Abstract
Background
The recent increase in cases of azole-resistant Aspergillus fumigatus (ARAf) infections is a major clinical concern owing to its treatment limitations. Patient-derived ARAf occurs after prolonged azole treatment in patients with aspergillosis and involves various cyp51A point mutations or non-cyp51A mutations. The prognosis of patients with chronic pulmonary aspergillosis (CPA) with patient-derived ARAf infection remains unclear. In this study, we reported the case of a patient with ARAf due to HapE mutation, as well as the virulence of the isolate.
Case presentation
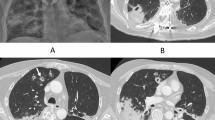
A 37-year-old male was presented with productive cough and low-grade fever. The patient was diagnosed with CPA based on the chronic course, presence of a fungus ball in the upper left lobe on chest computed tomography (CT), positivity for Aspergillus-precipitating antibody and denial of other diseases. The patient underwent left upper lobe and left S6 segment resection surgery because of repeated haemoptysis during voriconazole (VRC) treatment. The patient was postoperatively treated with VRC for 6 months. Since then, the patient was followed up without antifungal treatment but relapsed 4 years later, and VRC treatment was reinitiated. Although an azole-resistant isolate was isolated after VRC treatment, the patient did not show any disease progression in either respiratory symptoms or radiological findings. The ARAf isolated from this patient showed slow growth, decreased biomass and biofilm formation in vitro, and decreased virulence in the Galleria mellonella infection model compared with its parental strain. These phenotypes could be caused by the HapE splice site mutation.
Conclusions
This is the first to report a case demonstrating the clinical manifestation of a CPA patient infected with ARAf with a HapE splice site mutation, which was consistent with the in vitro and in vivo attenuated virulence of the ARAf isolate. These results imply that not all the ARAf infections in immunocompetent patients require antifungal treatment. Further studies on the virulence of non-cyp51A mutations in ARAf are warranted.
Similar content being viewed by others
Background
Aspergillus fumigatus is one of the most important opportunistic fungal pathogens in humans and causes aspergillosis, including invasive aspergillosis (IA), chronic pulmonary aspergillosis (CPA), and allergic bronchopulmonary aspergillosis (ABPA) [1]. Triazole antifungals are the first choice of treatment for aspergillosis [2, 3]. However, an increase in cases of azole-resistant A. fumigatus (ARAf) infections has been reported worldwide in the last 20 years, which is regarded as an emerging clinical problem due to its treatment limitations for this disease [4,5,6,7].
Infections due to azole-resistant strains are classified into the environmental and patient routes and occur through the mutation or upregulation of the cyp51A gene, which encodes 14-α sterol demethylase, a triazole target enzyme in A. fumigatus [6, 8, 9]. Environment-derived ARAf occurs after environmental exposure to fungicides and involves tandem repeats (TRs) in the promoter region of cyp51A coupled with point mutations such as TR34/L98H and TR46/Y121F/T289A, whereas patient-derived ARAf occurs after prolonged azole treatment in aspergillosis-infected patients and involves various cyp51A point mutations (G54, G138, and M220) or non-cyp51A mutations [9]. Although environment-derived ARAf infection has been associated with poor prognosis in patients with IA and CPA [7, 9,10,11], the prognosis of patient-derived ARAf infections remains unclear.
The proportion of ARAf exhibiting non-cyp51A mutations has increased over the last 5 years [12]. To date, non-cyp51A mutations, such as those in the cdr1B, hapE, and hmg1 genes, have been reported to cause azole resistance [13,14,15]. The cdr1B gene encodes an ATP-binding cassette (ABC) transporter, and constitutive expression of the cdr1B gene leads to a decrease in intracellular drug concentration, resulting in azole resistance [13]. The hapE gene is a subunit of the CCAAT-binding complex (CBC), and the amino acid substitution of HapE (P88L) leads to an increase in the cyp51A expression and consequent azole resistance [14, 16]. Mutation in the sterol-sensing domain of the 3-hydroxy-3-methyl-glutaryl-coenzyme A reductase-encoding gene, hmg1, results in the accumulation of ergosterol precursors in cells, leading to triazole resistance [15].
Although some of the resistance mechanisms of ARAf with non-cyp51A mutations have been well investigated [13, 15, 17], the causes of its change in virulence remain unclear. In this study, we evaluated the clinical course of with ARAf due to HapE mutation, as well as the virulence of the isolate.
