated PD-L1 induction [32].
PD-L1 CTC Assay Development (a) PD-L1-specific antibody and species-matched isotype control were tested in negative (Colo205) and high (H820) PD-L1-expressing cell lines. Individual cellular PD-L1 IF signal is quantified and plotted. No staining above background was seen with isotype control or in Colo205 stained with anti-PD-L1. b IFN-γ treatment increases PD-L1 expression in Colo205 and A549, while SU-DHL-1 is insensitive. c PD-L1 antibody was titrated in PD-L1 IF staining of high (H820), medium (H441), low (SU-DHL-1) and negative (Colo205, H23) PD-L1-expressing cell lines to determine assay sensitivity and dynamic range. At the optimal antibody concentration (1:2000 dilution), mean H820, H441 and SU-DHL-1 PD-L1 expression was determined to be 140-, 36- and 13-fold higher than mean background staining in negative controls. d Representative images of high, medium and negative PD-L1 expressing cell lines show membrane-localization of PD-L1 IF signal
PD-L1 assay development
Anti-PD-L1 titration curves were generated with cell line controls expressing high (H820), medium (H441), low (SU-DHL-1) and negative (H23, Colo205) levels of PD-L1 (Fig. 2a). At the optimal antibody condition (1:2000 dilution), relative mean IF PD-L1 signal in H820, H441, SU-DHL-1 was detected at 140-, 36-and 13-fold, respectively, over baseline levels in negative control Colo205 (Fig. 1c). As expected, PD-L1 staining in positive cell lines was observed to be enriched in the plasma membrane (Fig. 1d) [33].
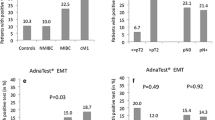
PD-L1 positive CTCs observable in patients with bladder cancer (a) Of the 7 total patients with PD-L1+ CTCs, two had exclusively CK+/PD-L1+ CTCs, four patients had CTCs that were exclusively CK−/PD-L1+, and one had both CK−and CK+/PD-L1+ CTCs. Further breakdown of CK+/PD-L1+ and CK−/PD-L1+ CTCs detected by tumor subtype and staging indicates that inclusion of CK− CTCs substantially increased sensitivity of PD-L1+ CTC detection. b Representative images of CK+/PD-L1+ and CK−/PD-L1+ patient CTCs are shown. c Representative FISH images of CK−/PD-L1+ cells demonstrate gross genomic instability and polyploidy using DNA probes for CEP3 (aqua), CEP7 (orange), CEP10 (green) and 5p15 (red). 29/33 (88 %) CK−/PD-L1+ cells assessed from one individual patient with high CTC burden were observed to have at least one abnormality determined by FISH
Patient characteristics
Demographics
Blood was obtained from 25 patients with bladder cancer from May 2014 to January 2016. Follow-up data for clinical outcomes was available for 19 patients. This cohort of patients represented a broad range of burden of disease. Overall, 17 men and 8 women participated in the study. The median age of all participants was 67 (range: 43 – 89), 4 patients had MIBC at the time of draw and 21 had mBCa. Fifteen of these patients had received prior chemotherapy, with at least 12 receiving one or more cisplatin-based regimens. Median time to blood draw from diagnosis was 1075 days. Please refer to Table 2 for median time to blood draw from diagnosis and prior therapies received. Of note, two patients who contributed samples had prior malignancies. One patient (B-026) had a history of BRCA2 positive breast cancer with no evidence of disease since 1993. The second patient (B-011) developed acute myeloid leukemia (AML) months after his CTCs were drawn. This patient is included in the analysis as the CTCs were CD45−, while cells from AML were found to be uniformly CD45+; FISH analysis performed on these CD45− CTCs confirmed genomic alterations consistent with bladder cancer, further discussed below. See Table 1 for summary of patient demographics.
CTC Detection
CK+ single CTCs were detected in 13/25 (52 %) patient samples. CK+ CTC Clusters were detected in 6/25 (24 %), apoptotic CTCs were detected in 13/25 (52 %), and CK− CTCs were detected in 14/25 (56 %) patient samples. Combined, 20/25 (80 %) patient samples had detectable CTCs of all subtypes. Interestingly, 3 of 4 patients with MIBC had detectable CTCs (one patient had CK−CTCs, one with CK+ CTCs, and one with apoptotic CTCs).
PD-L1 Expression on CTCs
Of the 20 patients with detectable CTCs, 7/20 (35 %) had CTCs with PD-L1 positivity. Of these, five patients had CK−/PDL1+ CTCs, with four of these patients having PD-L1 positivity exclusively on CK− CTCs. Patients with PD-L1 expression detectable on CTCs all had metastatic bladder cancer. See patient sample CTC summary in Table 2 and Fig. 2a. Thus far, seven patients have received PD-1 targeting therapy-one received it prior to CTC collection, the remaining started therapy after blood collection.
