Abstract
Background
Plasmodium falciparum dihydrofolate reductase (Pfdhfr) and dihydropteroate synthetase (Pfdhps) mutations compromise the effectiveness of sulfadoxine-pyrimethamine (SP) for treatment of uncomplicated malaria, and are likely to impair the efficiency of intermittent preventive treatment during pregnancy (IPTp). This study was conducted to determine the level of Pfdhfr-Pfdhps mutations, a decade since SP was limited for IPTp use in pregnant women in Tanzania.
Methods
P. falciparum genomic DNA was extracted from dried blood spots prepared from a finger prick. Extracted DNA were sequenced using a single MiSeq lane by combining all PCR products. Genotyping of Pfdhfr and Pfdhps mutations were done using bcftools whereas custom scripts were used to filter and translate genotypes into SP resistance haplotypes.
Results
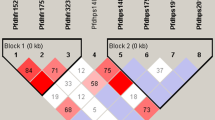
The Pfdhfr was analyzed from 445 samples, the wild type (WT) Pfdhfr haplotype NCSI was detected in 6 (1.3%) samples. Triple PfdhfrIRNI (mutations are bolded and underlined) haplotype was dominant, contributing to 84% (number [n] = 374) of haplotypes while 446 samples were studied for Pfdhps, WT for Pfdhps (SAKAA) was found in 6.7% (n = 30) in samples. Double Pfdhps haplotype (SGEAA) accounted for 83% of all mutations at Pfdhps gene. Of 447 Pfdhfr-Pfdhps combined genotypes, only 0.9% (n = 4) samples contained WT gene (SAKAA-NCSI). Quintuple (five) mutations, SGEAA-IRNI accounted for 71.4% (n = 319) whereas 0.2% (n = 1) had septuple (seven) mutations (AGKGS-IRNI). The overall prevalence of Pfdhfr K540E was 90.4% (n = 396) while Pfdhps A581G was 1.1% (n = 5).
Conclusions
This study found high prevalence of Pfdhfr–Pfdhps quintuple and presence of septuple mutations. Mutations at Pfdhfr K540E and Pfdhps A581G, major predictors for IPTp-SP failure were within the recommended WHO range. Abandonment of IPTp-SP is recommended in settings where the Pfdhfr K540E prevalence is > 95% and Pfdhps A581G is > 10% as SP is likely to be not effective. Nonetheless, saturation in Pfdhfr and Pfdhps haplotypes is alarming, a search for alternative antimalarial drug for IPTp in the study area is recommended.
Similar content being viewed by others
Background
Antimalarial drugs use and vector control are the commendable tools for global malaria prevention and control respectively [1]. However, resistance of malaria parasites to antimalarial drugs and resistance of Anopheles mosquitoes to insecticides impair the progressive fight against malaria [2]. Resistance of Plasmodium falciparum to previous generations of medicines, i.e., chloroquine (CQ) [3] and sulfadoxine-pyrimethamine (SP) [4] influenced the replacement of CQ with SP [5], then artemisinin-based combination therapy (ACT) as the first-line treatment for uncomplicated malaria in Tanzania [6].
The changes in malaria treatment policy were supported by evidence from molecular epidemiological resistance surveillance against SP [7], mainly P. falciparum dihydrofolate reductase (Pfdhfr) and dihydropteroate synthetase (Pfdhps) enzymes mutations [8, 9]. Mutations in the Pfdhps gene at codons serine 436 to alanine (S436A), alanine 437 to glycine (A437G), lysine 540 glutamic acid (K540E), alanine 581 to glycine (A581G) and alanine 613 to serine (A613S) predicts sulfadoxine resistance whereby mutations in the Pfdhfr gene at codons cysteine 50 to arginine (C50R), asparagine 51 to isoleucine (N51I), cysteine 59 to arginine (C59R), Serine 108 to asparagine/threonine (S108 N/T), and isoleucine 164 to leucine (I164L) are associated with pyrimethamine resistance [10].
Mutations in both the Pfdhfr/Pfdhps genes which greatly influence SP clinical treatment failures [11] resulted SP being unfit for first -line treatment of uncomplicated malaria in Tanzania [12]. However, in 2006 the drug was limited for intermittent preventive treatment during pregnancy (IPTp-SP), in areas with moderate to high malaria transmission [13]. Pregnant women regardless of the presence or absence of malaria should administer at least three curative doses of SP with an interval of at least 1 month between the two doses, starting in the second trimester of pregnancy [14], for preventing pregnancy associated malaria and improve pregnancy outcomes [13].