Case presentation
Case
A 37-year-old male presented with productive cough and low-grade fever. The patient had a history of thoracic surgery for recurrent pneumothorax with chronic obstructive pulmonary disease as the underlying condition. Chest computed tomography (CT) presented a fungus ball in the upper left lobe, and the patient was positive for Aspergillus-precipitating antibody. Although the patient was negative for fungal culture on bronchoscopy, other diseases such as mycobacterial infection and lung cancer were excluded. The patient was diagnosed with CPA based on the chronic course, presence of a fungus ball on chest CT, and an immunological response [18]. The patient was treated with itraconazole (ITC; 400 mg/day) from August 2009 to July 2010 and subsequently, with voriconazole (VRC; 400 mg/day) until December 2011; however, he had repeated haemoptysis during treatment. The patient then underwent left upper lobe and left S6 segment resection surgery in December 2011 and was treated postoperatively with VRC (400 mg/day) for 6 months. Since then, the patient was followed up without antifungal treatment. In June 2016, he had worsening respiratory symptoms and chest CT showed thickening of the cavity wall in the upper left lobe. The sensitive isolate MF-2046 was obtained from the sputum, and VRC treatment was reinitiated due to CPA relapse. Although the resistant isolate MF-2108 was obtained from the sputum in September 2016, the VRC treatment was continued until November 2018 as respiratory symptoms and radiological findings improved. Since then, the patient has not been treated, but no disease progression has been observed (Fig. 1).
Clinical course of a patient with CPA. The patient was treated with itraconazole (ITC) from August 2009 to July 2010 and with voriconazole (VRC) until December 2011, however, he had repeated haemoptysis during treatment. He underwent left upper lobe and left S6 segment resection surgery in December 2011 and was treated with VRC for 6 months. The patient was then followed up without antifungal treatment, but relapsed in June 2016, and treatment with VRC was initiated again. The sensitive isolate, MF-2046, was obtained from sputum before treatment with VRC, while the resistant isolate, MF-2108, was obtained from sputum during treatment with VRC. Although the MF-2108 isolate was isolated from sputum in September 2016, VRC treatment was continued until November 2018, as respiratory symptoms and radiological findings improved. Since then, the patient has not been treated, but no disease progression has been observed
Strains
The MF-2046 and MF-2108 isolates were obtained from the patient’s sputum in June and September of 2016, respectively (Fig. 1). Based on the macroscopic colony morphology, micromorphological characteristics, the ability to grow at 48 °C, and the sequences of the β-tubulin gene, these were identified as A. fumigatus sensu strico isolates [19, 20].
Drug susceptibility test
In vitro susceptibility testing of the isolates was performed as previously described [19]. Briefly, minimum inhibitory concentrations (MICs) of ITC, VCZ, and amphotericin B and minimum effective concentrations (MECs) of micafungin were determined using the Clinical and Laboratory Standards Institute reference method for broth microdilution, document M38-A2, with partial modifications using a yeast-like fungus DP plate (Eiken Chemical, Tokyo, Japan) [21]. The results were evaluated according to the European Committee on Antimicrobial Susceptibility (EUCAST) clinical breakpoints.
Short tandem repeats (STR) analysis and cyp51A mutation
STR analysis was performed as previously descibed [22]. Briefly, nine microsatellite regions were amplified by polymerase chain reaction (PCR) using the designed primer pairs and sequenced. The number of repeats in each region was counted from the sequences.
The cyp51A gene was amplified by PCR using the designed primer pairs and was sequenced. The results of the STR analysis and primers used in this study are shown in Table S1.
Preparation of A. fumigatus conidia from isolates
All isolates were incubated on a potato dextrose agar (PDA) (0.4% potato starch, 2% dextrose, and 1.5% agar) (Difco Laboratories, Detroit, MI) slanted at 30 °C for 1 week. The conidia were harvested with phosphate-buffered saline (PBS) containing 1% Tween 20 solution, and the conidial suspensions obtained were passed through a sterile 40-μm strainer to remove hyphal fragments. The number of conidia was counted using a haemocytometer.
Growth assay
A total of 1 × 104 conidia /A. fumigatus isolate were spotted on the PDA plates and incubated for 5 days at 30 °C. Colony diameters were measured after 72, 96, and 120 h and the average diameters were calculated from three independent experiments [23].
