Abstract
Background
Immunoglobulin A nephropathy (IgAN) is the most frequent primary glomerulopathy worldwide. The study aimed to provide potential molecular biomarkers for IgAN management.
Methods
The public gene expression profiling GSE58539 was utilized, which contained 17 monocytes samples (8 monocytes samples isolated from IgAN patients and 9 monocytes samples isolated from healthy blood donors). Firstly, differentially expressed genes (DEGs) between the two kinds of samples were identified by limma package. Afterwards, pathway enrichment analysis was implemented. Thereafter, protein-protein interaction (PPI) network was constructed and key nodes in PPI network were predicted using four network centrality analyses. Ultimately, gene functional interaction (FI) was constructed according to expressions in each sample, and then module network was extracted from FI network.
Results
A total of 678 DEGs were screened out, of these, 72 DEGs were identified as crucial nodes in PPI network that could well distinguish IgAN and healthy samples. In particular, IL6, TNF, IL1B, PRKACA and CCL20 were closely related to pathways such as hematopoietic cell lineage, apoptosis and Toll-like receptor (TLR) signaling pathway. Moreover, 12 genes in the FI network belonged to the 72 identified key nodes, such as CCL20, HDAC10, FPR2 and PRKACA, which were also key genes in 4 module networks.
Conclusions
Several crucial genes were identified in monocytes of IgAN patients, such as IL6, TNF, IL1B, CCL20, PRKACA, FPR2 and HDAC10. These genes might co-involve in pathways such as TLR and apoptosis signaling during IgAN progression.
Similar content being viewed by others
Background
Worldwide, immunoglobulin A nephropathy (IgAN) is the most frequent primary glomerulopathy. Reportedly, 20–50% of adults who suffered with IgAN would progress to end-stage renal diseases [1]. Therefore, it is pivotal for IgAN patients to identify predictors of prognosis. Numerous risk factors associated with IgAN progression have been reported. A study in Chinese population identifies three risk factors, including renal impairment, hypertension as well as advanced histological involvement [2]. Besides, another study reveals that expressions of renal leukocyte infiltrations and cytokines, such as leukocyte common antigen (LCA), CD3, CD68 and interleukin-1Beta (IL1B), are highly correlated with IgAN [3]. Currently, biochemical and genetic data indicate that aberrantly glycosylated IgA1 play significant roles in pathogenesis of IgAN [4,5,6]. Moreover, alteration on the glycan structure of IgA1 causes the deposition of nephritogenic immune complexes, which induce resident mesangial cells proliferation and extracellular matrix proteins expression, and subsequently lead to the loss of glomerular function [7]. Based on the pathogenesis, several biomarkers have been identified, such as levels of urinary secretory (sIgA) [8], serum galactose-deficient immunoglobulin A1 (Gd-IgA1) [9] and the tandem repeats polymorphism of MUC20 gene [10]. However, cellular events involved in the IgAN pathogenesis are unclear.
Recently, it is found that abnormality of IgAN disease is related to IgA immune system and peripheral blood leucocytes, especially the peripheral blood mononuclear cells [11, 12]. Monocytes, a kind of the phagocytes that formed in bone marrow, can differentiate into macrophages and dendritic cells (DCs) in peripheral tissues. Monocytes have a crucial part in immune response and may contribute to the pathogenesis of IgAN [13]. Thus, a guideline for target therapy of IgAN will be obtained through identifying gene alterations in monocytes of IgAN patients. Moreover, Cox et al. uncover that the altered genes in IgAN monocytes are mainly associated with apoptotic pathway and mitochondrial dysfunction [13]. In particular, the expression of NADH: ubiquinone oxidoreductase core subunit S3 (NDUFS3) and TNF receptor superfamily member 1A (TNFRSF1A) proteins are upregulated, thus verifying the altered mitochondrial respiratory system and death receptor homeostasis. Additionally, the TNF expression in monocytes of IgAN patients are reduced compared with those in healthy blood donors (HBDs) [13]. However, other critical genes and their interaction have not been investigated.
In the present study, we re-analyzed GSE58539 profiling using a more comprehensive bioinformatics. After identifying the differentially expressed genes (DEGs) in monocytes between IgAN patients and HBDs, functional enrichment and protein-protein interactions (PPIs) network analyses were carried out, followed by key nodes prediction of the network through four network centrality analyses. Notably, in order to reveal potential interactions of DEGs that involved in similar functions and pathways, gene functional interaction (FI) network and the module network analyses were performed based on gene expressions of each sample. The study aimed to further uncover the pathogenesis and progression of IgAN, and thus provide potential molecular biomarkers for the diagnosis and targeting therapy of IgAN.