The elevated Pfdhfr/Pfdhps protein mutations and saturation of genetic haplotypes such as quintuple (CIRNI-SGEAA) and sextuple (CIRNI-SGEGA) reported in Tanzania [10] pose risks against the effectiveness of SP in preventing placental malaria during pregnancy. In this regard, World Health Organization (WHO) recommends abandonment of IPTp-SP in settings where the Pfdhfr K540E prevalence is > 95% and Pfdhps A581G is > 10% because SP is more likely to be less effective [15].
For the purpose of guiding IPTp-SP policy and inform on the current SP resistance status, routinely surveillance of SP effectiveness using molecular markers should be implemented as recommended by WHO [15]. Therefore, a study was conducted in a general population to determine the status of mutations associated with SP resistance, more than one decade since SP was limited for IPTp use in pregnant women in Tanzania.
Methods
Study design
A hospital based surveillance of molecular markers for SP resistance was conducted for 4 months from April, 2019 at Kibiti Health Center (KHC) found in Eastern part of Tanzania. The study was conducted to determine the current antimalarial status of SP, more than a decade since SP were limited for IPTp for pregnant women in Tanzania. Tanzania is found in sub-Saharan region where malaria is endemic, adopted the use of SP for malaria prevention in pregnancy women for more than one decade [12].
Study area
KHC is found in Kibiti District, Pwani region. Pwani region is one of the five regions found along the coastal of Indian Ocean. Pwani region is bordered to north by Tanga region, to the east by Dar es Salaam region and the Indian Ocean, to the south by the Lindi region, and to the west by the Morogoro region. The coastal region experiences malaria transmission throughout the year with regional prevalence ranging between 3.1 and 14.8% [16].
Study population
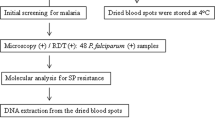
Patients attending clinic at KHC who presented with malaria infection related symptoms qualified for inclusion in the study. The suggestive symptoms example fever, general body weakness and headache were confirmed by the attending clinician [17]. Rapid malaria testing was performed using CareStart™ malaria HRP2/pLDH (Pf/pan) Combo test (Access Bio, Ethiopia). Samples tested positive by rapid tests qualified for blood smear (BS) microscopy for confirmation. A total of 489 positive BS microscopy qualified for further dried blood spot preparation.
DBS preparation and DNA extraction
Dried Blood Spots (DBS) were prepared as per MalariaGEN SpotMalaria, DBS collection protocol [18]. Briefly, a well sterilized middle patient’s finger was pricked, and 4 blood spots from each patient were dropped on a separate circle of the filter paper; two on each paper card. The spotted blood were dried and put in the desiccant sachet for storage. DNA from the DBS was extracted following QIAamp DNA Investigator Kit for isolation of total DNA from filter papers (Qiagen, Valencia, CA, USA) and as described by Oyola et al.,2016 [19].
Genotyping of Pfdhfr and Pfdhps mutations
Molecular genotyping of Pfdhfr and Pfdhps gene mutations associated with SP-resistance was performed by Wellcome Sanger Institute, UK as previously described but customized for Pfdhfr and Pfdhps mutations [20].
To summarize, targeted genotypes were spotted and multiplex PCR primers were designed using mPrimer software [21] and were run according to Wellcome Sanger Institute protocol (Goncalves, manuscript in preparation; https://www.malariagen.net/projects/spotmalaria). Primers were made to amplify products between 190 and 250 bp and combined into 3 pools. A multi steps protocol was used to first amplify the target regions of the P. falciparum genome, then PCR to combine sequencing and multiplexing adapters. About 384 samples were sequenced in one MiSeq lane by combining all PCR products. Samples were de-plexed using the multiplexing adapters and individual CRAM files were aligned to a modified amplicon reference genome for further analysis.
Statistical analysis
Statistical Package for Social Sciences version 25 (SPSS Software, Chicago Inc., USA) was used to import laboratory data from the Microsoft Excel sheet (Redmond, WA) for data analysis and interpretation. Genotyping was done using bcftools and custom scripts to filter and translate genotypes into Pfdhfr and Pfdhps resistance haplotypes. Haplotypes were expressed as frequencies and percentages.