Biomass measurement and biofilm assay
A total of 5.0 × 105 conidia / A. fumigatus isolate were incubated in 5 mL of yeast glucose medium at 37 °C for 24 h with shaking at 250 rpm. The precipitate obtained was then recovered by filtration, frozen at − 80 °C, lyophilized overnight, and weighed. The average biomass was calculated from three independent experiments.
Biofilm assay was performed as previously described [24, 25]. Briefly, a round-bottomed 96-well plate was inoculated with 100 μL of the conidial suspension at a density of 1 × 105 conidia/mL in a Brian medium and incubated at 37 °C for 24 h. The spent culture medium was removed from each well and the adherent cells were washed three times with distilled water (dH2O). Next, 100 μL of 0.1% (w/v) crystal violet solution was added to each well and incubated for 10 min. This solution was carefully removed and washed twice with dH2O. The biofilms were destained for 10 min by adding 125 μL of 100% ethanol to each well. The absorbance of the destaining solution was measured at 595 nm.
Galleria mellonella virulence assay
Healthy G. mellonella larvae (Oita General Service Co., Ltd., Japan) of the same size were selected for the assay. Groups of 10 larvae were inoculated with 1.0 × 106 conidia into the haemocoel using a Hamilton syringe through the last left pro-leg [26]. The inoculated larvae were incubated in the dark at 37 °C and survival was monitored daily for 7 days. Ten larvae were inoculated with PBS and used as controls, and no larvae died during this time. Virulence assays were repeated three times independently.
Whole-genome sequencing
A total of 5.0 × 105 conidia / A. fumigatus isolate were incubated in 5 mL of yeast extract-peptone-dextrose (1% yeast extract, 2% peptone, and 2% dextrose) (Difco Laboratories, Detroit, MI, USA) broth at 37 °C for 24 h with shaking at 250 rpm. Mycelium was recovered using a sterile 40-μm strainer, rapidly frozen using absolute ethanol and dry ice, and lyophilized overnight. The lyophilized mycelium was homogenized and used for DNA extraction. DNA was extracted using a MasterPure™ DNA Purification Kit (Epicentre, Madison, WI, USA) and purified using a QIAquick PCR Purification Kit (Qiagen, Hilden, Germany) according to the manufacturer’s instructions. Whole-genome sequencing was performed by a commercial vendor (Novogene Bioinformatics Technology Co. Ltd., Beijing, China) using NovaSeq 6000 (350-bp insert library with 150-bp paired-end sequencing; Illumina, San Diego, CA).
Sequence analysis
Adaptor sequences and low-quality reads were eliminated from paired-end sequence reads using the Cutadapt v1.16 software [27]. The sequence reads were mapped to the Af293 reference genome using Bowtie2 v2.3.4.1 [28, 29], and BAM files were processed with the MarkDuplicates program from Picard-tools v2.18.1 to generate analysis-ready BAM files [30]. Variants were then called to obtain multi-sample Variant Call Format file using the HaplotypeCaller program from GATK v4.1.0.0 [31]. Subsequently, the effects of the variants were predicted using the SnpEff software [32]. Single nucleotide mutations were reconfirmed using Sanger sequencing.
RNA isolation and cyp51A expression
Total RNA was extracted using the RNeasy Plant Mini Kit (Qiagen, Hilden, Germany) according to the manufacturer’s instructions. Complementary DNA (cDNA) was synthesised from 2.5 μg of the total RNA using a QuantiTect Reverse Transcription kit (Qiagen, Hilden, Germany), and cDNA was used as the template for PCR using a QuantiTect SYBR Green PCR kit (Qiagen, Hilden, Germany). Quantitative reverse-transcription PCR (qRT-PCR) was performed in triplicate using a 7500 Real-Time PCR System (Applied Biosystems, Foster City, CA, USA). The mRNA abundance of the target genes was normalised to 18SrRNA. The primers used for qRT-PCR are listed in Table S1.
Statistical analysis
Unpaired t-test was used to assess the differences in the colony diameter, and the biomass and biofilm amounts. Survival was plotted on a Kaplan–Meier curve for each strain, and log-rank (Mantel-Cox) test was used for pairwise comparison of percent survival using the GraphPad Prism 5 software (GraphPad Software, La Jolla, CA). Statistical significance was set at P < 0.05.