CK− PDL1+ CD45−CTCs have gross genomic aberrations
To further assess the specificity of the PD-L1 assay in patient samples, we analyzed CK−/CD45−/PD-L1+ CTCs from two metastatic bladder cancer patients for genomic abnormalities. Patient selection was determined by the ability to perform FISH analysis, which excluded those requiring PDL1 amplification during processing of the samples. Thirty-three CK−/CD45−/PD-L1+ CTCs detected in one mBCa patient (B-011) were analyzed by FISH for genetic alterations commonly associated with bladder cancer. As shown in Fig. 2c, the presence of these genetic abnormalities in identified CTCs is consistent with malignant origin. See detailed results in Table 3.
Nine CK−/CD45−/PD-L1+ CTCs detected in a different patient with metastatic disease (B-022) were further assessed for CNV using NGS. Read count analysis of these CTCs are provided in Additional file 1: Figure S1. Five of nine (56 %) CTCs demonstrated a significant number of genomic aberrations in chromosomal copy number changes, including chromosomes 1, 2, 6, 17, 18, 20, 21, X and Y (Fig. 3a–e). Figure 3f depicts the ploidy analysis of genomic aberrations seen in the CTC evaluated in Fig. 3b. As seen, this shows numerous amplifications and deletions within multiple arms and chromosomes. Interestingly, chromosome 6 and Y appear to be diploid, while the remainder of the chromosomes demonstrate some level of ploidy abnormality. Evaluating these results against copy number data available from The Cancer Genome Atlas (TCGA) cohort of 412 MIBC patients, we found some concordant losses in chromosomes 1, 2, 6, 17, and 18. These genomic aberrations provide supporting data that the cells identified were of malignant origin.
Genomic heterogeneity of bladder cancer CTCs. Plots of whole genome CNV profiles of five CK−/CD45−/PD-L1+ CTCs from patient B-022 (a-e). X axis: chromosomes displayed as from chromosome 1 to 22, X and Y (from left to right, shifted by red and blue color); Y axis: normalized log2 transformed ratio of copy number of test sample over that of WBC control. Five CTCs show various genomic aberrations a loss of chromosome 1, 2, 17, 18, and 20; b loss of chromosome 6; c gain of chromosome 21; d and e gain of chromosome X and loss of chromosome Y. f Ploidy analysis for genomic aberrations from NSG seen in CTC (b), predicted ploidy = 3.25
High circulating PD-L1+ CD45−CTC burden and overall survival
Follow-up survival data was available for 19 of 25 patients. High burden of PD-L1+ CTC is defined as >1 PD-L1+ CTC/mL (n = 4). Low burden of PD-L1+ CTC is defined as greater than 0, but less than 1 PD-L1+ CTC/mL (n = 3). PD-L1+ negative staining was defined as 0 PD-L1+ CTCs/mL (n = 12). Recent data presented by Boffa, et al. at the American Society of Clinical Oncology (ASCO 2016) meeting, which utilized the Epic PD-L1 assay to demonstrate higher burden of PD-L1+ CTCs is a poor prognostic factor in lung cancer compared those patients with the highest burden of PD-L1+ CTCs to low/negative PD-L1+ CTCs. Our analysis used a similar approach evaluating highest burden of PD-L1+ CTCs (>1/mL, n = 4) to the rest of the cohort (n = 15). While no statistically significant conclusions could be drawn from this data, patients with higher circulating PD-L1+ CTC burden had a shorter median overall survival (194 vs. 303 days, log-rank p = 0.416) compared to those with low circulating PD-L1+ CTC burden (Additional file 2: Figure S2A).
Of the seven patients who received PD-1 checkpoint immunotherapy, follow-up survival data was available for 5 patients. One patient passed away after one cycle of therapy. The remaining four patients received at least three cycles of therapy. Two of four patients had detectable CTCs (B-025 and B-028). Only patient B-025 exhibited PD-L1+ (1.3 %) CTCs. The patient demonstrated progression on PD-1 immunotherapy on radiographic assessment after cycle 5 of therapy and was discontinued. The other three patients who lacked PD-L1+ CTCs also demonstrated radiographic progression on PD-1 immunotherapy after 3, 6, and 8 cycles of therapy, respectively.