Results
Haplotypes for Pfdhfr and Pfdhps
The Pfdhfr was analyzed from 445 samples, the wild type (WT) Pfdhfr haplotype NCSI was detected in only 1.3% (number [n] = 6) samples. Triple PfdhfrIRNI haplotype was dominant, contributing to 84% (n = 374) of haplotypes. The total of 446 samples were studied for Pfdhps. The WT for Pfdhps was found in 6.7% (n = 30) of all samples detected. The most common mutation was the change of amino acid alanine to glycine (A437G) and lysine to glutamic acid (K540E). Double Pfdhps haplotype (SGEAA) accounted for 83% of mutations of the Pfdhps gene (Table 1). The overall prevalence of K540E was 90.4% (n = 396) while A581G was 1.1% (n = 5).
Combined Pfdhfr and Pfdhps haplotypes
Overall 447 (91.4%) genotypes were detected from 489 sequenced samples. The concomitant mutations (Pfdhfr-Pfdhps) were present in 99.1% (n = 443) of samples. Of 447 genotypes, only 4 samples (0.9%) were WT (SAKAA-NCSI). Concomitant mutations with quintuple haplotype (SGEAA-IRNI) dominated by 71.4% whereby sextuple haplotype (AGKGS-IRNI) was detected in one sample (0.2%) (Table 2).
Discussion
This study was conducted to determine the current SP resistance status, more than a decade since SP was limited for IPTp use in pregnant women in Tanzania.
The study found high level (99.1%) concomitant mutations (Pfdhfr-Pfdhps) where quintuple mutation (SGEAA-IRNI) dominated by 71.4%. Additionally, one sample detected septuple mutation (AGKGS-IRNI) and only 0.9% contained the WT (SAKAA-NCSI) protein. These results were close to the findings from the previous study which reported high-level of P. falciparum SP resistance with concomitant occurrence of septuple haplotype [10]. There was a high number of quintuple (SGEAA-IRNI) and few sextuple mutations. These haplotypes have been highly associated with sub-optimal IPTp-SP effectiveness in study conducted between 2008 and 2010 in Korogwe district, Tanzania [22]. Moreover, high prevalence of the Pfdhfr/Pfdhps sextuple haplotype was associated with reduced birth- weight [23,24,25].
In this study, triple PfdhfrIRNI haplotype mutation was dominant, contributing to 84% of all Pfdhfr haplotypes whereas double Pfdhps haplotype (SGEAA) accounted for 83% of mutations of the Pfdhps haplotypes. Compared to study by Baraka et al.,2015 [10], PfdhfrIRNI haplotype were predominant in all sites with significantly higher frequencies at Muheza (93.3%) compared to Muleba (75.0%) and Nachingwea districts (70.6%). In this regard, there was 13.4% increase in PfdhfrIRNI haplotype prevalence when Nanchingwea (70.6%) and Kibiti district (84%) both found along the coastal region were compared. Additionally, the previous study found that double Pfdhps haplotype SGEAA was significantly high at Muheza (27.2%) and Muleba (20.8%) while none (0%) was detected at Nachingwea while the current study at Kibiti district detected 83% Pfdhps SGEAA mutations. Meaning that, from 2015 to 2019 there is an increase of 0 to 83% Pfdhps SGEAA mutation in the coastal region, respectively.
WHO malaria report of 2019 has clearly mentioned pregnant women as the most affected group and use of effective IPTp should be highly implemented to combat risks associated with pregnancy malaria [26]. On top of that, a recent study by Mikomangwa et al., [27] concluded that pregnant woman who was malaria positive had 11 times more risk of giving birth to low birth weight (LBW) child when compared to those who tested malaria negative.
Nevertheless, Pfdhfr K540E and Pfdhps A581G, major predictors for IPTp-SP failure, had the prevalence of 90.4% for Pfdhfr K540E and 1.1% for Pfdhps A581G. This guarantee the continued use of IPTp-SP in the study area. WHO recommends stoppage of SP for IPTp if Pfdhfr K540E prevalence > 95% and Pfdhps A581G > 10% [15]. However, rapid increase and saturation in Pfdhfr and Pfdhps mutations with spread of sextuples and septuple is of great concern. The effort to search for alternative drug (s) for IPTp in the study areas should be prioritized. The current study was conducted in the general population, hence limited data on clinical, birth outcomes and malaria infection status in pregnant women in relation to SP resistance genotypes.