Results
Clinical course and characterisation of A. fumigatus isolates
The sensitive isolate, MF-2046, was obtained from the sputum of the patient in June 2016, 4 years after VRC and ITC treatment and surgery, before retreatment with VRC. The resistant isolate, MF-2108, was isolated from the sputum of the same patient in September 2016 during VRC treatment (Fig. 1). Microsatellite analysis showed that the two isolates obtained from the patient exhibited identical genetic backgrounds (Table 1). The MF-2046 isolate was susceptible to azole antifungal drugs (MICs: itraconazole, 0.5 mg/L and voriconazole, 0.5 mg/L), whereas the MF-2108 isolate showed resistance to azoles (MICs: itraconazole, > 8 mg/L and voriconazole, 4 mg/L). The azole-resistant isolate did not harbour a point mutation of cyp51A or TRs in the promoter region of cyp51A (Table 1).
Although the MF-2108 was isolated from the sputum in September 2016, VRC treatment continued until November 2018 as respiratory symptoms and radiological findings improved. Since then, the patient has not been treated, but no disease progression has been observed (Fig. 1).
Differences in phenotype and virulence of A. fumigatus isolates
Although the colony morphologies of the two isolates were almost similar, the colony colours were different. The azole-susceptible MF-2046 isolate exhibited green colonies, whereas the azole-resistant MF-2108 isolate exhibited slightly white-in-green-coloured colonies (Fig. 2a). In the growth assay, MF-2108 colonies showed slower growth than that of the azole-susceptible MF-2046 (Fig. 2b). In addition, MF-2108 showed significantly decreased biomass and biofilm formation compared with those of MF-2046 (Fig. 2c, d). To evaluate the virulence of both isolates, a virulence assay was performed using the G. mellonella infection model. The MF-2108 isolate showed significantly decreased virulence compared with that of MF-2046 (Fig. 3).
Phenotype of Aspergillus fumigatus isolates. a A total of 1 × 104 conidia /A. fumigatus isolate were spotted on the PDA plates at 30 °C. Colony morphology was observed after 96 h incubation. b A total of 1 × 104 conidia /A. fumigatus isolate were spotted on the PDA plates at 30 °C. Colony diameters were measured after 72, 96, and 120 h and average diameters were calculated from three independent experiments. Error bars represent standard deviations. *P < 0.001, Unpaired t-test. c A total of 5.0 × 105 conidia /A. fumigatus isolate were incubated in 5 mL of yeast glucose (YG) media at 37 °C for 24 h with shaking at 250 rpm. The precipitates obtained were recovered by filtration, frozen at − 80 °C, lyophilized overnight, and weighed. The average biomass was calculated from three independent experiments. Error bars represent standard deviations. P = 0.0286, Unpaired t-test. d A round-bottomed 96-well plate was inoculated with 100 μL of the conidial suspension at a density of 1 × 105 conidia/mL in a Brian medium and incubated at 37 °C for 24 h. The spent culture medium was removed from each well and the adherent cells were washed three times with distilled water (dH2O). Biofilms were stained with 100 μL of 0.1% (w/v) crystal violet solution and washed and destained with 125 μL of 100% ethanol. The absorbance of the destaining solution was measured at 595 nm. Error bars represent standard deviations. *P < 0.001, Unpaired t-test
Virulence assay using the Galleria mellonella infection model. Groups of 10 larvae were inoculated with 1.0 × 106 conidia into the haemocoel and incubated in the dark at 37 °C, and survival was monitored daily for 7 days. Kaplan–Meier curves were generated and compared using log rank (Mantel–Cox) test. *P < 0.0001
Whole-genome comparisons between A. fumigatus isolates
To investigate the azole-resistance mechanisms of the isolate, we compared the genome sequences of both isolates using next-generation sequencing. Compared with that in MF-2046, MF-2108 exhibited one splice site, two nonsynonymous, and two synonymous mutations (Table 2). The two nonsynonymous mutations were identified as Y622S in Afu1g09770 and F551L in Afu2g02120, which encode uncharacterised proteins and proteins exhibiting phosphopentomutase activity, respectively. Two synonymous mutations were identified in Afu2g00910 and Afu2g03450. In addition, one splice site mutation was identified in the gene encoding HapE. The mutation from G to A was observed at the last base of the third intron of the hapE gene. It is presumed that this mutation resulted in the intron remaining in the mature mRNA and the translation stopping at the 57th amino acid, thereby leading to the production of abnormal HapE protein (Fig. S1).