Apoptotic CTCs and overall survival
Thirteen out of 25 patients (52 %) were detected to have apoptotic CTCs. Two of four patients with MIBC had detectable apoptotic CTCs (B-003 = 1.8, B-015 = 1.5) and 11/16 (69 %) mBCa patients with detectable CTCs had apoptotic CTCs. Kaplan-Meier analysis of all 13 patients with apoptotic CTCs suggests a trend towards improved overall survival in patients with apoptotic CTCs (Hazard ratio = 0.4, 0.12–1.31; p = 0.159). See Additional file 2: Figure S2B.
Discussion
In this study, we demonstrate the ability to detect PD-L1 positivity both in cell lines spiked into human blood as well as in bladder cancer CTCs processed on the Epic Sciences platform. Pre-clinically, using cell lines with known PD-L1 expression, we observed assay specificity for PD-L1 expression by IF staining. Furthermore, expression was found to be 140-fold higher in H820 (high expressing) cells as compared to negative controls, indicative of high dynamic range and assay sensitivity. This is further supported by detection of upregulated PD-L1 expression on Colo205 and A549 cell lines treated with IFN-γ.
We evaluated the clinical utility and feasibility of the Epic Sciences PD-L1 CTC assay using 25 bladder cancer patient samples of various stages (MIBC and mBCa). While CTCs were detected in 3 of the 4 patients with MIBC, PD-L1 expression was not identified in this small sub-cohort of patients. PD-L1+ CTCs were detected in 7/20 (35 %) patients with mBCa, and four of these patients expressed PD-L1 exclusively on CK− CTCs. To confirm that these cells were truly CTCs, CK−/PD-L1+ CTCs from one patient were evaluated using a bladder cancer FISH probe panel and CK−/PD-L1+ CTCs from another patient were assessed for CNV by NGS. Both of these methods found genomic aberrations in CTCs consistent with malignant origin. CNV analysis on 5 of 9 CTCs that underwent NGS showed marked chromosomal copy number variations with ploidy analysis of one cell revealing a high level of aberrancy. Four of the nine cells did not exhibit a large number of chromosomal copy number variations (see Additional file 2: Figure S2). The heterogeneity of aberrancies found in these cells is consistent with prior findings of intratumoral DNA ploidy heterogeneity described in various tumor types, including bladder cancer [34–36] and highly supports malignant origin. The finding of patients with exclusively CK−/PD-L1+ cells is intriguing as it could suggest that cells undergoing mesenchymal differentiation and metastasis may escape immune surveillance potentially via expression of PD-L1. However, this finding requires confirmation in a larger cohort. This also points to the utility of using a CTC enrichment technique that does not require CK expression.
It has previously been demonstrated in bladder cancer and other solid tumors using tissue biopsy staining that patients fare worse if their tumors are able to evade the immune system [37]. While these patient samples represent a small, cross-sectional cohort rather than a prospective controlled trial, it is worth noting that those with the highest PD-L1+ CTC burden (>1/mL) had a shorter overall survival from the time of the CTC draw. Furthermore, evaluation of apoptotic CTC counts demonstrated a trend toward shorter survival for those with fewer apoptotic CTCs. Fewer apoptotic cells have been observed in patients with metastatic breast cancer compared to those with early stage disease, suggesting a correlation between lack of apoptosis with cell survival and an aggressive phenotype [38]. Of note, the time of CTC draw was highly variable across our patient population and represented snapshots of a wide range of burden of disease and prior therapies. Some patients were actively undergoing chemotherapy at the time of their draw, which could influence the burden of total and apoptotic CTCs detected. The aim of this study was to demonstrate feasibility and consistency in detecting PD-L1+CTCs in bladder cancer and was not powered to evaluate survival benefits. Further evaluation and optimization in a larger and more uniform cohort with appropriate power and design is warranted to better evaluate the association of PD-L1 expression and apoptotic CTCs with survival.