Conclusions
This study found high prevalence of Pfdhfr–Pfdhps quintuple mutations. Mutations at Pfdhfr K540E and Pfdhps A581G, major predictors for IPTp-SP failure were within the recommended WHO range. Abandonment of IPTp-SP is recommended in areas where the Pfdhfr K540E prevalence is > 95% and Pfdhps A581G is > 10% because SP is likely to be not effective. Saturation in Pfdhfr and Pfdhps haplotypes is alarming, a search for alternative antimalarial drug for IPTp, the study area in particular is recommended.
Availability of data and materials
The datasets generated and/or analysed during the current study are available from the corresponding author on reasonable request.
Abbreviations
- SP:
-
Sulfadoxine-pyrimethamine
- WT:
-
Wild type
- IPTp:
-
Intermittent preventive treatment during pregnancy
- Pfdhfr :
-
Plasmodium falciparum dihydrofolate reductase
- Pfdhps :
-
Plasmodium falciparum dihydropteroate synthetase
- WHO:
-
World Health Organization
References
Patouillard E, Griffin J, Bhatt S, Ghani A, Cibulskis R. Global investment targets for malaria control and elimination between 2016 and 2030. BMJ Glob Health. 2017;2(2):e000176.
WHO. Global report on antimalarial drug efficacy and drug resistance: 2000–2010, vol. 115: Who; 2010.
Mohammed A, et al. Trends in chloroquine resistance marker , Pfcrt-K76T mutation ten years after chloroquine withdrawal in Tanzania. Malar J. 2013;12(1):1.
NDEJEMBI M, et al. Therapeutic efficacy of Sulfadoxine-Pyrimethamine and prevalence of resistance markers in Tanzania Prior to revision of malaria treatment policy: Plasmodium falciparum Dihydrofolate Reductase and Dihydropteroate synthase mutations in monitoring in vivo re. Am J Trop Med Hyg. 2018;71(6):696–702.
Mubyazi GM, Gonzalez-Block MA. Research influence on antimalarial drug policy change in Tanzania: case study of replacing chloroquine with sulfadoxine-pyrimethamine as the first-line drug. Malar J. 2005;4:1–13.
Y. P., C. J.L., A. S., A. J., L. P.S., and M. J. Trends in availability and prices of subsidized ACT over the first year of the AMFm: Evidence from remote regions of Tanzania. Malar J. 2012;11(299).
Mugittu K, et al. Efficacy of sulfadoxine-pyrimethamine in Tanzania after two years as first-line drug for uncomplicated malaria: assessment protocol and implication for treatment policy strategies. Malar J. 2005;4.
Brooks DR, Wang P, Read M, Watkins WM, Sims PFG, Hyde JE. Sequence variation of the Hydroxymethyldihydropterin Pyrophosphokinase: Dihydropteroate synthase gene in lines of the human malaria parasite, Plasmodium falciparum, with differing resistance to Sulfadoxine. Eur J Biochem. 1994;224(2):397–405.
Triglia T, Menting JGT, Wilson C, Cowman AF. Mutations in dihydropteroate synthase are responsible for sulfone and sulfonamide resistance in Plasmodium falciparum. Proc Natl Acad Sci U S A. 1997;94(25):13944–9.
Baraka V, et al. High-level Plasmodium falciparum sulfadoxine-pyrimethamine resistance with the concomitant occurrence of septuple haplotype in Tanzania. Malar J. 2015;14(1):1–9.
Mugittu K, et al. Therapeutic efficacy of sulfadoxine-pyrimethamine and prevalence of resistance markers in Tanzania prior to revision of malaria treatment policy: Plasmodium falciparum dihydrofolate reductase and dihydropteroate synthase mutations in monitoring in vivo re. Am J Trop Med Hyg. 2004;71(6):696–702.
Kavishe RA, et al. Molecular monitoring of Plasmodium falciparum super-resistance to sulfadoxine-pyrimethamine in Tanzania. Malar J. 2016;15(1):1–8.
“IT Committee: This Friday we have two items on our agenda. 1. Al Wysocki,” p. 226, 2017.
Rogerson SJ, Mwapasa V, Meshnick SR. Malaria in pregnancy: linking immunity and pathogenesis to prevention. Am J Trop Med Hyg. 2007;77(SUPPL. 6):14–22.
R. Gana, F. Leke, and L. Slutsker, “WHO Evidence Review Group on Intermittent Screening and Treatment (ISTp) and ACT Treatment of Malaria in Pregnancy,” September, pp. 1–17, 2015.
