Abstract
Introduction
Understanding the proportion of patients with COVID-19 who have respiratory bacterial co-infections and the responsible pathogens is important for managing COVID-19 effectively while ensuring responsible antibiotic use.
Objective
To estimate the frequency of bacterial co-infection in COVID-19 hospitalized patients and of antibiotic prescribing during the early pandemic period and to appraise the use of antibiotic stewardship criteria.
Methods
Systematic review and meta-analysis was performed using major databases up to May 5, 2021. We included studies that reported proportion/prevalence of bacterial co-infection in hospitalized COVID-19 patients and use of antibiotics. Where available, data on duration and type of antibiotics, adverse events, and any information about antibiotic stewardship policies were also collected.
Results
We retrieved 6,798 studies and included 85 studies with data from more than 30,000 patients. The overall prevalence of bacterial co-infection was 11% (95% CI 8% to 16%; 70 studies). When only confirmed bacterial co-infections were included the prevalence was 4% (95% CI 3% to 6%; 20 studies). Overall antibiotic use was 60% (95% CI 52% to 68%; 52 studies). Empirical antibiotic use rate was 62% (95% CI 55% to 69%; 11 studies). Few studies described criteria for stopping antibiotics.
Conclusion
There is currently insufficient evidence to support widespread empirical use of antibiotics in most hospitalised patients with COVID-19, as the overall proportion of bacterial co-infection is low. Furthermore, as the use of antibiotics during the study period appears to have been largely empirical, clinical guidelines to promote and support more targeted administration of antibiotics in patients admitted to hospital with COVID-19 are required.
Similar content being viewed by others
Introduction
The COVID-19 pandemic has impacted health systems worldwide, with SARS-CoV-2 infection being implicated in more than 6 million deaths to date [1, 2]. Some clinical guidelines have recommended empirical antibiotic therapy to treat suspected bacterial respiratory co-infection in COVID-19 patients, and tools to support and promote antibiotic stewardship in this population are therefore needed [3, 4].
Distinguishing between viral pneumonia and bacterial co-infection at presentation and during the course of COVID-19 disease can be challenging due to various similarities, including characteristically high inflammatory markers and the frequent presence of pulmonary infiltrates on chest X-ray or computed tomography (CT) imaging [5]. There is therefore potential for considerable overuse of antibiotics in the management of COVID-19 pneumonia, with the attendant risk of an increase in the prevalence of antimicrobial resistance in affected populations. Given the current pandemic context, the implications of this for public health and health systems are likely to be considerable. Clinical guidelines to support the most effective treatment for patients while promoting the responsible use of antibiotics should be informed by an understanding of what proportion of patients admitted to hospital with COVID-19 pneumonia have confirmed acute respiratory bacterial co-infection and of the commonly associated pathogens.
We performed a systematic review to estimate the frequency of confirmed bacterial co-infection in patients admitted to hospital with COVID-19 pneumonitis, the frequency of empirical antibiotic use in this patient group, and to identify any antibiotic stewardship criteria that have been used during the COVID-19 pandemic to date.
Methods
We registered the review protocol at the PROSPERO international prospective register of systematic reviews (CRD 42020181215). We followed the method for the elaboration of systematic reviews recommended by the Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA) statement [6]. Although the PRISMA statement is mainly used in systematic reviews of intervention studies, several domains are also applicable to systematic reviews of prevalence [7]. As PRISMA is the most widely used tool for the reporting of systematic reviews, we used it in the present work. The PRISMA checklist for this study is presented in Additional file 1: Material S1.