The cyp51A expression in the azole-resistant isolate
The cyp51A expression levels in both isolates were evaluated using qRT-PCR. The expression of cyp51A was increased by approximately six fold in the azole-resistant isolate (MF-2108) compared with that in the azole-susceptible isolate (MF-2046) (Fig. 4).
Discussion and conclusions
To the best of our knowledge, this is the first to report a case demonstrating the clinical phenotype and virulence of an ARAf isolate with a HapE splice site mutation. In this study, we demonstrated that ARAf isolated from a patient with CPA showed slow growth, decreased biomass and biofilm formation, and decreased virulence, and these phenotypes may be caused by a HapE splice site mutation. In addition, the virulence of the isolated ARAf was consistent with the patient’s clinical course, and the patient did not exhibit any disease progression in respiratory symptoms or radiological findings, despite continued treatment with VRC to which the isolate with the HapE mutation was resistant.
A previous study reported that ARAf with HapE P88L substitution showed increased cyp51A expression levels and slow growth in an in vitro growth assay [14]. In addition, another study reported that each deletion strain of the HapB, HapC, and HapE subunits, which confer the CBC-acquired resistance to triazole antifungals, showed slow growth and decreased virulence of aspergillosis in a pulmonary and systemic mouse model [16]. Here, an ARAf isolate with a HapE splice site mutation showed slow growth and increased cyp51A expression levels, and these in vitro phenotypes were similar to those with HapE P88L mutation and a CBC mutations. These results suggest that the decreased HapE function due to the HapE splice site mutation may affect the in vitro and in vivo phenotypes, although other mutations may also affect these phenotypes. Clinically, infection with ARAf exhibiting a HapE P88L mutation has been reported to cause death in a patient with chronic granulomatous disease, which is caused by a primary immunodeficiency associated with phagocytic cell abnormalities, despite a clear decrease in virulence in vitro [14]. However, our patient did not exhibit any disease progression in either respiratory symptoms or radiological findings after infection with ARAf with a HapE splice site mutation. The patient had no apparent systemic immunodeficiency in this case, and these prognostic differences may have been caused by differences in host immune status.
In bacteria, long-term antibiotic exposure has been reported to cause resistance mutation, resulting in decreased growth and virulence. This evolution of a microorganism is termed the fitness costs of antibiotic resistance [33, 34]. Similarly, in fungi, fitness losses in patient-derived ARAf have also been frequently reported [35,36,37]. The cyp51A-mediated resistance mechanisms are not thought to be associated with fitness costs because these mutated strains do not show slow growth [36, 38]. However, in our study, the ARAf isolate with the HapE splice site mutation showed slow growth and decreased virulence. This adaptation of the strain due to the HapE mutation was considered a fitness cost.
ARAf infection has been suggested to be associated with poor prognosis in IA patients with attenuated immunity due to leukaemia, solid organ transplantation, and hematopoietic stem cell transplantation [39]. It has also been associated with poor prognosis in patients with CPA [11]. There are many cases of infections with ARAf exhibiting a cyp51A mutation that does not affect growth and virulence [39]. Our study results suggest that ARAf with non-cyp51A mutation may not necessarily need to be treated in patients who are not severely immunocompromised, depending on their clinical course.
Although we present the phenotype of an ARAf isolate with a HapE splice site mutation, our study has some limitations. First, clinically derived ARAf isolates have multiple mutations, and phenotypic changes are not necessarily due to a single gene change. Second, our report describes the result of the analysis of a single strain with a HapE mutation in one case.
In conclusion, this is the first to report a case demonstrating the clinical manifestation of a patient with CPA infected with an ARAf isolate with a HapE splice site mutation, which was consistent with the in vitro and in vivo attenuated virulence of the ARAf isolate. Our results imply that not all ARAf isolates obtained from immunocompetent patients should be considered targets for antifungal treatment. Further studies on the virulence of non-cyp51A mutations are warranted to better understand the resistance mechanisms in A. fumigatus.
Availability of data and materials
The sequenced genome data were deposited with the DDBJ Sequence Read Archive under accession no. DRA010499. All other data and materials are included in the manuscript.
