Abstract
Background
Wilson’s disease (WD) is a rare autosomal recessive metabolic disease caused by ATP7B gene mutations tat cause excessively high copper levels, particularly in the liver and brain. The WD phenotype varies in terms of its clinical presentation and intensity. Diagnosing this metabolic disorder is important as a lifelong treatment, based on the use of copper chelating agents or zinc salts, is more effective if it’s started early. Worldwide prevalence of WD is variable, with an average of 1/30,000. In France, a recent study based on French health insurance data estimated the clinical prevalence of the disease to be around 3/200,000.
Methods
To estimate the genetic prevalence of WD in France, we analysed the ATP7B gene by Next Generation Sequencing from a large French cohort of indiscriminate subjects.
Results
We observed a high heterozygous carrier frequency of ATP7B in France. Among the 697 subjects studied, 18 variants classified as pathogenic or probably pathogenic were found at heterozygous level in 22 subjects (22 alleles/1394 alleles), yielding a prevalence of 0.032 or 1/31 subjects.
Conclusions
This considerable and unexplained discrepancy between the heterozygous carrier frequency and the clinical prevalence of WD may be explained by the clinical variability, the incomplete penetrance and the existence of modifiers genes. It suggests that the molecular analysis of ATP7B should be interpreted with caution, always alongside copper assays (ceruloplasmin, relative exchangeable copper, 24 h-urinary copper excretion) with particular respect to exome sequencing.
Similar content being viewed by others
Background
Wilson’s disease (WD) is a rare autosomal recessive disorder caused by mutations to the ATP7B gene (13q14.3). ATP7B encodes a member of the P-type cation transport ATPase family that has functions in exporting copper out of cells, such as the efflux of hepatic copper into bile. WD is an inherited metabolic disorder which results in excess copper levels, particularly in the liver and brain. The initial symptoms may vary; in more than half of patients they may include hepatic abnormalities such as chronic active hepatitis, cirrhosis and more rarely fulminant hepatic failure, whereas others might display different neuropsychiatric manifestations such as tremor, Parkinsonism, dystonia or psychiatric abnormalities [1, 2]. The clinical signs of WD can appear at different ages (usually between 8 and 30 years) but they may also occur at extreme ages (1 to 80 years) [3, 4]. The outcome of WD is generally fatal without treatment based on copper chelating agents (such as trientine and D-penicillamine), zinc salts or a liver graft in the most severe hepatic cases.
Early studies had indicated the prevalence of WD as being around 30 per one million [5], and based on the Hardy-Weinberg equilibrium, a heterozygous carrier frequency of about 1/90. Subsequent epidemiological studies have suggested a lower clinical prevalence of WD in European countries with a low degree of consanguinity, estimated at between 12 and 20 per one million [6]. Nevertheless, genetic prevalence appears to be higher than clinical prevalence, as shown by two recent British and Korean studies which produced heterozygous carrier frequencies of 1/25 and 1/53, respectively [7, 8].
In France, WD patients are followed by the National Reference Centre for Wilson’s disease. According to the French National Health Insurance Information System database, the clinical prevalence of WD is around 3/200,000 [9]. In order to determine the heterozygous carrier frequency of ATP7B in France, we studied a large cohort in non-WD individuals by sequencing the entire coding regions of ATP7B associated with its 5’UTR regions and promoter.
Methods
NGS sequencing and genetic analysis
Our retrospective study included anonymised data on 697 individuals between January 2015 and May 2017. These patients who had no family history of WD were French residents treated in 18 French university hospitals and had various indications other than WD, hepatic or neurological diseases. After automated DNA extraction (Qiagen, Courtaboeuf, France), the DNA samples were screened by NGS using a panel of various genes which included ATP7B. Library preparations for NGS were established using the surelectQXT kit (Agilent, Les Ulis, France) based on a hybrid capture system for sequencing on the Miseq sequencer (Illumina, Paris, France). The bioinformatics pipeline included the miseq-reporter followed by Fastq alignments with SeqNext and visual variant control (JSI Medical Systems, Ettenheim, Germany). The variants were included if the depth of lecture was greater than 30X with a variant frequency ranging from 0.40 to 0.60. The design included the promoter, the 5’UTR region, the 3’UTR region and the coding region of the ATP7B gene (10,000 pb) with Refseq for the nomenclature: ATP7B (NM_000053.3) for the exonic regions and intronic-exonic boundaries; ATP7B (NG_008806.1) for exon numbering and the intronic region. Studies of missense variant pathogenicity involved the use of predictive software programs such as Polyphen-2 (http://genetics.bwh.harvard.edu/pph2/), PhyloP (http://compgen.cshl.edu/phast/), and CADD (http://cadd.gs.washington.edu) and MutationTaster (http://www.mutationtaster.org), while splicing variant pathogenicity was determined used predictive software such as MaxEntScan (http://genes.mit.edu/burgelab/maxent), Human Splicing Finder (http://www.umd.be/HSF3/) and NNSPLICE (http://www.fruitfly.org/seq_tools/splice.html).