TMIS, “Tanzania Malaria Indicator Survey (TMIS) 2017,” 2017.
T. H. E. United and R. Of, “The UNITED republic of Tanzania standard treatment guidelines and essential medicines list,” 2013.
U. MalariaGEN Resource Centre members based at the Wellcome Trust Sanger Institute, “Protocol for collecting dried blood spots from malaria patients, for the purposes of sequencing Plasmodium parasites.,” https://www.malariagen.net/network/capacity-building/methods#DBS .
Oyola SO, et al. Whole genome sequencing of Plasmodium falciparum from dried blood spots using selective whole genome amplification. Malar J. 2016;15(1):1–12.
Bwire GM, Ngasala B, Mikomangwa WP, Kilonzi M, Kamuhabwa AAR. Detection of mutations associated with artemisinin resistance at k13-propeller gene and a near complete return of chloroquine susceptible falciparum malaria in southeast of Tanzania. Sci Rep. 2020;10(1):1–7.
Shen Z, et al. MPprimer: A program for reliable multiplex PCR primer design. BMC Bioinformatics. 2010;11.
Minja DTR, et al. Plasmodium falciparum mutant haplotype infection during pregnancy associated with reduced birthweight, Tanzania. Emerg Infect Dis. 2013;19(9):1446–54.
McCollum AM, Mueller K, Villegas L, Udhayakumar V, Escalante AA. Common origin and fixation of Plasmodium falciparum dhfr and dhps mutations associated with sulfadoxine-pyrimethamine resistance in a low-transmission area in South America. Antimicrob Agents Chemother. 2007;51(6):2085–91.
Naidoo I, Roper C. Mapping ‘partially resistant’, ‘fully resistant’, and ‘super resistant’ malaria. Trends Parasitol. 2013;29(10):505–15.
Harrington WE, Morrison R, Fried M, Duffy PE. Intermittent preventive treatment in pregnant women is associated with increased risk of severe malaria in their offspring. PLoS One. 2013;8(2):2–7.
World Health Organization (WHO), “World Malaria Report,” 2019.
Mikomangwa WP, Oms M, Aklillu E, Kamuhabwa AAR. Adverse birth outcomes among mothers who received intermittent preventive treatment with Sulphadoxine-Pyrimethamine in the low malaria transmission region. BMC Pregnancy Childbirth. 2019;19(1):1–11.
Acknowledgements
This publication uses data generated using Tanzanian samples in collaboration with MalariaGEN SpotMalaria Project (https://www.malariagen.net/projects/spotmalaria); the project is coordinated by the MalariaGEN Resource Centre with funding from Wellcome (206194, 090770). The authors would like to thank the staff of Wellcome Sanger Institute Sample Management, Genotyping, Sequencing and Informatics teams for their contribution. The author (GMB) acknowledge the training on antimalarial resistance markers when he attended Global Health Fellowship 2018, Novartis Institutes for Biomedical Research, Emeryville, CA, USA. Special thanks to Prof. A. Kamuhabwa and Dr. B. Ngasala for their mentorship support.
Funding
This study was funded by Swedish International Development Cooperation Agency (Sida), Sweden through Muhimbili University of Health and Allied Sciences. The funder did not participate in the design of the study, data collection, analysis, interpretation, and manuscript preparation.
Author information
Authors and Affiliations
Contributions
GMB participated in conception; study design, data collection, analysis and manuscript writing, MK, WPM participated in data analysis and manuscript writing. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Ethical approval to conduct this study was obtained from Muhimbili University of Health and Allied Sciences Ethical Review Board (Ref. DA.282/298/01A.C/) and National Institute for Medical Research (Ref. NIMR/HQ/R.8A/Vol.IX/3107). Permission to conduct the study at KHC was obtained from both Kibiti District Medical Officer and KHC Medical Officer In-charge. Written informed consents after explaining the purpose of the study were requested before enrollment.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Bwire, G.M., Mikomangwa, W.P. & Kilonzi, M. Occurrence of septuple and elevated Pfdhfr-Pfdhps quintuple mutations in a general population threatens the use of sulfadoxine-pyrimethamine for malaria prevention during pregnancy in eastern-coast of Tanzania. BMC Infect Dis 20, 530 (2020). https://doi.org/10.1186/s12879-020-05253-7
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12879-020-05253-7