Selection criteria and search strategy
We included studies with patients admitted to a hospital setting with suspected lower respiratory tract infection (LRTI) and with SARS-CoV-2 infection confirmed by PCR. Due to the high number of publications, we only included original studies with at least 10 participants and which provided enough information to appraise the methods used. Randomised and non-randomised studies that presented at least one of the following outcomes of interest were included: (a) prevalence of bacterial co-infection in patients with confirmed SARS-CoV-2 infection; (b) the proportion of patients with confirmed SARS-CoV-2 infection that were commenced on empirical antibiotic treatment. Where available, we collected information on the duration and type of antibiotics and on any related adverse events. In cases receiving specific treatment for COVID-19 as part of a clinical trial, we only included standard-of-care comparator arms. We excluded antibiotic use for indications other than bacterial LRTI (e.g., azithromycin used as specific therapy for SARS-CoV-2 at the beginning of the pandemic was excluded). In order for our findings to be readily generalisable, we excluded pregnant women and patients with chronic immunosuppressive conditions, these being specific populations with different and increased infection risk profiles. We also excluded studies that mentioned bacterial co-infection rates but did not provide clinical details (e.g., cost-effectiveness analyses or modelling studies). Given that many authors provided only limited descriptions of antibiotic use, we performed two sub-analyses: one of studies clearly stating bacterial co-infection confirmed by cultures taken less than 48 h from point of admission, and another including only studies that clearly stated the empirical use of antibiotics. In the latter, we also describe any antibiotic stewardship strategies.
We also performed sub-group analyses of any available data on critically ill patients, defined as those patients identified by study authors as requiring admission to high-dependency or intensive care. Definitions of bacterial co-infection provided by study authors were accepted.
We searched the following databases up to May 5, 2021: Pubmed, LILACS, Embase, Web of Science and Cochrane Library. Our search strategy is given in Additional file 1: Material S2. Searches were limited to papers written in English, German, Russian, French, Spanish, or Portuguese. Reference lists from all included articles were also scrutinised to identify additional studies of potential interest.
Screening and data extraction
We used a two-stage screening process to identify publications that would be eligible for inclusion: title and abstract, followed by full text review. Any original manuscripts referenced by systematic reviews but not identified by the initial search were also included if they were eligible. All publications were then screened in duplicate and independently by reviewers working in pairs (MC, GG, AG, LK, AM, DC); any disagreements in screening were resolved by a third, independent reviewer (EH or BP). Data from eligible papers were extracted by two independent reviewers into separate, piloted and standardised Microsoft Excel spreadsheets; the third reviewer was then asked to resolve any discrepancies and a single consensus dataset was produced after discussion.
Data analysis
We present the results of all included studies according to the selected outcomes of interest. We analysed our data using a proportion meta-analysis. We applied an arc-sine transformation to stabilise the variance of proportions (Freeman-Tukey variant of the arc-sine square-root of transformed proportions method), where y = arcsine[√(r/(n + 1))] + arcsine[√(r/(n + 1)/(n + 1)], with a variance of 1/(n + 1), with n being the population size. The pooled proportion was calculated as the back-transformation of the weighted mean of the transformed proportions, using inverse arcsine variance weights for the fixed and random effects models. Where heterogeneity between studies was found we applied DerSimonian-Laird weights for the random effects model. We calculated the I2 statistic as a measure of the overall variation in the proportion that was attributable to between-study heterogeneity. STATA 17.0 was used for all analyses.
Study quality assessment
To describe the quality of the prevalence data extracted from the included studies, we used The Joanna Briggs Institute (JBI) Critical Appraisal Checklist for cross-sectional/prevalence data [8]. This is a tool that has been developed acknowledging that prevalence data can come from different study designs, as in our case.
Quality of included studies were assessed independently by two investigators; any disagreements were resolved by a third senior investigator.
Results
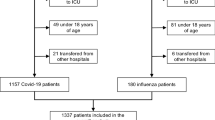
Database searches identified 6798 studies. After removing duplicates and reviewing the secondary reference lists from included papers we screened a total of 4,132 studies for title and abstract. Of these, 162 (3.9%) went to full text review and 85 (2.1%) were selected for data extraction (Fig. 1).
Flowchart of included studies. Moher D, Liberati A, Tetzlaff J, Altman DG, The PRISMA Group (2009). Preferred Reporting Items for Systematic Reviews and Meta-Analyses: The PRISMA Statement. PLoS Med 6(7): e1000097. https://doi.org/10.1371/journal.pmed1000097. For more information, visit www.prisma-statement.org
The independent assessment of the quality of included papers is described in the Additional file 1: Material S3. Using the selected quality assessment tool, we identified that the majority of included studies had appropriate samples for their specific objectives, adequate description of participants and diagnosis of condition.