Methods
Data resource
The microarray data GSE58539 [13] was downloaded from Gene Expression Omnibus (GEO, http://www.ncbi.nlm.nih.gov/geo) database. This dataset contained 17 monocytes samples, including 8 monocytes samples isolated from IgAN patients (IgAN group) and 9 monocytes samples isolated from HBDs (healthy group). The platform of the dataset was Illumina HumanHT-12 V4.0 expression beadchip (Illumina, San Diego, California, USA).
Data preprocessing
We used the robust multi-array average (RMA) method in Linear Models for Microarray Analysis (limma, http://www.bioconductor.org/packages/release/bioc/html/limma.html) package of R [14] to preprocess the non-normalized raw data by performing background correction, quantile normalization and microarray data condensation. Afterwards, the probe identification numbers (IDs) were transformed into gene symbols utilizing illuminaHumanv4.db [15] and annotate [16] software in R package and the probes were eliminated which did not correspond to gene symbols. Finally, the average value of different probes would serve as the final expression of the gene if different probes were mapped to the same gene.
DEGs identification
Non-paired t-test method in limma package was utilized to calculate significance p-value of the DEGs between IgAN and healthy samples. The thresholds for DEG selection were p-value < 0.05 and log2|fold change| ≥ 0.58. Subsequently, coupled two-way clustering analysis (CTWC) was conducted using gplots tools [17] in R package.
Enrichment analysis of the DEGs
The Database for Annotation, Visualization and Integration Discovery (DAVID, http://david.abcc.Ncifcrf.gov/) [18] tool was used to conduct Gene ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG, http://www.genome.jp/kegg/pathway.html) [19] pathway enrichment analyses for DEGs. The number of enrichment genes (count number) ≥ 2 and p-value < 0.05 were chosen as cut-off criteria.
Construction of the PPI network
The Search Tool for the Retrieval of Interacting Genes (STRING, http://string-db.org/) [20] database was used to predict potential interactions among proteins encoded by the DEGs. Relevant parameters were as follows: species was “Homo”, the input genes were DEGs and the PPI score (referred to medium confidence) was set as 0.4. A protein in the PPI network serves as a node. The network was visualized by the Cytoscape (http://cytoscape.org/) software [21].
Prediction of key nodes in the PPI network
The topological property of PPI network makes it possible to investigate key genes in the network. Here, four network centrality analyses were performed to explore the key genes, including degree centrality, betweenness centrality, subgraph centrality and closeness centrality of key genes. Generally, degree was used for describing the importance of protein nodes in the network, and betweenness centrality is a kind of indicator that describes the global topological properties of the network. Besides, subgraph centrality was used to measure the importance of nodes in the network based on the combination of network topology and protein complex information. Closely centricity represented the closely connection degree of a certain node and all other nodes [22,23,24,25].
A cytoscape plug-in, CytoNCA [26], was used to perform the above analyses. Nodes with high values in the above four network centrality analyses were screened out to predict key genes, and the genes influence on sample clustering were observed using gplots packages. Detailed steps for the selection of key genes were: (1) the top ten genes with high values calculated by each network centrality analysis were selected and then were integrated; (2) if these integrated genes could not well distinguish the IgAN and healthy samples, more nodes were gradationally selected based on their ranked values to conduct the clustering analysis till they could distinguish completely the two kinds of samples. These key genes were then defined as feature genes of IgAN and healthy samples.
FI network analysis
Based on gene expression value of each sample, the gene FI network was established using Cytoscape app-ReactomeFI [27]. The input dataset was the expression matrix of all DEGs. The FI network was analyzed utilizing ReactomeFI and the gene functional interaction in the PATHWAY of the Reactome database, thereafter modules from the FI network were obtained through Monte Carlo Localization clustering algorithm [28]. In addition, co-expression relationships of genes in each module were determined according to their expression value. The selection parameters in ReactomeFI network were module size ≥7 and average correlation ≥0.25. Subsequently, pathway enrichment analysis was carried out for each functional module to identify potential biological pathways associated with genes in each module, and the threshold for significant pathway selection was false discovery rate (FDR) < 0.05.