The frequencies of all variants were searched for in Exome Aggregation Consortium (ExAc, exac.broadinstitute.org), Exome Sequencing Project (ESP, evs.gs.washington.edu) and dbSNP (https://www.ncbi.nlm.nih.gov/projects/SNP). We retained variants with an allelic frequency lower than 0.06% and variants already described in the literature and reported in the Professional Human Gene Mutation Database (HGMD, http://www.hgmd.cf.ac.uk/ac/index.php), the Wilson Mutation Database, University of Alberta, Canada (http://www.wilsondisease.med.ualberta.ca) and in our Lariboisière Wilson’s disease Database (Paris), which includes 496 index cases.
All the variants identified were classified according to ACMG guidelines including data as Minor Allele Frequency (MAF) in the general population, results of the predictive software programs and descriptions in the literature. Variants were considered to be:
-
pathogenic when they had been described in the literature without any ambiguities as to their interpretation;
-
likely pathogenic when they had not been described in the literature or professional HGMD, but were classified as deleterious based on all subsequent in silico predictive algorithms and having MAF < 0.06%.
-
Variant with Uncertain Significance (VUS), when is conflicting: the criteria for benign and pathogenic are contradictory by using ACMG guidelines.
Results
In order to determine the genetic prevalence of Wilson’s disease in French population, the promoter, the 5’UTR region, the 3’UTR region, all coding region and exon-intron junctions of ATP7B gene were sequenced in 697 DNA samples. We identified 4222 variants in coding ATP7B across the 697 subjects. Of these, 64 variants have a MAF < 1% and were seen in 94 subjects. The average number of ATP7B variants with MAF below 1% identified per subject was 0.142. No Copy Number Variation was detected by NGS.
Among the 697 subjects studied, 23 heterozygous variants were classified as pathogenic, likely pathogenic and VUS (Table 1). These 23 heterozygous variants were found in 37 individuals, leading to a ATP7B variant carrier frequency of 0.0265 (37/1394 alleles) or a heterozygous carrier frequency of 1/19 subjects.
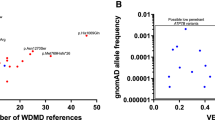
In a second step and to be more stringent, we excluded five variants reported in the literature on WD and that we have classified as VUS according to ACMG guidelines [10,11,12]. The c.4301C > T (p.Thr1434Met) variant was predicted to be “probably damaging” according to Polyphen-2, but was found in nine subjects which produced a variant frequency of 0.00645 (9/1394 alleles) in our study, and moreover was never found in our large WD database. Five other subjects harboured four VUS (p.Val536Ala, p.Glu541Lys, p.Met665Ile, p.Val1297Ile) that were predicted to be benign by predictive softwares and were not present in our WD database. Moreover, the p.Val536Ala variant (c.1607C > T) (with a MAF = T at 0.45% in the European population) reached a high frequency (1.29%) in the Finnish population according to Exac. In addition p.Val1297Ile (c.3889G > A) displayed a high MAF = A at 1.5% in the Asian population [10, 13] (Table 1). Consequently, ATP7B heterozygous carrier frequency was recalculated to include only heterozygous variants classified as pathogenic [14,15,16,17,18,19] and likely pathogenic. Eighteen heterozygous mutations were thus found in 22 subjects, yielding a variant frequency of 0.01578 (22/1394 alleles) or a heterozygous carrier frequency of 1/31 subjects (Table 1). All these mutations were deleterious according to the predictive softwares. At a more detailed level, ten missense or splicing mutations were identified in 14 subjects, all of whom were present in HGMD and in our WD database. Among the eight other subjects, one novel deletion, p.(Lys1028Serfs*40) causing a frameshift mutation in exon 14, one splicing variant in intron 7, and six probably pathogenic missense variants were identified. Of the six missense variants, three are novel (p.Asp829His p.Lys1238Glu, p.Ser1272Cys) and three have a MAF < 0.011% (p.Val731Leu, p.Arg1224Leu, p.Gly1259Glu) (Table 1).
Discussion
Our study therefore confirmed the high heterozygous carrier frequency of ATP7B in the French population with one in 31 subjects harbouring a pathogenic variant at a heterozygous level. Our NGS approach was similar to that used by Coffey et al. [7] and Jang et al. [8]. We only included variants with probably pathogenic or pathogenic deleterious effects. Consequently, we were able to conclude that the heterozygous carrier frequency of ATP7B in our French cohort (1/31) was higher than described in Korean (1/53) population and slightly lower than observed in the British population (1/25).