Data derived from a total of 31,123 individuals were included for analysis. The study designs of all included papers comprised case series, cohorts, registries, and clinical trials. The majority of papers were from China (29, 34.1%) and USA (16, 18.8%). The main characteristics of the included studies are shown in Table 1. Full references of included studies are provided in Additional file 1: Material S4.
Bacterial co-infection prevalence
We included 70 studies that reported on the prevalence of bacterial co-infection (including critically ill patients and not critically ill patients) (Table 2). Meta-analysis of these studies showed an overall prevalence of bacterial co-infection of 12% (51 studies, 95% CI 8% to 16%; I2 99.2%) (Fig. 2A); subgroup meta-analysis of critically ill patients showed a prevalence of 23% (21 studies, 95% CI 16 to 31%; I2 94.6%) (Fig. 2B).
Twenty studies (31.4%) gave a clear definition of bacterial co-infection, stating that this was diagnosed within 48 h from admission. All of them included cultures, urinary antigen and PCR for definitions of bacterial co-infection. We performed a meta-analysis of this subgroup that showed a prevalence of 4% (15 studies, 95% CI 3% to 6%; I2 94.2%) in the overall population (Fig. 3A) and a bacterial coinfection prevalence of 12% (5 studies, 95% CI 4% to 22%; I2 91.2%) in critically ill patients. (Fig. 3B).
Antibiotic use
Fifty-two (61.2%) studies were included in the analysis of antibiotic use (Table 3). Meta-analysis showed an overall prevalence of antibiotic use of 60% (38 studies, 95% CI 52% to 76%; I2 98.8%) (Fig. 4A); sub-group analysis restricted to critically ill patients identified a prevalence of antibiotic usage of 86% (19 studies, 95% CI 78% to 92%; I2 93.2%) (Fig. 4B).
Eleven studies (12.9%) clearly described empirical antibiotic use. A sub-analysis of these papers found that overall empirical antibiotic use was 62% (eight studies, 95% CI 55 to 69%; I2 95.1%) (Fig. 5A) and in critically ill patients was 66% (six studies, 95% CI 58 to 73%; I2 96.6%) (Fig. 5B).
Antibiotic stewardship
Eleven studies specifically stated that empirical antibiotics were commenced of which five described decision-making processes regarding antibiotics.
Cheng et al. [9] mentioned that 52/147 (35%) patients received empirical antibiotics and that 19 (37%) received antibiotics for more than a week despite negative cultures. The median length of course of empirical antibiotics was seven (IQR = 5 to 12) days.
Rothe et al. [10] described the implementation of an antibiotic stewardship standard operational procedure in their institution in which initiation of antibiotic therapy was recommended only in cases of clinically suspected infection (narrow spectrum aminopenicillin/beta-lactamase inhibitor combination). However, decisions regarding stewardship were at the clinician’s discretion. The most used antibiotic scheme during the observation period were ampicillin/sulbactam (41.5%) and piperacillin/ tazobactam (19.3%) with or without azithromycin. Median duration of were variable being longer in the case of piperacillin/ tazobactam 10 (range 3 to 26) days. Interestingly, azithromycin was not included in the guidelines, although it was used in 43 patients (31.9%) as combination therapy.
Townsend et al. [11] described 84 patients treated empirically for respiratory bacterial co-infection of which 78 (92.9%) received monotherapy. All treatment was initially intravenous, and an oral switch took place in only 34 (40.5%) cases. The median durations of intravenous and oral therapies were five days (range 1 to 14) and three days (range 1 to 4) respectively.
Karami et al. [12] described the adherence to local guidelines on empiric antibiotic therapy in their institution. Mean adherence was 60.3% (range 45.3% to 74.7%) on the first day of admission showing that 556 of 925 (60.1%) patients were prescribed empirical antibiotics. However, the rate of antibiotic prescribing increased after seven days of admission to 669 (72.3%). Confirmed bacterial co-infection was confirmed only in 12/925 (1.2%) patients. Regarding length of antibiotics use, 467 of 555 (84.1%) had five days of antibiotics. Intravenous antibiotics exceeded 48 h in 413 patients who started antibiotic treatment on the first day of admission and oral switched were performed in 9.9% of those.