Results
DEGs identification
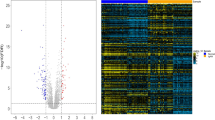
Here, we obtained a total of 453 up-regulated and 225 down-regulated DEGs. As indicated in the clustering heat map (Fig. 1), these DEGs could well distinguish the IgAN and healthy samples completely.
Pathway enrichment analysis of the DEGs
Unfortunately, the up-regulated DEGs were not enriched in any pathways. However, the down-regulated DEGs were significantly enriched in nine pathways. The enriched pathways were listed in Table 1, including hematopoietic cell lineage (pathway, p-value = 7.92 × 10− 4; which involved interleukin-6 (IL6), tumor necrosis factor (TNF) and interleukin 1 beta (IL1B)), NOD-like receptor signaling pathway (pathway, p-value = 8.22 × 10− 3; which involved IL6, TNF and IL1B), cytokine-cytokine receptor interaction (pathway, p-value = 1.91 × 10− 2; which involved IL6, TNF, IL1B and C-C motif chemokine ligand 20 (CCL20)), intestinal immune network for IgA production (pathway, p-value = 2.52 × 10− 2; which involved IL6) and apoptosis (pathway, p-value = 2.58 × 10− 2; which involved TNF and IL1B).
PPI network of the DEGs
As presented in Fig. 2, the PPI network with 379 nodes and 692 interactions was constructed. The hub nodes (whose degree > 10) mainly included TNF (degree = 31), PRKACA (degree = 26), IL6 (degree = 23), YWHAZ (degree = 19), MYB (degree = 15), TYK2 (degree = 14), FPR2 (degree = 13), IL1B (degree = 13), CCL20 (degree = 12), GNA11 (degree = 11).
Key nodes in the PPI network
Combined with the integrating results by four network centrality analyses, nodes with higher degree were used to cluster the two different kinds of samples. As a result, a total of 72 genes were identified that could well distinguish IgAN and healthy samples (Fig. 3). Among them, genes such as IL6, TNF, IL1B, PRKACA, TYK2 and CCL20 were closely related to five the pathways, including NOD-like receptor signaling pathway, cytokine-cytokine receptor interaction, hematopoietic cell lineage, apoptosis, and Toll-like receptor signaling pathway (Table 2).
FI network analysis
The FI network of the DEGs was constructed utilizing ReactomeFI, which included 42 genes and 71 interaction edges (Fig. 4). Moreover, five modules (module a-e) were extracted from the FI network, and the absolute average correlation of genes in module a-e was 0.58, 0.4222, 0.5069, 0.4709 and 0.4275, respectively. Genes such as PRKACA in module a was enriched in integration of energy metabolism, morphine addiction and glutamatergic synapse pathways; in module b, genes such as CCL20, FPR2 and GNA11 were related to GPCR ligand binding, GPCR downstream signaling and Gastrin-CREB signaling pathway via PKC and MAPK pathways; in module c, the gene NFYB was highly associated with RNA binding-related pathways; while in module d, the gene HDAC10 was significantly enriched in two pathways, alcoholism and chromatin modifying enzymes (Table 3) . Genes in module e were not enriched in any pathways.
Notably, we found that 12 genes in the FI network also belonged to the hub genes, such as CCL20, FPR2, and PRKACA.
Discussion
In the present study, a total of 72 crucial nodes in the PPI network were identified via re-analyzing the dataset GSE58539, which could well distinguish the IgAN and healthy samples. Among which, genes such as IL6, TNF, IL1B, PRKACA, and CCL20 were closely related to the following pathways: NOD-like receptor signaling pathway, cytokine-cytokine receptor interaction, hematopoietic cell lineage, apoptosis and Toll-like receptor signaling pathway. Moreover, 12 genes in the FI network belonged to the 72 identified key nodes, such as CCL20, HDAC10, FPR2 and PRKACA. Besides, the 12 genes were also the key genes in 4 module networks correlating with pathways of integration of energy metabolism (module a), GPCR-related pathways (module b), RNA binding-related pathways (module c), alcoholism and chromatin modifying enzymes (module d).