Biomarkers to predict response to PD-1-directed therapies are far from established. Higher PD-L1 expression in solid tumor biopsy samples is associated with response to pembrolizumab in non-small cell lung cancer [20] and correlates with response to therapy in other indications [14, 39, 40]. However, a significant portion of PD-L1− patients also respond to therapy. Recent multifocal genetic and proteomic analyses of regions within tumors have revealed levels of spatial heterogeneity in several cancer types that might limit the interpretation of solid tumor biopsies [41–45]. Due to tumor heterogeneity, smaller sample size or intratumoral location of the biopsy site may yield a false negative tissue assessment of PD-L1. Similarly, PD-L1 expression is thought to be dynamic and can vary in response to interferon levels and possibly other factors, making the reliability of a static assessment on a limited tumor tissue sample questionable. While it would be challenging to monitor expression via serial tumor tissue biopsies, it is much more feasible to monitor expression of this dynamic marker via serial CTC assessment from whole blood. It may even be possible to detect PD-L1+ CTCs in patients who are PD-L1−on tissue biopsy. The correlation of CTC PD-L1 status with paired tumor tissue samples and the implications of discordant expression was not pursued in this study, but will be important for future assessment. Similarly, five evaluable patients received PD-1 targeting therapy after blood was drawn for analysis of CTCs. One patient had a rapid decline after therapy and the remaining 4 patients demonstrated radiographic progression after 3, 6, and 8 cycles of PD-1 checkpoint immune therapy. Serial blood draws were not performed in this study. Therefore there was an insufficient sample size to assess the prognostic value of PD-L1+ CTCs in patients receiving checkpoint inhibitors. This would be best explored in a prospective study with a larger cohort of patients with similar disease burden receiving PD-1 or PD-L1 checkpoint therapy.
Another limiting factor in the analysis of PD-L1 expression in solid tumor samples is the lack of standardization of PD-L1 immunohistochemical assays and their respective positivity thresholds [13, 14]. CTCs provide a minimally-invasive sampling method that could prove useful for prognostication of therapeutic benefit through longitudinal monitoring and measurement of pharmacodynamic changes in CTC counts and/or changes of CTC PD-L1 expression. The Epic Sciences CTC platform utilizes a central laboratory for consistent quality with a central biorepository for retrospective analyses of biomarkers on morphologically intact CTCs. Analytical validation studies of Epic’s CTC platform have been published [26]. In addition, this platform has previously been compared to the FDA approved CellSearch platform (Janssen Diagnostics, NJ, USA) demonstrating consistent, if not increased sensitivity in the detection of CTCs [27]. Using this technology, repeat sampling of patients utilizing CTCs is both feasible and amenable to pharmacodynamic biomarker development to identify not only patients that might respond, but those who are responding to therapy.
Conclusions
Our findings demonstrate the ability to detect and quantify PD-L1 protein on bladder cancer patients’ CTCs using an assay with specificity and sensitivity demonstrated in CTC surrogate cell lines. Exploratory analysis of survival data suggests a trend towards improved survival in those with low PD-L1 expression or with higher burden of apoptotic CTCs. While the data presented here are compelling, it should be emphasized that this study is descriptive, represents a small sample size, and requires validation with a larger, prospective study encompassing a broader patient population that is appropriately powered to evaluate survival benefits. Nonetheless, these data provide initial support for broader development of CTC PD-L1 expression. With further study, PD-L1 expression on CTCs isolated from peripheral circulation has the potential to become a new prognostic and predictive biomarker with which to stratify treatments for patients and possibly predict response to immunotherapy in bladder cancer.
Abbreviations
- BAM:
-
Binary Alignment Map
- CK:
-
Cytokeratin
- CLC:
-
Cell line control
- CNV:
-
Copy number variation
- CTC:
-
Circulating tumor cell
- FISH:
-
Fluorescence in situ hybridization
- HD:
-
Healthy donor
- IF:
-
Immunofluorescence
- IFN-γ:
-
Interferon gamma
- mBCa:
-
Metastatic bladder cancer
- MIBC:
-
Muscle-invasive bladder cancer
- NGS:
-
Next generation sequencing
- PD-1:
-
Programmed death 1
- PD-L1:
-
Programmed death ligand 1
- TCGA:
-
The cancer genome atlas
- WBC:
-
White blood cell
- WGA:
-
Whole genome amplification
References
Ploeg M, Aben KK, Kiemeney LA. The present and future burden of urinary bladder cancer in the world. World J Urol. 2009;27(3):289–93.
Siegel R, Ma J, Zou Z, Jemal A. Cancer statistics, 2014. CA Cancer J Clin. 2014;64(1):9–29.
Bellmunt J, von der Maase H, Mead GM, Skoneczna I, De Santis M, Daugaard G, Boehle A, Chevreau C, Paz-Ares L, Laufman LR, Winquist E, Raghavan D, Marreaud S, Collette S, Sylvester R, De Wit R. Randomized phase III study comparing paclitaxel/cisplatin/gemcitabine and gemcitabine/cisplatin in patients with locally advanced or metastatic urothelial cancer without prior systemic therapy: EORTC Intergroup Study 30987. J Clin Oncol. 2012;30(10):1107–13.