When based on the VUS parameter, the heterozygous prevalence of ATP7B (1/19) was found to be similar than that described in the British population (1/18) [8]. Among the five VUS described during our study (p.Val536Ala, p.Glu541Lys, p.Met665Ile, p.Val1297Ile and p.Thr1434Met), three (p.Val536Ala, p.Glu541Lys, p.Met665Ile) had been reported in Coffey’s publication as being benign. However, unlike our British colleagues, we considered that one of the VUS (p.Met645Arg) was pathogenic. Indeed, the frequency of this mutation was 1.8% (11/604 patients) in the Lariboisière WD cohort, and various publications have reported this mutation in the literature since the 1990s [12, 16]. Furthermore, this is the most frequent mutation reported in Spanish WD patients [20].
p.Thr1434Met and p.Val1297Ile were not found during Coffey’s study. p.Val1297Ile displays a high frequency in the Asian population (1.5%), so it would be preferable to consider it as VUS even though it has already been described in the literature [11, 13]. Also, p.Thr1434Met should be definitively reclassified as benign, despite the first Turkish publication 18 years ago [10]. Indeed, its variant frequency was elevated in our study (0.645%) and in dbSNP databases, its frequency reaching 4% in the African population. Moreover, neither variant was ever found among the 604 WD patients in the Lariboisière WD database.
The high heterozygous prevalence of ATP7B observed in France raises the question of the approach to be adopted in the event of a genetic WD finding. Firstly, account needs to be taken of the discrepancies between the clinical and genetic frequencies seen both in France [9] and worldwide [7, 8], for different reasons. The diagnosis is certainly underestimated, or the pathology may be misdiagnosed. Indeed, in fulminant liver or tardive forms, or with a mild neurological presentation, a diagnosis of WD may not be suggested and the cause of death incorrectly determined. In addition, asymptomatic forms may be due to incomplete gene penetrance or the presence of disease-modifying genes (COMMD1, ATOX1, XIAP, HFE, prion protein, methylenetetrahydrofolate MTHF reductase, apolipoprotein E) [21].
Therefore, in order to determine the clinical status of WD in cases of genetic WD, a precise exploration of the copper balance (ceruloplasmin, relative exchangeable copper and 24 h urinary copper excretion) needs to be performed and patients monitored carefully so as to determine the clinical course of an asymptomatic form [22].
Furthermore, because the heterozygous carrier frequency of the ATP7B mutation is higher than previously thought, the guidelines concerning familial screening need to be revaluated in the near future. The families of WD patients should more frequently be offered not only a copper balance evaluation but sequencing of the whole coding region of ATP7B in order to improve genetic counselling [23].
Our study highlighted the difficulties of interpreting genetic results, particularly in the case of Exome sequencing analysis. Indeed, the ATP7B gene is on the list of genes that should be reported as incidental or secondary findings according to the recommendations of American College of Medical Genetics and Genomics (ACMG), notably because treatments for WD are available and effective when administered prior to the onset of symptoms [24].
Conclusions
Our study highlighted the major difference between the high heterozygous carrier frequency and the low clinical prevalence of WD. This discrepancy may be explained by the clinical variability, the incomplete penetrance and the existence of modifiers genes. Also, the molecular analysis of ATP7B should be interpreted with caution, always alongside copper assays (ceruloplasmin, relative exchangeable copper, 24 h-urinary copper excretion) with particular respect to exome sequencing. The variability in the major phenotype, the potential for reduced penetrance and genetic prevalence combine to hamper the diagnosis of WD. However, the existence of effective therapies urges us to detect a maximum of individuals with genetic WD before any symptoms appear.
References
Poujois A, Devedjian JC, Moreau C, Devos D, Chaine P, Woimant F, et al. Bioavailable trace metals in neurological diseases. Curr Treat Options Neurol. 2016;18(10):46.
Woimant F, Trocello J-M. Disorders of heavy metals. Handb Clin Neurol Elsevier. 2014;120:851–64.
Ferenci P, Czlonkowska A, Merle U, Ferenc S, Gromadzka G, Yurdaydin C, et al. Late-onset Wilson’s disease. Gastroenterology. 2007;32:1294–8.
Kim JW, Kim JH, Seo JK, Ko JS, Chang JY, Yang HR, et al. Genetically confirmed Wilson disease in a 9-month old boy with elevations of aminotransferases. World J Hepatol. 2013;5:156–9.
Scheinberg IH. Wilson’s disease. J Rheumatol Suppl. 1981;7:90–3.
Hoogenraad T. Wilson’s disease. Amsterdam-Rotterdam: interned Medical Publishers; 2001.
Coffey AJ, Durkie M, Hague S, McLay K, Emmerson J, Lo C, et al. A genetic study of Wilson's disease in the United Kingdom. Brain. 2013;136:1476–8.
Jang JY, Lee T, Bang S, Kim YE, Cho EH. Carrier frequency of Wilson’s disease in the Korean population: a DNA-based approach. J Hum Genet. 2017;62:815–8.