The cytokine encoded by IL6 has great roles in inflammation and regulation of immune response [29]. Toll-like receptors (TLRs) are major factors that initiate the immune reaction. Most TLRs promote immune response (including innate and adaptive) via inducing expression of proinflammatory cytokines [30]. Increased TLRs, such as TLR-4, has been detected in circulating monocytes of patients with IgAN [31]. Expression of IL6 protein is also increased in mouse proximal tubular epithelial cells, accompanying by the upregulation of TLR4 mRNA [32]. IL1B, encoded by IL1B gene, is a member of interleukin 1 cytokine family and crucial for the regulation of inflammatory response [33]. In response to the external infections, gene expressions of the proinflammatory cytokines (e. g. IL1A, IL1B and IL6) are always upregulated simultaneously [34, 35]. In particular, IL1B is implicated in the TLR-4 induced immune response in chronic pain [36]. In our study, IL6 and IL1B were both downregulated and enriched in TLR signaling pathway. These results suggested that IL6 and IL1B might be co-regulated in TLR signaling pathway and contribute to the abnormality of the immune response in monocytes of IgAN patients.
TNF is a multifunctional proinflammatory cytokine. Reportedly, TNF expression is dramatically increased in Mycoplasma penetrans-infected IgAN mice model, and the protein is proposed to involve in the induction of renal damage in IgAN [37]. Moreover, levels of serum TNF receptors are also elevated in IgAN patients compared with healthy control [38]. However, Cox et al. uncover that TNF expression is obviously reduced in monocytes of IgAN patients, compared with those of HBDs [13]. The finding indicates the downregulated TNF may lead to the monocytes apoptosis. Moreover, the inhibition of TNF-α is proposed as a causative factor of IgAN [39]. Therefore, it might be speculated that the apoptosis of monocytes induced by downregulation of TNF contribute to IgAN progression.
CCL20 is a small cytokine that also involves in immune regulation and inflammation [40]. Combination of CCL20 with CCR6 (the CCL20 receptor) cause the recruitment of leukocyte subsets, which finally promote immune-mediated kidney damage [41]. Additionally, CCL20 is one of the chemokines that take part in the host response to pathogens invasions by activating inflammatory cells, and it has the similar effects on monocytes [42]. Therefore, the downregulated CCL20 in monocytes of IgAN patients might cause alteration in immune response, and thereby influence the IgAN development.
Three novel genes, protein kinase, CAMP-dependent, catalytic, alpha (PRKACA), formyl peptide receptor 2 (FPR2) and histone deacetylase 10 (HDAC10) were firstly predicted in monocytes of IgAN. PRKACA protein encoded by PRKACA gene is one subunit of protein kinase A that participates in apoptosis. At present, PRKACA amplification is served as a method for identifying genetic defect correlated with Cushing’s syndrome [43]. Somatic mutations of PRKACA have been detected in adenomas of the adrenal cortex [44]. Moreover, PRKACA mediates apoptosis-related signaling pathways in many cancer diseases, such as breast cancer and follicular thyroid cancer cells [45, 46]. In the present study, PRKACA was up-regulated and significantly enriched in apoptosis pathway, suggesting it might exert its function in monocytes via regulating apoptosis during IgAN progression. FPR2 is known to activate the G-protein coupled receptor and N-formyl peptide receptor. FPR2 is found in adipose tissues as a receptor for the pro-resolving mediators [47], which contribute to the restoration of in adipose inflammation and treatment of obesity-related glomerulopathy [48]. HDAC10, containing two catalytic sites, is highly expressed in numerous human tissues such as kidney [49]. In lung cancer, decreased HDAC10 is associated with the advanced stage and adverse outcome [50]. However, there are rare reports on the relationship of IgAN and HDAC10. In the current study, FPR2 and HDAC10 were hub up-regulated genes in both PPI network and FI network, implying they might co-function in monocytes of IgAN patients. One limitation of this study is the lack of expression validation. However, we will do more experiments to verify our conclusions once we collect the samples in the future.
Conclusions
In conclusion, several crucial genes were identified in monocytes of IgAN patients, such as IL6, TNF, IL1B, CCL20, PRKACA, FPR2 and HDAC10. They might have co-functions and their dysregulations might alter activities of pathways such as TLR and apoptosis signaling, which might finally promote IgAN progression. The study is of great value for the prediction of key regulators in monocytes of IgAN and the identification of targeting therapeutic management for IgAN.