Sternberg CN, De Mulder P, Schornagel JH, Theodore C, Fossa SD, Van Oosterom AT, Witjes JA, Spina M, Van Groeningen CJ, Duclos B, Roberts JT, De Balincourt C, Collette L, EORTC Genito-Urinary Cancer Group. Seven year update of an EORTC phase III trial of high-dose intensity M-VAC chemotherapy and G-CSF versus classic M-VAC in advanced urothelial tract tumours. Eur J Cancer. 2006;42(1):50–4.
von der Maase H, Hansen SW, Roberts JT, Dogliotti L, Oliver T, Moore MJ, Bodrogi I, Albers P, Knuth A, Lippert CM, Kerbrat P, Sanchez Rovira P, Wersall P, Cleall SP, Roychowdhury DF, Tomlin I, Visseren-Grul CM, Conte PF. Gemcitabine and cisplatin versus methotrexate, vinblastine, doxorubicin, and cisplatin in advanced or metastatic bladder cancer: results of a large, randomized, multinational, multicenter, phase III study. J Clin Oncol. 2000;18(17):3068–77.
International Collaboration of Trialists, Medical Research Council Advanced Bladder Cancer Working Party (now the National Cancer Research Institute Bladder Cancer Clinical Studies Group), European Organisation for Research and Treatment of Cancer Genito-Urinary Tract Cancer Group, Australian Bladder Cancer Study Group, National Cancer Institute of Canada Clinical Trials Group, Finnbladder, Norwegian Bladder Cancer Study Group, Club Urologico Espanol de Tratamiento Oncologico Group, Griffiths G, Hall R, Sylvester R, Raghavan D, Parmar MK. International phase III trial assessing neoadjuvant cisplatin, methotrexate, and vinblastine chemotherapy for muscle-invasive bladder cancer: long-term results of the BA06 30894 trial. J Clin Oncol. 2011;29(16):2171–7.
Powles T, Eder JP, Fine GD, Braiteh FS, Loriot Y, Cruz C, Bellmunt J, Burris HA, Petrylak DP, Teng SL, Shen X, Boyd Z, Hegde PS, Chen DS, Vogelzang NJ. MPDL3280A (anti-PD-L1) treatment leads to clinical activity in metastatic bladder cancer. Nature. 2014;515(7528):558–62.
Okazaki T, Chikuma S, Iwai Y, Fagarasan S, Honjo T. A rheostat for immune responses: the unique properties of PD-1 and their advantages for clinical application. Nat Immunol. 2013;14(12):1212–8.
Pedoeem A, Azoulay-Alfaguter I, Strazza M, Silverman GJ, Mor A. Programmed death-1 pathway in cancer and autoimmunity. Clin Immunol. 2014;153(1):145–52.
Perez-Gracia JL, Labiano S, Rodriguez-Ruiz ME, Sanmamed MF, Melero I. Orchestrating immune check-point blockade for cancer immunotherapy in combinations. Curr Opin Immunol. 2014;27:89–97.
Razzak M. From ASCO-targeted therapies: Anti-PD-1 approaches--important steps forward in metastatic melanoma. Nat Rev Clin Oncol. 2013;10(7):365.
Drake CG, Lipson EJ, Brahmer JR. Breathing new life into immunotherapy: review of melanoma, lung and kidney cancer. Nat Rev Clin Oncol. 2014;11(1):24–37.
Patel SP, Kurzrock R. PD-L1 Expression as a Predictive Biomarker in Cancer Immunotherapy. Mol Cancer Ther. 2015;14(4):847–56.
Philips GK, Atkins M. Therapeutic uses of anti-PD-1 and anti-PD-L1 antibodies. Int Immunol. 2015;27(1):39–46.
Lipson EJ, Sharfman WH, Drake CG, Wollner I, Taube JM, Anders RA, Xu H, Yao S, Pons A, Chen L, Pardoll DM, Brahmer JR, Topalian SL. Durable cancer regression off-treatment and effective reinduction therapy with an anti-PD-1 antibody. Clin Cancer Res. 2013;19(2):462–8.
Robert C, Thomas L, Bondarenko I, O’Day S, Weber J, Garbe C, Lebbe C, Baurain JF, Testori A, Grob JJ, Davidson N, Richards J, Maio M, Hauschild A, Miller Jr WH, Gascon P, Lotem M, Harmankaya K, Ibrahim R, Francis S, Chen TT, Humphrey R, Hoos A, Wolchok JD. Ipilimumab plus dacarbazine for previously untreated metastatic melanoma. N Engl J Med. 2011;364(26):2517–26.