Vaugh et al. [13] described that of the patients who received empiric antibiotic therapy (N = 965), the majority (612, 63.4%) received antibiotics targeting community-acquired microorganism. The median of duration of inpatient antibiotic was three days (IQR, 2 to 6 days) in the patients receiving antibiotics. Total days of inpatient antibiotic therapy was 4158 days/1000 patients.
The remainder of these studies (Seaton et al. [14], Baskaraban et al. [15], Goncalves et al. [16], Karaba et al. [17], Asmarawati et al. [18], D’onofrio et al. [19] and Elabbadi [20]) did not describe empirical antibiotic duration or any specific criteria for stopping treatment, although they do state that local guidelines for empirical antibiotic use in COVID-19 pneumonitis should be applied.
Discussion
In the absence of clear guidance on when to give empirical antibiotic therapy to patients admitted to hospital with COVID-19 pneumonia, clinicians face a dilemma. In the context of a global pandemic and given the potential risks of antibiotic treatment to patients and to public health, it is essential that the best available evidence is used to support clinicians on the front line to appropriately balance risks to patients and to the wider public.
We analysed bacterial co-infection in different ways in order to evaluate how estimates may vary depending on authors’ definitions. Based only on author descriptions, we found a prevalence of bacterial co-infection of 12% (95% CI 8 to 16%) in the overall population. Interestingly, we observed that bacterial co-infection was lower when including only studies with clear definitions of bacterial co-infection (overall population 4% (95% CI 3 to 6%), critically ill patients 12% (95% CI 4 to 22%)). Our results are similar to that found by other authors who have evaluated co-infections in patients with COVID-19. For example, Rawson et al. [21] conducted a meta-analysis which found a prevalence of bacterial and fungal coinfection of 8%. Langford et al. evaluated bacterial co-infection at presentation and after presentation of COVID-19, finding a prevalence of 3.5% (95% CI 0.4 to 6.7%) for primary co-infection and 14.3% (95% CI 9.6 to 18.9%) for secondary (nosocomial) co-infection [22].
It is important to acknowledge that our estimates of the prevalence of bacterial co-infection prevalence were derived from a number of different definitions, as provided by the authors of the source papers. This is relevant, as although microbiological cultures are the gold standard for diagnosis, these are neither quick nor universally available tools on which to base prescribing decisions, particularly in patients with severe disease.
Our study also finds that, as expected, the overall use of antibiotics in patients with COVID-19 is high compared to the estimated prevalence of bacterial co-infection. We identified a prevalence of empirical antibiotic use of 62% (95%CI 55 to 29%). These estimates are similar to those of Langford et al. [22], who found an overall prevalence of antibiotic use of 71.9% (95% CI 56.1 to 87.7%). Our slightly lower estimates may be explained by having retrieved studies nearly one and a half years after the start of the pandemic. This could reflect changes in empirical practice through increased experience in managing COVID-19, coupled with more data being available to inform evidence-based practice regarding antibiotic use. Furthermore, the previous study provided estimates of antibiotic use based only on patients with culture confirmed bacterial co-infections, while we included all COVID-19 patients that were considered to have an infection in our estimate, regardless of whether bacterial co-infections were ultimately confirmed. In doing so, we have sought to reflect real world practice, and we suggest that estimates of overall empirical antibiotic use that are not restricted to patients with confirmed infections are important to understanding the need for, and potential impact of, antimicrobial stewardship tools and strategies as part of the response to the COVID-19 pandemic.