Abbreviations
- DCs:
-
Dendritic cells
- DEGs:
-
Differentially expressed genes
- FDR:
-
False discovery rate
- FI:
-
Functional interaction
- FPR2 :
-
Formyl peptide receptor 2
- Gd-IgA1:
-
Galactose-deficient immunoglobulin A1
- GEO:
-
Gene Expression Omnibus
- HBDs:
-
Healthy blood donors
- HDAC10 :
-
Histone deacetylase 10
- IDs:
-
Identification numbers
- IgAN:
-
Immunoglobulin A nephropathy
- IL1B:
-
interleukin-1Beta
- IL6 :
-
Interleukin-6
- KEGG:
-
Kyoto encyclopedia of genes and genomes
- LCA:
-
Leukocyte common antigen
- PPI:
-
Protein-protein interaction
- RMA:
-
Robust multi-array average
- STRING:
-
Search tool for the retrieval of interacting genes
- TLRs:
-
Toll-like receptors
- TNF:
-
Protein Kinase, CAMP-Dependent, Catalytic, Alpha
- TNF:
-
Tumor necrosis factor
References
Nakanishi K, Yoshikawa N. Immunoglobulin a nephropathy. Springer Berlin Heidelberg. 2009;7(4):275–398.
Lv J, Zhang H, Zhou Y, Li G, Zou W, Wang H. Natural history of immunoglobulin a nephropathy and predictive factors of prognosis: a long-term follow up of 204 cases in China. Nephrology. 2008;13(3):242–6.
Myllymäki J, Honkanen T, Syrjänen J, Helin H, Rantala I, Pasternack A, Mustonen J. Severity of tubulointerstitial inflammation and prognosis in immunoglobulin a nephropathy. Kidney Int. 2007;71(4):343–8.
Kiryluk K, Novak J, Gharavi AG. Pathogenesis of immunoglobulin a nephropathy: recent insight from genetic studies. Annu Rev Med. 2013;64:339.
Novak J, Julian BA, Tomana M, Mestecky J. IgA glycosylation and IgA immune complexes in the pathogenesis of IgA nephropathy. Seminars in nephrology. 2008;28(1):78–87.
Wyatt RJ, Julian BA. IgA nephropathy. N Engl J Med. 2013;368(25):2402–14.
Mestecky J, Raska M, Julian BA, Gharavi AG, Renfrow MB, Moldoveanu Z, Novak L, Matousovic K, Novak J. IgA nephropathy: molecular mechanisms of the disease. Annu Rev Pathol. 2013;8:217–40.
Tan Y, Zhang JJ, Liu G, Zhang H, Zhao MH. The level of urinary secretory immunoglobulin A (sIgA) of patients with IgA nephropathy is elevated and associated with pathological phenotypes. Clin Exp Immunol. 2009;156(1):111–6.
Kiryluk K, Moldoveanu Z, Sanders JT, Eison TM, Suzuki H, Julian BA, Novak J, Gharavi AG, Wyatt RJ. Aberrant glycosylation of IgA1 is inherited in both pediatric IgA nephropathy and Henoch–Schönlein purpura nephritis. Kidney Int. 2011;80(1):79–87.
Li G, Zhang H, Lv J, Hou P, Wang H. Tandem repeats polymorphism of MUC20 is an independent factor for the progression of immunoglobulin a nephropathy. Am J Nephrol. 2006;26(1):43–9.
Canaud G, Audard V, Kofman T, Lang P, Legendre C, Grimbert P. Recurrence from primary and secondary glomerulopathy after renal transplant. Transpl Int. 2012;25(8):812–24.
Coppo R, Camilla R, Alfarano A, Balegno S, Mancuso D, Peruzzi L, Amore A, Dal Canton A, Sepe V, Tovo P. Upregulation of the immunoproteasome in peripheral blood mononuclear cells of patients with IgA nephropathy. Kidney Int. 2009;75(5):536–41.
Cox SN, Serino G, Sallustio F, Blasi A, Rossini M, Pesce F, Schena FP. Altered monocyte expression and expansion of non-classical monocyte subset in IgA nephropathy patients. Nephrol Dial Transplant. 2015;30(7):1122–232.
Smyth GK. Limma: linear models for microarray data. Bioinformatics and computational biology solutions using R and Bioconductor. 2005. p. 397–420.
Dunning M, Lynch A, Eldridge M: illuminaHumanv4. db: Illumina HumanHT12v4 annotation data (chip illuminaHumanv4). R package version. 2013, 2(0).