Change history
16 September 2021
A Correction to this paper has been published: https://doi.org/10.1186/s12879-021-06667-7
Abbreviations
- ARAf :
-
Azole-resistant Aspergillus fumigatus
- CBC:
-
CCAAT-binding complex
- cDNA:
-
Complementary DNA
- CPA:
-
Chronic pulmonary aspergillosis
- CT:
-
Computed tomography
- dH2O:
-
Distilled water
- IA:
-
Invasive aspergillosis
- ITC:
-
Itraconazole
- MEC:
-
Minimum effective concentration
- MIC:
-
Minimum inhibitory concentration
- PBS:
-
Phosphate-buffered saline
- PCR:
-
Polymerase chain reaction
- PDA:
-
Potato dextrose agar
- qRT-PCR:
-
Quantitative reverse-transcription polymerase chain reaction
- STR:
-
Short tandem repeat
- TR:
-
Tandem repeat
- VRC:
-
Voriconazole
References
Latge JP, Chamilos G. Aspergillus fumigatus and Aspergillosis in 2019. Clin Microbiol Rev. 2020;33(1):e00140–18.
Verweij PE, Ananda-Rajah M, Andes D, Arendrup MC, Bruggemann RJ, Chowdhary A, et al. International expert opinion on the management of infection caused by azole-resistant Aspergillus fumigatus. Drug Resist Updat. 2015;21-22:30–40. https://doi.org/10.1016/j.drup.2015.08.001.
Patterson TF, Thompson GR 3rd, Denning DW, Fishman JA, Hadley S, Herbrecht R, et al. Practice Guidelines for the Diagnosis and Management of Aspergillosis: 2016 Update by the Infectious Diseases Society of America. Clin Infect Dis. 2016;63(4):e1–e60. https://doi.org/10.1093/cid/ciw326.
Denning DW, Venkateswarlu K, Oakley KL, Anderson MJ, Manning NJ, Stevens DA, et al. Itraconazole resistance in Aspergillus fumigatus. Antimicrob Agents Chemother. 1997;41(6):1364–8. https://doi.org/10.1128/AAC.41.6.1364.
Buil JB, Snelders E, Denardi LB, Melchers WJG, Verweij PE. Trends in azole resistance in Aspergillus fumigatus, the Netherlands, 1994-2016. Emerg Infect Dis. 2019;25(1):176–8. https://doi.org/10.3201/eid2501.171925.
Resendiz Sharpe A, Lagrou K, Meis JF, Chowdhary A, Lockhart SR, Verweij PE, et al. Triazole resistance surveillance in Aspergillus fumigatus. Med Mycol. 2018;56(suppl_1):83–92.
Rivero-Menendez O, Alastruey-Izquierdo A, Mellado E, Cuenca-Estrella M. Triazole resistance in aspergillus spp.: a worldwide problem? J Fungi (Basel). 2016;2(3):21.
Hagiwara D, Watanabe A, Kamei K, Goldman GH. Epidemiological and genomic landscape of azole resistance mechanisms in Aspergillus Fungi. Front Microbiol. 2016;7:1382.
Meis JF, Chowdhary A, Rhodes JL, Fisher MC, Verweij PE. Clinical implications of globally emerging azole resistance in Aspergillus fumigatus. Philos Trans R Soc Lond Ser B Biol Sci. 2016;371(1709):20150460.
Lestrade PP, Bentvelsen RG, Schauwvlieghe A, Schalekamp S, van der Velden W, Kuiper EJ, et al. Voriconazole resistance and mortality in invasive Aspergillosis: a multicenter retrospective cohort study. Clin Infect Dis. 2019;68(9):1463–71. https://doi.org/10.1093/cid/ciy859.
Lowes D, Al-Shair K, Newton PJ, Morris J, Harris C, Rautemaa-Richardson R, et al. Predictors of mortality in chronic pulmonary aspergillosis. Eur Respir J. 2017;49:1601062.
Lestrade PPA, Buil JB, van der Beek MT, Kuijper EJ, van Dijk K, Kampinga GA, et al. Paradoxal trends in azole-resistant Aspergillus fumigatus in a National Multicenter Surveillance Program, the Netherlands, 2013-2018. Emerg Infect Dis. 2020;26(7):1447–55. https://doi.org/10.3201/eid2607.200088.
Fraczek MG, Bromley M, Buied A, Moore CB, Rajendran R, Rautemaa R, et al. The cdr1B efflux transporter is associated with non-cyp51a-mediated itraconazole resistance in Aspergillus fumigatus. J Antimicrob Chemother. 2013;68(7):1486–96. https://doi.org/10.1093/jac/dkt075.