The final aim of our study was to identify to what degree decisions to stop empirically prescribed antibiotics were being made according to any defined criteria. This aspect has not been addressed previously in published systematic reviews and meta-analyses. Despite terms related to stewardship being specifically included in our search strategy and despite meticulously reading all included citations in full, including discussion sections, we found very little information on stewardship measures. We found this absence of information particularly notable given that antimicrobial resistance is widely acknowledged as being one of the most serious public health challenges of our times [23,24,25]. Whilst we acknowledge that case reports and series are generally more concerned with describing the clinical and demographic characteristics of their patients, it is nevertheless disappointing that the large observed differences between confirmed bacterial co-infection and frequency of antibiotic use does not prompt authors to consider this matter more prominently in their discussions. Despite these deficits in the current literature, we assert that it is of fundamental importance to preserve any goals and achievements relating to antibiotic stewardship established prior to the COVID-19 pandemic. Several antibiotic stewardship programs such as ARK (Antibiotic Review Kit) and TARGET (Treat Antibiotics Responsibly, Guidance, Education, Tools) have been shown to be both feasible and acceptable in supporting the safe discontinuation of antibiotics post-prescription in acute hospital settings [26, 27]. These are just two examples of efforts that must be continued, particularly in the current climate of highly prevalent empirical use of antibiotics during a viral pandemic in which the prevalence of confirmed bacterial co-infection appears to be low.
Our study has some important limitations. The most important being that the COVID-19 pandemic has given rise to an unprecedented situation in the scientific world in terms of a seemingly exponential increase in the volume of related publications over a very short time. Thus, at the time of writing there are likely to be additional studies that would have qualified for inclusion. This rapidity of publication would necessitate updating searches and analysis on as much as a weekly basis, which we suggest would be unrealistic for a piece of peer-reviewed work such as this. It is reassuring to know, however, that other groups pursuing similar research questions [21, 22, 28] have found similar results despite not having included the same studies or conducting searches that cover the same dates. To the best of our knowledge, the present systematic review is the currently most up to date systematic review of this subject, presenting data from more than 30,000 patients from studies identified through an exhaustive search strategy. Another important point to highlight is that most of the included studies in this systematic review are from high income countries, and caution should therefore be exercised when generalising from our results to other settings. Further studies should analyse how COVID-19 has affected antibiotic use in low- and middle-income countries, where the burden of drug-resistant infections is greatest [24].
Another important limitation is one that is inherent to this type of analysis. There is a consensus that the methodology for systematic reviews of prevalence data is not well developed, with a notable lack of methodological and reporting guidance for systematic reviews of prevalence data [29, 30]. Thus, in most cases authors present adapted or de novo tools to assess the quality of the prevalence data that will be included in the analysis, regardless of the study design [22, 28, 31]. In our case, we used a tool that has been developed acknowledging that prevalence data can come from different study designs, however we cannot make an overall assessment of risk of bias [8]. Prevalence metanalysis have also the risk of presenting high level of heterogeneity. We have sought to address the high level of heterogeneity by using statistical correction as well as performing subgroup analyses. Nevertheless, caution should be exercised with extrapolation to specific contexts.
Conclusion
In this study we have reported bacterial co-infection and antibiotic use during the first 18 months of the SARS-CoV-2 pandemic. This work can help clinicians to reflect on and understand the initial response to a global pandemic of a novel respiratory virus. Our results show that there is currently insufficient evidence to support the use of empirical use of antibiotics in most hospitalised patients with COVID-19, as the overall proportion of bacterial co-infection in these patients is low. Furthermore, as the use of antibiotics in COVID-19 appears to have been largely empirical, it is necessary to identify clinical and laboratory markers and to formulate guidelines to promote more targeted administration of antibiotics in patients admitted to hospital with COVID-19.
Availability of data and materials
All data generated or analysed during this study are included in this published article [and its supplementary information files].
References
Ibn-Mohammed T, Mustapha KB, Godsell J, Adamu Z, Babatunde KA, Akintade DD, et al. A critical analysis of the impacts of COVID-19 on the global economy and ecosystems and opportunities for circular economy strategies. Resour Conserv Recycl. 2021;164:105169.
WHO Coronavirus (COVID-19) Dashboard. Geneva: World Health Organization; 2022. https://covid19.who.int.
COVID-19 rapid guideline: antibiotics for pneumonia in adults in hospital. United Kingdom: National Institute of Clinical Excellence; 2020. https://www.nice.org.uk/guidance/ng173/chapter/3-Initial-approach-to-antibiotic-treatment-choices.
Gautret P, Lagier JC, Parola P, Hoang VT, Meddeb L, Mailhe M, et al. Hydroxychloroquine and azithromycin as a treatment of COVID-19: results of an open-label non-randomized clinical trial. Int J Antimicrob Agents. 2020;56:105949.