Wolchok JD, Kluger H, Callahan MK, Postow MA, Rizvi NA, Lesokhin AM, Segal NH, Ariyan CE, Gordon RA, Reed K, Burke MM, Caldwell A, Kronenberg SA, Agunwamba BU, Zhang X, Lowy I, Inzunza HD, Feely W, Horak CE, Hong Q, Korman AJ, Wigginton JM, Gupta A, Sznol M. Nivolumab plus ipilimumab in advanced melanoma. N Engl J Med. 2013;369(2):122–33.
Topalian SL, Hodi FS, Brahmer JR, Gettinger SN, Smith DC, McDermott DF, Powderly JD, Carvajal RD, Sosman JA, Atkins MB, Leming PD, Spigel DR, Antonia SJ, Horn L, Drake CG, Pardoll DM, Chen L, Sharfman WH, Anders RA, Taube JM, McMiller TL, Xu H, Korman AJ, Jure-Kunkel M, Agrawal S, McDonald D, Kollia GD, Gupta A, Wigginton JM, Sznol M. Safety, activity, and immune correlates of anti-PD-1 antibody in cancer. N Engl J Med. 2012;366(26):2443–54.
Momtaz P, Postow MA. Immunologic checkpoints in cancer therapy: focus on the programmed death-1 (PD-1) receptor pathway. Pharmgenomics Pers Med. 2014;7:357–65.
Garon EB, Rizvi NA, Hui R, Leighl N, Balmanoukian AS, Eder JP, Patnaik A, Aggarwal C, Gubens M, Horn L, Carcereny E, Ahn MJ, Felip E, Lee JS, Hellmann MD, Hamid O, Goldman JW, Soria JC, Dolled-Filhart M, Rutledge RZ, Zhang J, Lunceford JK, Rangwala R, Lubiniecki GM, Roach C, Emancipator K, Gandhi L, KEYNOTE-001 Investigators. Pembrolizumab for the treatment of non-small-cell lung cancer. N Engl J Med. 2015;372(21):2018–28.
Brahmer J, Reckamp KL, Baas P, Crino L, Eberhardt WE, Poddubskaya E, Antonia S, Pluzanski A, Vokes EE, Holgado E, Waterhouse D, Ready N, Gainor J, Aren Frontera O, Havel L, Steins M, Garassino MC, Aerts JG, Domine M, Paz-Ares L, Reck M, Baudelet C, Harbison CT, Lestini B, Spigel DR. Nivolumab versus Docetaxel in Advanced Squamous-Cell Non-Small-Cell Lung Cancer. N Engl J Med. 2015;373(2):123–35.
Borghaei H, Paz-Ares L, Horn L, Spigel DR, Steins M, Ready NE, Chow LQ, Vokes EE, Felip E, Holgado E, Barlesi F, Kohlhaufl M, Arrieta O, Burgio MA, Fayette J, Lena H, Poddubskaya E, Gerber DE, Gettinger SN, Rudin CM, Rizvi N, Crino L, Blumenschein Jr GR, Antonia SJ, Dorange C, Harbison CT, Graf Finckenstein F, Brahmer JR. Nivolumab versus Docetaxel in Advanced Nonsquamous Non-Small-Cell Lung Cancer. N Engl J Med. 2015;373(17):1627–39.
Motzer RJ, Escudier B, McDermott DF, George S, Hammers HJ, Srinivas S, Tykodi SS, Sosman JA, Procopio G, Plimack ER, Castellano D, Choueiri TK, Gurney H, Donskov F, Bono P, Wagstaff J, Gauler TC, Ueda T, Tomita Y, Schutz FA, Kollmannsberger C, Larkin J, Ravaud A, Simon JS, Xu LA, Waxman IM, Sharma P, CheckMate 025 Investigators. Nivolumab versus Everolimus in Advanced Renal-Cell Carcinoma. N Engl J Med. 2015;373(19):1803–13.
Rosenberg JE, Hoffman-Censits J, Powles T, van der Heijden MS, Balar AV, Necchi A, Dawson N, O’Donnell PH, Balmanoukian A, Loriot Y, Srinivas S, Retz MM, Grivas P, Joseph RW, Galsky MD, Fleming MT, Petrylak DP, Perez-Gracia JL, Burris HA, Castellano D, Canil C, Bellmunt J, Bajorin D, Nickles D, Bourgon R, Frampton GM, Cui N, Mariathasan S, Abidoye O, Fine GD, Dreicer R. Atezolizumab in patients with locally advanced and metastatic urothelial carcinoma who have progressed following treatment with platinum-based chemotherapy: a single-arm, multicentre, phase 2 trial. Lancet. 2016;387(10031):1909–20.
Errico A. Immunotherapy: PD-1-PD-L1 axis: efficient checkpoint blockade against cancer. Nat Rev Clin Oncol. 2015;12(2):63.