Camps SM, Dutilh BE, Arendrup MC, Rijs AJ, Snelders E, Huynen MA, et al. Discovery of a HapE mutation that causes azole resistance in Aspergillus fumigatus through whole genome sequencing and sexual crossing. PLoS One. 2012;7(11):e50034. https://doi.org/10.1371/journal.pone.0050034.
Rybak JM, Ge W, Wiederhold NP, Parker JE, Kelly SL, Rogers PD, et al. Mutations in hmg1, challenging the paradigm of clinical triazole resistance in aspergillus fumigatus. MBio. 2019;10(2):e00437–19.
Gsaller F, Hortschansky P, Furukawa T, Carr PD, Rash B, Capilla J, et al. Sterol biosynthesis and azole tolerance is governed by the opposing actions of SrbA and the CCAAT binding complex. PLoS Pathog. 2016;12(7):e1005775. https://doi.org/10.1371/journal.ppat.1005775.
Hagiwara D, Arai T, Takahashi H, Kusuya Y, Watanabe A, Kamei K. Non-cyp51A azole-resistant Aspergillus fumigatus isolates with mutation in HMG-CoA Reductase. Emerg Infect Dis. 2018;24(10):1889–97. https://doi.org/10.3201/eid2410.180730.
Denning DW, Cadranel J, Beigelman-Aubry C, Ader F, Chakrabarti A, Blot S, et al. Chronic pulmonary aspergillosis: rationale and clinical guidelines for diagnosis and management. Eur Respir J. 2016;47(1):45–68. https://doi.org/10.1183/13993003.00583-2015.
Tashiro M, Izumikawa K, Minematsu A, Hirano K, Iwanaga N, Ide S, et al. Antifungal susceptibilities of Aspergillus fumigatus clinical isolates obtained in Nagasaki, Japan. Antimicrob Agents Chemother. 2012;56(1):584–7. https://doi.org/10.1128/AAC.05394-11.
Takazono T, Ito Y, Tashiro M, Nakano Y, Hirayama T, Hosogaya N, et al. Transition of triazole-resistant Aspergillus fumigatus isolates in a Japanese tertiary hospital and subsequent genetic analysis. J Infect Chemother. 2021;27(3):537–9. https://doi.org/10.1016/j.jiac.2020.11.027.
Clinical and Laboratory Standards Institute. Reference method for broth dilution antifungal susceptibility testing for filamentous fungi; approved standard. Document M38-A2. Wayne: Clinical and Laboratory Standards Institute; 2008.
de Valk HA, Meis JF, Curfs IM, Muehlethaler K, Mouton JW, Klaassen CH. Use of a novel panel of nine short tandem repeats for exact and high-resolution fingerprinting of Aspergillus fumigatus isolates. J Clin Microbiol. 2005;43(8):4112–20. https://doi.org/10.1128/JCM.43.8.4112-4120.2005.
Ballard E, Melchers WJG, Zoll J, Brown AJP, Verweij PE, Warris A. In-host microevolution of Aspergillus fumigatus: a phenotypic and genotypic analysis. Fungal Genet Biol. 2018;113:1–13. https://doi.org/10.1016/j.fgb.2018.02.003.
Gravelat FN, Beauvais A, Liu H, Lee MJ, Snarr BD, Chen D, et al. Aspergillus galactosaminogalactan mediates adherence to host constituents and conceals hyphal beta-glucan from the immune system. PLoS Pathog. 2013;9(8):e1003575. https://doi.org/10.1371/journal.ppat.1003575.
Gravelat FN, Ejzykowicz DE, Chiang LY, Chabot JC, Urb M, Macdonald KD, et al. Aspergillus fumigatus MedA governs adherence, host cell interactions and virulence. Cell Microbiol. 2010;12(4):473–88. https://doi.org/10.1111/j.1462-5822.2009.01408.x.
Maurer E, Browne N, Surlis C, Jukic E, Moser P, Kavanagh K, et al. Galleria mellonella as a host model to study Aspergillus terreus virulence and amphotericin B resistance. Virulence. 2015;6(6):591–8. https://doi.org/10.1080/21505594.2015.1045183.
Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet journal. 2011;17(1):10–2. https://doi.org/10.14806/ej.17.1.200.
Langmead B, Salzberg SL. Fast gapped-read alignment with bowtie 2. Nat Methods. 2012;9(4):357–9. https://doi.org/10.1038/nmeth.1923.