Feldman C, Anderson R. The role of co-infections and secondary infections in patients with COVID-19. Pneumonia. 2021;13(1):5.
Liberati A, Altman DG, Tetzlaff J, Mulrow C, Gøtzsche PC, Ioannidis JP, et al. The PRISMA statement for reporting systematic reviews and meta-analyses of studies that evaluate healthcare interventions: explanation and elaboration. BMJ. 2009;339: b2700.
Page MJ, McKenzie JE, Bossuyt PM, Boutron I, Hoffmann TC, Mulrow CD, et al. The PRISMA 2020 statement: an updated guideline for reporting systematic reviews. Syst Rev. 2021;10(1):89.
Munn Z, Moola S, Riitano D, Lisy K. The development of a critical appraisal tool for use in systematic reviews addressing questions of prevalence. Int J Health Policy Manag. 2014;3(3):123–8.
Cheng LS-K, Chau SK-Y, Tso EY-K, Tsang SW-C, Li IY-F, Wong BK-C, et al. Bacterial co-infections and antibiotic prescribing practice in adults with COVID-19: experience from a single hospital cluster. Ther Adv Infect Dis. 2020;7:20499.
Rothe K, Feihl S, Schneider J, Wallnöfer F, Wurst M, Lukas M, et al. Rates of bacterial co-infections and antimicrobial use in COVID-19 patients: a retrospective cohort study in light of antibiotic stewardship. Eur J Clin Microbiol Infect Dis. 2021;40(4):859–69.
Townsend L, Hughes G, Kerr C, Kelly M, O’Connor R, Sweeney E, et al. Bacterial pneumonia coinfection and antimicrobial therapy duration in SARS-CoV-2 (COVID-19) infection. JAC Antimicrob Resist. 2020;2(3):dlaa071.
Karami Z, Knoop BT, Dofferhoff ASM, Blaauw MJT, Janssen NA, van Apeldoorn M, et al. Few bacterial co-infections but frequent empiric antibiotic use in the early phase of hospitalized patients with COVID-19: results from a multicentre retrospective cohort study in The Netherlands. Infect Dis. 2021;53(2):102–10.
Vaughn VM, Gandhi TN, Petty LA, Patel PK, Prescott HC, Malani AN, et al. Empiric antibacterial therapy and community-onset bacterial coinfection in patients hospitalized with coronavirus disease 2019 (COVID-19): a multi-hospital cohort study. Clin Infect Dis. 2021;72(10):e533–41.
Seaton RA, Gibbons CL, Cooper L, Malcolm W, McKinney R, Dundas S, et al. Survey of antibiotic and antifungal prescribing in patients with suspected and confirmed COVID-19 in Scottish hospitals. J Infect. 2020;81(6):952–60.
Baskaran V, Lawrence H, Lansbury LE, Webb K, Safavi S, Zainuddin NI, et al. Co-infection in critically ill patients with COVID-19: an observational cohort study from England. J Med Microbiol. 2021;70(4):2068.
Goncalves Mendes Neto A, Lo KB, Wattoo A, Salacup G, Pelayo J, DeJoy R 3rd, et al. Bacterial infections and patterns of antibiotic use in patients with COVID-19. J Med Virol. 2021;93(3):1489–95.
Karaba SM, Jones G, Helsel T, Smith LL, Avery R, Dzintars K, et al. Prevalence of Co-infection at the time of hospital admission in COVID-19 patients, a multicenter study. Open Forum Infect Dis. 2021;8(1):ofaa578.
Asmarawati TP, Rosyid AN, Suryantoro SD, Mahdi BA, Windradi C, Wulaningrum PA, et al. The clinical impact of bacterial co-infection among moderate, severe and critically ill COVID-19 patients in the second referral hospital in Surabaya. F1000Res. 2021;10:113.
D’Onofrio V, Van Steenkiste E, Meersman A, Waumans L, Cartuyvels R, Van Halem K, et al. Differentiating influenza from COVID-19 in patients presenting with suspected sepsis. Eur J Clin Microbiol Infect Dis. 2021;40(5):987–95.