Werner SG RP, Landers M, Valenta DT, Schroeder M, Greene S, Bales N, Dittamore R, Marrinucci D. Analytical Validation and Capabilities of the Epic CTC Platform: Enrichment-Free Circulating Tumour Cell Detection and Characterization. J Circulating Biomarkers. 2015;4:3.
Punnoose EA, et al. PTEN loss in circulating tumour cells correlates with PTEN loss in fresh tumour tissue from castration-resistant prostate cancer patients. Br J Cancer. 2015.
Cho EH, Wendel M, Luttgen M, Yoshioka C, Marrinucci D, Lazar D, Schram E, Nieva J, Bazhenova L, Morgan A, Ko AH, Korn WM, Kolatkar A, Bethel K, Kuhn P: Characterization of circulating tumor cell aggregates identified in patients with epithelial tumors. Phys Biol 2012, 9(1):016001-3975/9/1/016001. Epub 2012 Feb 3.
Marrinucci D, Bethel K, Kolatkar A, Luttgen MS, Malchiodi M, Baehring F, Voigt K, Lazar D, Nieva J, Bazhenova L, Ko AH, Korn WM, Schram E, Coward M, Yang X, Metzner T, Lamy R, Honnatti M, Yoshioka C, Kunken J, Petrova Y, Sok D, Nelson D, Kuhn P: Fluid biopsy in patients with metastatic prostate, pancreatic and breast cancers. Phys Biol 2012, 9(1):016003-3975/9/1/016003. Epub 2012 Feb 3.
Wickman G, Julian L, Olson MF. How apoptotic cells aid in the removal of their own cold dead bodies. Cell Death Differ. 2012;19(5):735–42.
Haile ST, Bosch JJ, Agu NI, Zeender AM, Somasundaram P, Srivastava MK, Britting S, Wolf JB, Ksander BR, Ostrand-Rosenberg S. Tumor cell programmed death ligand 1-mediated T cell suppression is overcome by coexpression of CD80. J Immunol. 2011;186(12):6822–9.
Cho-Vega JH, Rassidakis GZ, Amin HM, Tsioli P, Spurgers K, Remache YK, Vega F, Goy AH, Gilles F, Medeiros LJ. Suppressor of cytokine signaling 3 expression in anaplastic large cell lymphoma. Leukemia. 2004;18(11):1872–8.
Fay AP, Signoretti S, Callea M, Telomicron GH, McKay RR, Song J, Carvo I, Lampron ME, Kaymakcalan MD, Poli-de-Figueiredo CE, Bellmunt J, Hodi FS, Freeman GJ, Elfiky A, Choueiri TK. Programmed death ligand-1 expression in adrenocortical carcinoma: an exploratory biomarker study. J Immunother Cancer. 2015;3:3-015–0047-3. eCollection.
Sasaki K, Hamano K, Kinjo M, Hara S. Intratumoral heterogeneity in DNA ploidy of bladder carcinomas. Oncology. 1992;49(3):219–22.
Furuya T, Uchiyama T, Murakami T, Adachi A, Kawauchi S, Oga A, Hirano T, Sasaki K. Relationship between chromosomal instability and intratumoral regional DNA ploidy heterogeneity in primary gastric cancers. Clin Cancer Res. 2000;6(7):2815–20.
Schvimer M, Lash RH, Katzin WE. Intratumoral heterogeneity of DNA ploidy in breast carcinomas: a flow cytometric assessment of sampling techniques. Cytometry. 1995;22(4):292–6.
Wu P, Wu D, Li L, Chai Y, Huang J. PD-L1 and Survival in Solid Tumors: A Meta-Analysis. PLoS ONE. 2015;10(6), e0131403.
Kallergi G, Konstantinidis G, Markomanolaki H, Papadaki MA, Mavroudis D, Stournaras C, Georgoulias V, Agelaki S. Apoptotic circulating tumor cells in early and metastatic breast cancer patients. Mol Cancer Ther. 2013;12(9):1886–95.
Herbst RS, Soria JC, Kowanetz M, Fine GD, Hamid O, Gordon MS, Sosman JA, McDermott DF, Powderly JD, Gettinger SN, Kohrt HE, Horn L, Lawrence DP, Rost S, Leabman M, Xiao Y, Mokatrin A, Koeppen H, Hegde PS, Mellman I, Chen DS, Hodi FS. Predictive correlates of response to the anti-PD-L1 antibody MPDL3280A in cancer patients. Nature. 2014;515(7528):563–7.