Gentleman R: annotate: Annotation for microarrays. R package version. 2003, 1(0):19.
Warnes GR, Bolker B, Bonebakker L, Gentleman R, Huber W, Liaw A, Lumley T, Maechler M, Magnusson A, Moeller S: gplots: Various R programming tools for plotting data. R package version. 2009, 2(4).
Huang DW, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4(1):44.
Kanehisa M, Goto S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28(1):27–30.
Szklarczyk D, Franceschini A, Kuhn M, Simonovic M, Roth A, Minguez P, Doerks T, Stark M, Muller J, Bork P. The STRING database in 2011: functional interaction networks of proteins, globally integrated and scored. Nucleic Acids Research. 2011;39(suppl_1):561–8.
Smoot ME, Ono K, Ruscheinski J, Wang P-L, Ideker T. Cytoscape 2.8: new features for data integration and network visualization. Bioinformatics. 2011;27(3):431–2.
Latora V, Marchiori M. A measure of centrality based on network efficiency. New J Phys. 2007;9(6):188.
Estrada E. Virtual identification of essential proteins within the protein interaction network of yeast. Proteomics. 2006;6(1):35–40.
Estrada E, Rodriguez-Velazquez JA. Subgraph centrality in complex networks. Phys Rev E. 2005;71(5):056103.
Amitai G, Shemesh A, Sitbon E, Shklar M, Netanely D, Venger I, Pietrokovski S. Network analysis of protein structures identifies functional residues. J Mol Biol. 2004;344(4):1135–46.
Tang Y, Li M, Wang J, Pan Y, Wu F-X. CytoNCA: a cytoscape plugin for centrality analysis and evaluation of protein interaction networks. Biosystems. 2015;127:67–72.
Wu G, Dawson E, Duong A, Haw R, Stein L. ReactomeFIViz: a Cytoscape app for pathway and network-based data analysis. F1000Research. 2014;3:146.
Röfer T, Jüngel M. Vision-based fast and reactive Monte-Carlo localization. In: IEEE international conference on robotics and automation: 2003: IEEE; 1999; 2003. p. 856–61.
Rossignol J, Boyer C, Thinard R, Remy S, Dugast AS, Dubayle D, Dey ND, Boeffard F, Delecrin J, Heymann D. Mesenchymal stem cells induce a weak immune response in the rat striatum after allo or xenotransplantation. J Cell Mol Med. 2009;13(8b):2547–58.
Akira S, Uematsu S, Takeuchi O. Pathogen recognition and innate immunity. Cell. 2006;124(4):783–801.
Coppo R, Camilla R, Amore A, Peruzzi L, Daprà V, Loiacono E, Vatrano S, Rollino C, Sepe V, Rampino T. Toll-like receptor 4 expression is increased in circulating mononuclear cells of patients with immunoglobulin a nephropathy. Clin Exp Immunol. 2010;159(1):73–81.
Zhu X-L, Wang Y-J, Yang Y-Ζ, Yang R-C, Zhu B, Zhang YΗ, Lin Y, Lu Y, Li X-F, O'Byrne KT. Suppression of lipopolysaccharide-induced upregulation of toll-like receptor 4 by emodin in mouse proximal tubular epithelial cells. Mol Med Rep. 2012;6(3):493–500.
Dai D, Wang L, Xu L, Tang L, Xu X, Ye H, Zhou X, Chen C, Pan G, Ru P. A comprehensive meta-analysis of the association between three 1B polymorphisms and rheumatoid arthritis. Adv Biosci Biotechnol. 2014;5(2):108–16.
Günther J, Esch K, Poschadel N, Petzl W, Zerbe H, Mitterhuemer S, Blum H, Seyfert H-M. Comparative kinetics of Escherichia coli-and Staphylococcus aureus-specific activation of key immune pathways in mammary epithelial cells demonstrates that S. aureus elicits a delayed response dominated by interleukin-6 (IL-6) but not by IL-1A or tumor necrosis factor alpha. Infect Immun. 2011;79(2):695–707.
Chen C-J, Ou Y-C, Lin S-Y, Raung S-L, Liao S-L, Lai C-Y, Chen S-Y, Chen J-H. Glial activation involvement in neuronal death by Japanese encephalitis virus infection. J Gen Virol. 2010;91(4):1028–37.