Garcia-Alcalde F, Okonechnikov K, Carbonell J, Cruz LM, Gotz S, Tarazona S, et al. Qualimap: evaluating next-generation sequencing alignment data. Bioinformatics. 2012;28(20):2678–9. https://doi.org/10.1093/bioinformatics/bts503.
Nierman WC, Pain A, Anderson MJ, Wortman JR, Kim HS, Arroyo J, et al. Genomic sequence of the pathogenic and allergenic filamentous fungus Aspergillus fumigatus. Nature. 2005;438(7071):1151–6. https://doi.org/10.1038/nature04332.
McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, et al. The genome analysis toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20(9):1297–303. https://doi.org/10.1101/gr.107524.110.
Cingolani P, Platts A, Wang LL, Coon M, Nguyen T, Wang L, et al. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly (Austin). 2012;6(2):80–92. https://doi.org/10.4161/fly.19695.
Beceiro A, Tomas M, Bou G. Antimicrobial resistance and virulence: a successful or deleterious association in the bacterial world? Clin Microbiol Rev. 2013;26(2):185–230. https://doi.org/10.1128/CMR.00059-12.
Melnyk AH, Wong A, Kassen R. The fitness costs of antibiotic resistance mutations. Evol Appl. 2015;8(3):273–83. https://doi.org/10.1111/eva.12196.
Hagiwara D, Takahashi H, Watanabe A, Takahashi-Nakaguchi A, Kawamoto S, Kamei K, et al. Whole-genome comparison of Aspergillus fumigatus strains serially isolated from patients with aspergillosis. J Clin Microbiol. 2014;52(12):4202–9. https://doi.org/10.1128/JCM.01105-14.
Valsecchi I, Mellado E, Beau R, Raj S, Latge JP. Fitness studies of azole-resistant strains of Aspergillus fumigatus. Antimicrob Agents Chemother. 2015;59(12):7866–9. https://doi.org/10.1128/AAC.01594-15.
Steinbach WJ, Arendrup MC, Mavridou E, Mortensen KL, Snelders E, Frimodt-Møller N, et al. Development of azole resistance in aspergillus fumigatus during azole therapy associated with change in virulence. PLoS One. 2010;5(4):e10080.
Lackner M, Rambach G, Jukic E, Sartori B, Fritz J, Seger C, et al. Azole-resistant and -susceptible Aspergillus fumigatus isolates show comparable fitness and azole treatment outcome in immunocompetent mice. Med Mycol. 2018;56(6):703–10. https://doi.org/10.1093/mmy/myx109.
Chowdhary A, Sharma C, Meis JF. Azole-Resistant Aspergillosis: Epidemiology, Molecular Mechanisms, and Treatment. J Infect Dis. 2017;216(suppl_3):S436–44.
Acknowledgments
The authors would like to thank all members of the co-author, patient, Kazuyo Ishida, technical assistant, involved in this article.
Funding
This research received no specific grant from any funding agency in the public, commercial, or not-for-profit sectors.
Author information
Authors and Affiliations
Contributions
Y.I., T.T., and K.I. developed study design. Y.I., S.K., T.S., and T.T. collected clinical information. Y.I. and T.T. performed most esperiments. Y.N. and M.T. contributed to STR analysis and drug susceptibility test. N.A., T.H., K.Y., Y.I., T.M., and K.Y. assisted with Biomass measurement and biofilm assay, Galleria mellonella virulence assay, and Whole-genome sequencing. All the authors interpreted data. K.I. and H.M. provided supervision throughout this project. Y.I. and T.T. wrote the manuscript. All authors read, reviewed and approved the manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Written informed consent was obtained from the patient for publication of this case report and any accompanying images. A copy of the written consent is available for review by the Editor of this journal upon request.
Competing interests
The authors report no conflicts of interest. The authors alone are responsible for the content and the writing of the paper.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
The authors identified an error in Additional file 1. The correct file is given hereby attached.
Supplementary Information
Additional file 1: Table S1
. Primers used in this study. Fig. S1. HapE splice site mutation.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Ito, Y., Takazono, T., Koga, S. et al. Clinical and experimental phenotype of azole-resistant Aspergillus fumigatus with a HapE splice site mutation: a case report. BMC Infect Dis 21, 573 (2021). https://doi.org/10.1186/s12879-021-06279-1
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12879-021-06279-1