Tumeh PC, Harview CL, Yearley JH, Shintaku IP, Taylor EJ, Robert L, Chmielowski B, Spasic M, Henry G, Ciobanu V, West AN, Carmona M, Kivork C, Seja E, Cherry G, Gutierrez AJ, Grogan TR, Mateus C, Tomasic G, Glaspy JA, Emerson RO, Robins H, Pierce RH, Elashoff DA, Robert C, Ribas A. PD-1 blockade induces responses by inhibiting adaptive immune resistance. Nature. 2014;515(7528):568–71.
Gerlinger M, Rowan AJ, Horswell S, Larkin J, Endesfelder D, Gronroos E, Martinez P, Matthews N, Stewart A, Tarpey P, Varela I, Phillimore B, Begum S, McDonald NQ, Butler A, Jones D, Raine K, Latimer C, Santos CR, Nohadani M, Eklund AC, Spencer-Dene B, Clark G, Pickering L, Stamp G, Gore M, Szallasi Z, Downward J, Futreal PA, Swanton C. Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. N Engl J Med. 2012;366(10):883–92.
Meacham CE, Morrison SJ. Tumour heterogeneity and cancer cell plasticity. Nature. 2013;501(7467):328–37.
Zhang J, Fujimoto J, Zhang J, Wedge DC, Song X, Zhang J, Seth S, Chow CW, Cao Y, Gumbs C, Gold KA, Kalhor N, Little L, Mahadeshwar H, Moran C, Protopopov A, Sun H, Tang J, Wu X, Ye Y, William WN, Lee JJ, Heymach JV, Hong WK, Swisher S, Wistuba II, Futreal PA. Intratumor heterogeneity in localized lung adenocarcinomas delineated by multiregion sequencing. Science. 2014;346(6206):256–9.
Bedard PL, Hansen AR, Ratain MJ, Siu LL. Tumour heterogeneity in the clinic. Nature. 2013;501(7467):355–64.
Crockford A, Jamal-Hanjani M, Hicks J, Swanton C. Implications of intratumour heterogeneity for treatment stratification. J Pathol. 2014;232(2):264–73.
Acknowledgements
We would like to thank Andrew Phillips and Bernard Schwartz for funding support, and Stephanie Greene, Angel Rodriguez, Jerry Lee, Mark Landers, and for their assistance with the CTC sequencing.
Funding
The UCSF philanthropic fund was the primary source of funding for this study.
Availability of data and materials
The data collected is not publically available, but could be made so upon request.
Authors’ contributions
AA, TWF, DL, RK, GP, JH, ME, RP, KL,RG, AJ, JL, LD, SB, YW, RD, and PP contributed to data collection. DL, RK, GP, KL, RG, AJ, and JL contributed to assay development, processing collected specimens, and all analyses related to CTC characterization, staining, and statistics. DL, RK, RG, AJ, JW, LD, YP, and RD contributed to PD1 assay development. GP and KL contributed to analysis of genetic studies. JH, ME, and RP were responsible for the database lock, patient sample collection, and data gathering. AA, TWF, and PP contributed to data analysis, data interpretation, and writing of the manuscript. YP, SB, and RD contributed to editing the report and oversight of author review of the report. AA, TF, RG, SB, YP, and RD contributed to the design of the figures. AA, TWF, YW, RD, PP, and the other authors were involved in data analysis and interpretation; the drafting, review, and approval of the report; and the decision to submit for publication. All read and approved the final manuscript.
Competing interests
DL, RK, RG, AJ, JL, LD, SB, YW and RB are employees of Epic Sciences.
Consent for publication
This manuscript does not contain any individual person data.
Ethics approval and consent to participate
This is a study approved by the UCSF Committee on Human Research. All patients who contributed samples signed a consent to participate in this study after going through a consenting packet with a physician, who expressed their right to refuse participation.
Author information
Authors and Affiliations
Corresponding author
Additional files
Additional file 1:
Sequencing read counts for 10 CTCs from two patients with metastatic bladder cancer undergoing NGS. (PPTX 48 kb)
Additional file 2:
(A) Kaplan-Meier curve of OS for patients with high (solid line) and low (dotted line) PD-L1+ CTC burden (high burden ≥ 1 PD-L1+ CTC/mL). (B) Kaplan-Meier curve of OS for patients with apoptotic CTCs (dotted line) and without apoptotic CTCs (solid line). (PPTX 71 kb)
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Anantharaman, A., Friedlander, T., Lu, D. et al. Programmed death-ligand 1 (PD-L1) characterization of circulating tumor cells (CTCs) in muscle invasive and metastatic bladder cancer patients. BMC Cancer 16, 744 (2016). https://doi.org/10.1186/s12885-016-2758-3
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12885-016-2758-3