Elabbadi A, Turpin M, Gerotziafas GT, Teulier M, Voiriot G, Fartoukh M. Bacterial coinfection in critically ill COVID-19 patients with severe pneumonia. Infection. 2021;49(3):559–62.
Rawson TM, Moore LSP, Zhu N, Ranganathan N, Skolimowska K, Gilchrist M, et al. Bacterial and fungal coinfection in individuals with coronavirus: a rapid review to support COVID-19 antimicrobial prescribing. Clin Infect Dis. 2020;71(9):2459–68.
Langford BJ, So M, Raybardhan S, Leung V, Westwood D, MacFadden DR, et al. Bacterial co-infection and secondary infection in patients with COVID-19: a living rapid review and meta-analysis. Clin Microbiol Infect. 2020;26(12):1622–9.
Antibiotic resistance. Geneva: World Health Organization; 2020. https://www.who.int/news-room/fact-sheets/detail/antibiotic-resistance.
O’Neill J. Review on Antimicrobial resistance: tackling a crisis for the health and wealth of nations. London, UK2014. https://amr-review.org/sites/default/files/AMR%20Review%20Paper%20-%20Tackling%20a%20crisis%20for%20the%20health%20and%20wealth%20of%20nations_1.pdf.
Antibiotic/Antimicrobial Resistance (AR/AMR). USA: Center of Disease Control and Prevention; 2020. https://www.cdc.gov/drugresistance/index.html.
Cross ELA, Sivyer K, Islam J, Santillo M, Mowbray F, Peto TEA, et al. Adaptation and implementation of the ARK (Antibiotic Review Kit) intervention to safely and substantially reduce antibiotic use in hospitals: a feasibility study. J Hosp Infect. 2019;103(3):268–75.
Jones LF, Hawking MKD, Owens R, Lecky D, Francis NA, Butler C, et al. An evaluation of the TARGET (Treat Antibiotics Responsibly; Guidance, Education, Tools) Antibiotics Toolkit to improve antimicrobial stewardship in primary care-is it fit for purpose? Fam Pract. 2018;35(4):461–7.
Lansbury L, Lim B, Baskaran V, Lim WS. Co-infections in people with COVID-19: a systematic review and meta-analysis. J Infect. 2020;81(2):266–75.
Borges Migliavaca C, Stein C, Colpani V, Barker TH, Munn Z, Falavigna M. How are systematic reviews of prevalence conducted? A methodological study. BMC Med Res Methodol. 2020;20(1):96.
Migliavaca CB, Stein C, Colpani V, Munn Z, Falavigna M. Quality assessment of prevalence studies: a systematic review. J Clin Epidemiol. 2020;127:59–68.
Joanna Briggs Institute Reviewers Manual 2014. The Systematic Review of Prevalence and Incidence Data. Australia: Joanna Briggs Institute. University of Adelaide; 2015. https://nursing.lsuhsc.edu/JBI/docs/ReviewersManuals/Prevalence-and-Incidence-Data.pdf.
Acknowledgements
None.
Funding
BP was funded in part by the Wellcome Trust [109975/Z/15/Z]. For the purpose of open access, the authors have applied a CC BY public copyright licence to any Author Accepted Manuscript version arising from this submission. All other authors—none to declare.
Author information
Authors and Affiliations
Contributions
MC, GG and EH participated in the conception and design of the study. MC, GG, AG, AM, LK, EH participated in the acquisition of data and analysis. MC, GG, BP and EH participated in the interpretation of data. MC and GG participated drafting the article. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Not applicable to the current study.
Consent for publication
Not applicable to the current study.
Competing interests
Authors declare no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Additional file 1. Material S1.
PRISMA check list. Material S2. Search Strategy. Material S3. Quality Assessment of included studies. Material S4. Reference list of included studies.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Calderon, M., Gysin, G., Gujjar, A. et al. Bacterial co-infection and antibiotic stewardship in patients with COVID-19: a systematic review and meta-analysis. BMC Infect Dis 23, 14 (2023). https://doi.org/10.1186/s12879-022-07942-x
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12879-022-07942-x