Guo L-H, Schluesener H. The innate immunity of the central nervous system in chronic pain: the role of toll-like receptors. Cell Mol Life Sci. 2007;64(9):1128–36.
Jiang X, Lv Y-Q, Zhang J-N, Shi Y-L, Xu F-F. Mycoplasma penetrans infection is a potential cause of immunoglobulin a nephropathy: a new animal model. J Nephrol. 2013;26:470–5.
Sonoda Y, Gohda T, Suzuki Y, Omote K, Ishizaka M, Matsuoka J, Tomino Y. Circulating TNF receptors 1 and 2 are associated with the severity of renal interstitial fibrosis in IgA nephropathy. PLoS One. 2015;10(4):e0122212.
Wei S, Sinniah R. Adalimumab (TNFα inhibitor) therapy exacerbates IgA glomerulonephritis acute renal injury and induces lupus autoantibodies in a psoriasis patient. Case Reports Nephrol. 2013;2013(4):1–4.
Starner TD, Barker CK, Jia HP, Kang Y, Jr MCP. CCL20 is an inducible product of human airway epithelia with innate immune properties. Am J Respir Cell Mol Biol. 2003;29(5):627.
Meng T, Li X, Ao X, Zhong Y, Tang R, Peng W, Yang J, Zou M, Zhou Q. Hemolytic Streptococcus may exacerbate kidney damage in IgA nephropathy through CCL20 response to the effect of Th17 cells. PLoS One. 2014;9(9):e108723.
Buonaguro L, Monaco A, Aricò E, Wang E, Tornesello ML, Lewis GK, Marincola FM, Buonaguro FM. Gene expression profile of peripheral blood mononuclear cells in response to HIV-VLPs stimulation. BMC Bioinformatics. 2008;9(2):1.
Lodish MB, Yuan B, Levy I, Braunstein GD, Lyssikatos C, Salpea P, Szarek E, Karageorgiadis AS, Belyavskaya E, Raygada M. Germline PRKACA amplification causes variable phenotypes that may depend on the extent of the genomic defect: molecular mechanisms and clinical presentations. Eur J Endocrinol. 2015;172(6):803–11.
Beuschlein F, Fassnacht M, Assié G, Calebiro D, Stratakis CA, Osswald A, Ronchi CL, Wieland T, Sbiera S, Faucz FR. Constitutive activation of PKA catalytic subunit in adrenal Cushing's syndrome. N Engl J Med. 2014;370(11):1019–28.
Moody SE, Schinzel AC, Singh S, Izzo F, Strickland MR, Luo L, Thomas SR, Boehm JS, Kim SY, Wang ZC. PRKACA mediates resistance to HER2-targeted therapy in breast cancer cells and restores anti-apoptotic signaling. Oncogene. 2015;34(16):2061–71.
Grosse J, Warnke E, Wehland M, Pietsch J, Pohl F, Wise P, Magnusson NE, Eilles C, Grimm D. Mechanisms of apoptosis in irradiated and sunitinib-treated follicular thyroid cancer cells. Apoptosis. 2014;19(3):480–90.
Clària J, Dalli J, Yacoubian S, Gao F, Serhan CN. Resolvin D1 and resolvin D2 govern local inflammatory tone in obese fat. J Immunol. 2012;189(5):2597–605.
Nolan E, O'Meara YM, Godson C. Lipid mediators of inflammation in obesity-related glomerulopathy. Nephrol Dial Transplant. 2013;28(suppl 4):iv22–9.
Tong JJ, Liu J, Bertos NR, Yang X-J. Identification of HDAC10, a novel class II human histone deacetylase containing a leucine-rich domain. Nucleic Acids Res. 2002;30(5):1114–23.
Yao H, Rahman I. Current concepts on the role of inflammation in COPD and lung cancer. Curr Opin Pharmacol. 2009;9(4):375–83.
Author information
Authors and Affiliations
Contributions
YG and WG designed the research and drafted the manuscript. ZL draft the manuscript and revised manuscript for important intellectual content. DW and WL acquired data, analyze data and Statistical analysis. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Guo, Y., Gao, W., Wang, D. et al. Gene alterations in monocytes are pathogenic factors for immunoglobulin a nephropathy by bioinformatics analysis of microarray data. BMC Nephrol 19, 184 (2018). https://doi.org/10.1186/s12882-018-0944-z
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12882-018-0944-z