Poujois A, Woimant F, Samson S, Chaine P, Girardot-Tinant N, Tuppin P. Characteristics and prevalence of Wilson's disease: a 2013 observational population-based study in France. Clin Res Hepatol Gastroenterol. 2017:S2210–7401.
Loudianos G, Dessi V, Lovicu M, et al. Mutation analysis in patients of Mediterranean descent with Wilson disease: identification of 19 novel mutations. J Med Genet. 1999;36:833–6.
Davies LP, Macintyre G, Cox DW. New mutations in the Wilson disease gene, ATP7B: implications for molecular testing. Genet Test. 2008;12:139–45.
Loudianos G, Dessi V, Lovicu M, et al. Further delineation of the molecular pathology of Wilson disease in the Mediterranean population. Hum Mutat. 1998;12:89–94.
Li XH, Lu Y, Ling Y, Fu QC, Xu J, Zang GQ, Zhou F, De-Min Y, Han Y, Zhang DH, Gong QM, Lu ZM, Kong XF, Wang JS, Zhang XX. Clinical and molecular characterization of Wilson's disease in China: identification of 14 novel mutations. BMC Med Genet. 2011;11(12):6.
Deguti MM, Genschel J, Cancado EL, et al. Wilson disease: novel mutations in the ATP7B gene and clinical correlation in Brazilian patients. Hum Mutat. 2004;23:398.
Cox DW, Prat L, Walshe JM, et al. Twenty-four novel mutations in Wilson disease patients of predominantly European ancestry. Hum Mutat. 2005;26:280.
Lepori MB, Lovicu M, Dessi V, et al. Twenty-four novel mutations in Wilson disease patients of predominantly Italian origin. Genet Test. 2007;11:328–32.
Gromadzka G, Schmidt HH, Genschel J, et al. Frameshift and nonsense mutations in the gene for ATPase7B are associated with severe impairment of copper metabolism and with an early clinical manifestation of Wilson's disease. Clin Genet. 2005;68:524–32.
Yuan ZF1, Wu W, Yu YL, Shen J, Mao SS, Gao F, Xia ZZ. Novel mutations of the ATP7B gene in Han Chinese families with pre-symptomatic Wilson's disease. World J Pediatr. 2015;11(3):255–60.
Santhosh S, Shaji RV, Eapen CE, et al. ATP7B mutation in families in a predominantly Southern Indian cohort of Wilson's disease patients. Indian J Gastroenterol. 2006;25:277–82.
Margarit E, Bach V, Gomez D, Bruguera M, Jara P, Queralt R, et al. Mutation analysis of Wilson disease in the Spanish population - identification of a prevalent substitution and eight novel mutations in the ATP7B gene. Clin Genet. 2005;68:61–8.
Bost M, Piguet-Lacroix G, Parant F, Wilson CM. Molecular analysis of Wilson patients: direct sequencing and MLPA analysis in the ATP7B gene and Atox1 and COMMD1 gene analysis. J Trace Elem Med Biol. 2012;26:97–101.
Balkhi El S, Trocello J-M, Poupon J, Chappuis P, Massicot F, Girardot-Tinant N, et al. Relative exchangeable copper: a new highly sensitive and highly specific biomarker for Wilson's disease diagnosis. Clin Chim Acta 2011; 412:2254–2260.
Trocello J-M, Balkhi El S, Woimant F, Girardot-Tinant N, Chappuis P, Lloyd C, et al. Relative exchangeable copper: a promising tool for family screening in Wilson disease. Mov Disord 2014;29:558–562.
Kalia SS, Adelman K, Bale SJ, Chung WK, Eng C, Evans JP, et al. Recommendations for reporting of secondary findings in clinical exome and genome sequencing, 2016 update (ACMG SF v2.0): a policy statement of the American College of Medical Genetics and Genomics. Genet Med. 2017;19:249–2.
Availability of data and materials
The datasets used and/or analysed during the current study are available from the corresponding author on reasonable request.
Author information
Authors and Affiliations
Contributions
CC, AP, and FW designed the study. CC, JP and HM analysed sequence data. CC, AP, JLL and FW conducted formal analysis and wrote the manuscript. Manuscript Approval: All authors have read and approved the manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Written informed consent was obtained from all the study participants. The study was approved by the local Ethics Committee of Lariboisiere Hospital belongs to the comité de protection des personnes Ile-de-France IV. The reported investigations have been carried out in accordance with the principles of the Declaration of Helsinki.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Collet, C., Laplanche, JL., Page, J. et al. High genetic carrier frequency of Wilson’s disease in France: discrepancies with clinical prevalence. BMC Med Genet 19, 143 (2018). https://doi.org/10.1186/s12881-018-0660-3
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12881-018-0660-3